

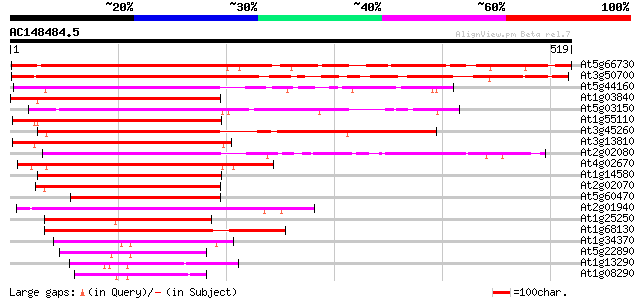
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148484.5 - phase: 0
(519 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g66730 zinc finger protein 541 e-154
At3g50700 zinc finger protein 533 e-152
At5g44160 unknown protein 333 1e-91
At1g03840 putative DNA-binding protein 326 2e-89
At5g03150 putative protein 325 5e-89
At1g55110 putative zinc finger protein 321 7e-88
At3g45260 zinc finger like protein 319 2e-87
At3g13810 zinc finger protein, putative 318 3e-87
At2g02080 C2H2-type zinc finger protein 313 2e-85
At4g02670 putative zinc finger protein 311 4e-85
At1g14580 putative zinc finger protein 311 6e-85
At2g02070 putative C2H2-type zinc finger protein 311 7e-85
At5g60470 putative zinc finger protein 259 2e-69
At2g01940 putative C2H2-type zinc finger protein 239 2e-63
At1g25250 Indeterminate 1 like Zn finger containing protein 234 1e-61
At1g68130 putative C2H2-type zinc finger protein 231 6e-61
At1g34370 zinc finger protein, putative 102 7e-22
At5g22890 unknown protein 101 9e-22
At1g13290 hypothetical protein 97 3e-20
At1g08290 unknown protein 89 8e-18
>At5g66730 zinc finger protein
Length = 500
Score = 541 bits (1395), Expect = e-154
Identities = 313/545 (57%), Positives = 371/545 (67%), Gaps = 74/545 (13%)
Query: 2 VDLDNVSTASGEASISSSGNNNIQSPIPKPT-KKKRNLPGMPDPEAEVIALSPTTLLATN 60
VDLDN ST SG+AS+SS+GN N+ PK KKKRNLPGMPDP+AEVIALSP TL+ATN
Sbjct: 3 VDLDNSSTVSGDASVSSTGNQNLT---PKSVGKKKRNLPGMPDPDAEVIALSPKTLMATN 59
Query: 61 RFVCEICNKGFQRDQNLQLHRRGHNLPWKLRQRSSKEIRKRVYVCPEPTCVHHDPSRALG 120
RFVCEICNKGFQRDQNLQLHRRGHNLPWKLRQRS+KE+RK+VYVCP CVHHDPSRALG
Sbjct: 60 RFVCEICNKGFQRDQNLQLHRRGHNLPWKLRQRSTKEVRKKVYVCPVSGCVHHDPSRALG 119
Query: 121 DLTGIKKHFCRKHGEKKWKCEKCSKKYAVQSDWKAHSKVCGSREYKCDCGTVFSRRDSFI 180
DLTGIKKHFCRKHGEKKWKCEKCSKKYAVQSDWKAHSK+CG++EYKCDCGT+FSRRDSFI
Sbjct: 120 DLTGIKKHFCRKHGEKKWKCEKCSKKYAVQSDWKAHSKICGTKEYKCDCGTLFSRRDSFI 179
Query: 181 THRAFCDALAEENAKSQNQA-------VGKANSESDSK---------VLTGDSLPAVITT 224
THRAFCDALAEE+AK+ Q+ V + N E + K L S P+V
Sbjct: 180 THRAFCDALAEESAKNHTQSKKLYPETVTRKNPEIEQKSPAAVESSPSLPPSSPPSVAIA 239
Query: 225 TAAAAATTPQSNSGVSSALETQKLDLP-ENPPQIIE---EPQVVVTTTASALNASCSSST 280
A A + +S +SS++ LP +N P+ E P+V++ + + + SSS
Sbjct: 240 PAPAISVETESVKIISSSV------LPIQNSPESQENNNHPEVIIEEASRTIGFNVSSSD 293
Query: 281 SSKSNGCAATSTGVFASLFASSTASASASLQPQAPAFSDLIRSMGCTDPRPTDFSAPPSS 340
S + + + G +A LF SSTAS S A P+ F+ S
Sbjct: 294 LSNDH---SNNNGGYAGLFVSSTASPSLYASSTA---------------SPSLFAPSSSM 335
Query: 341 EAISLCLSTSHGSSIFGTGGQECRQYV-PTHQPPAMSATALLQKAAQMGAAATNASLLRG 399
E ISLCLST+ S+FG ++ ++ P PAMSATALLQKAAQMG+ + SLLRG
Sbjct: 336 EPISLCLSTN--PSLFGPTIRDPPHFLTPLPPQPAMSATALLQKAAQMGSTGSGGSLLRG 393
Query: 400 LGIVSSSASTSSGQQDSLHWGLGPMEPEGSSLVSAGLGLGLPCD---ADSGLKELMLGTP 456
LGIVS +TSS + S H L ++ GLGLGLPC + SGLKELM+G
Sbjct: 394 LGIVS---TTSSSMELSNHDALS---------LAPGLGLGLPCSSGGSGSGLKELMMGNS 441
Query: 457 SMFGPKQTTLDFLGLGMAA--GGSAGGGLSALITSIGGGSGLDVTAAAASFGNGEFSGKD 514
S+FGPKQTTLDFLGLG A GG+ GGGLSAL+TSIGGG G+D+ FG+GEFSGKD
Sbjct: 442 SVFGPKQTTLDFLGLGRAVGNGGNTGGGLSALLTSIGGGGGIDL------FGSGEFSGKD 495
Query: 515 IGRSS 519
IGRSS
Sbjct: 496 IGRSS 500
>At3g50700 zinc finger protein
Length = 452
Score = 533 bits (1373), Expect = e-152
Identities = 299/520 (57%), Positives = 356/520 (67%), Gaps = 77/520 (14%)
Query: 2 VDLDNVSTASGEASISSSGNNNIQSPIPKPT-KKKRNLPGMPDPEAEVIALSPTTLLATN 60
VDLDN ST SGEAS+S S N Q+P+P T KKKRNLPGMPDPE+EVIALSP TLLATN
Sbjct: 3 VDLDNSSTVSGEASVSISSTGN-QNPLPNSTGKKKRNLPGMPDPESEVIALSPKTLLATN 61
Query: 61 RFVCEICNKGFQRDQNLQLHRRGHNLPWKLRQRSSKEIRKRVYVCPEPTCVHHDPSRALG 120
RFVCEICNKGFQRDQNLQLHRRGHNLPWKLRQ+S+KE++K+VYVCPE +CVHHDPSRALG
Sbjct: 62 RFVCEICNKGFQRDQNLQLHRRGHNLPWKLRQKSNKEVKKKVYVCPEVSCVHHDPSRALG 121
Query: 121 DLTGIKKHFCRKHGEKKWKCEKCSKKYAVQSDWKAHSKVCGSREYKCDCGTVFSRRDSFI 180
DLTGIKKHFCRKHGEKKWKC+KCSKKYAVQSDWKAHSK+CG++EYKCDCGT+FSRRDSFI
Sbjct: 122 DLTGIKKHFCRKHGEKKWKCDKCSKKYAVQSDWKAHSKICGTKEYKCDCGTLFSRRDSFI 181
Query: 181 THRAFCDALAEENAKSQNQAVGKANSE-SDSKVLTGDSLPAVITTTAAAAATTPQSNSGV 239
THRAFCDALAEENA+S + K N E K + +PA + T +A +
Sbjct: 182 THRAFCDALAEENARSHHSQSKKQNPEILTRKNPVPNPVPAPVDTESAKIKS-------- 233
Query: 240 SSALETQKLDLPENPPQIIEEPQVVVTTTASALNASCSSSTSSKSNGCAATSTGVFASLF 299
SS L ++ + P+ PP+I++E ++LN TS GVFA LF
Sbjct: 234 SSTLTIKQSESPKTPPEIVQE-----APKPTSLN--------------VVTSNGVFAGLF 274
Query: 300 ASSTASASASLQPQAPAFSDLIRSMGCTDPRPTDFSAPPSSEAISLCLSTSHGSSIFGTG 359
SS+AS P S +S+ F++ S E ISL LSTSHGSS G+
Sbjct: 275 ESSSAS------PSIYTTSSSSKSL---------FASSSSIEPISLGLSTSHGSSFLGSN 319
Query: 360 GQECRQYVPTHQPPAMSATALLQKAAQMGAAATNASLLRGLGIVSSSASTSSGQQDSLHW 419
H PAMSATALLQKAAQMGAA++ SLL GLGIVSS++++
Sbjct: 320 --------RFHAQPAMSATALLQKAAQMGAASSGGSLLHGLGIVSSTSTSI--------- 362
Query: 420 GLGPMEPEGSSLVSAGLGLGLPC--DADSGLKELMLGTPSMFGPKQTTLDFLGLGMAAGG 477
++V GLGLGLPC ++ SGLKELM+G S+FGPKQTTLDFLGLG A G
Sbjct: 363 ---------DAIVPHGLGLGLPCGGESSSGLKELMMGNSSVFGPKQTTLDFLGLGRAV-G 412
Query: 478 SAGGGLSALITSIGGGSGLDVTAAAASFGNGEFSGKDIGR 517
+ G + L T +GGG+G+D+ A +FG+GEFSGKDI R
Sbjct: 413 NGNGPSNGLSTLVGGGTGIDM---ATTFGSGEFSGKDISR 449
>At5g44160 unknown protein
Length = 466
Score = 333 bits (855), Expect = 1e-91
Identities = 207/422 (49%), Positives = 246/422 (58%), Gaps = 64/422 (15%)
Query: 4 LDNVSTASGEASISSSGNNNIQSPIPKP--TKKKRNLPGMPDPEAEVIALSPTTLLATNR 61
L +S+ SG A SS + + P KKKRNLPG PDPEAEVIALSPTTL+ATNR
Sbjct: 6 LQTISSGSGFAQPQSSSTLDHDESLINPPLVKKKRNLPGNPDPEAEVIALSPTTLMATNR 65
Query: 62 FVCEICNKGFQRDQNLQLHRRGHNLPWKLRQRSSKEIRKRVYVCPEPTCVHHDPSRALGD 121
F+CE+C KGFQRDQNLQLHRRGHNLPWKL+QR+SKE+RKRVYVCPE TCVHH SRALGD
Sbjct: 66 FLCEVCGKGFQRDQNLQLHRRGHNLPWKLKQRTSKEVRKRVYVCPEKTCVHHHSSRALGD 125
Query: 122 LTGIKKHFCRKHGEKKWKCEKCSKKYAVQSDWKAHSKVCGSREYKCDCGTVFSRRDSFIT 181
LTGIKKHFCRKHGEKKW CEKC+K+YAVQSDWKAHSK CG+REY+CDCGT+FSRRDSFIT
Sbjct: 126 LTGIKKHFCRKHGEKKWTCEKCAKRYAVQSDWKAHSKTCGTREYRCDCGTIFSRRDSFIT 185
Query: 182 HRAFCDALAEENAKSQNQAVGKANSESDSKVLTGDSLPAVITTTAAAAATTPQSNSGVSS 241
HRAFCDALAEE AK + AV AAA P S +
Sbjct: 186 HRAFCDALAEETAK----------------------INAVSHLNGLAAAGAPGSVN---- 219
Query: 242 ALETQKLDLPENPP--QIIEEPQVVVTTTASALNASCSSSTSSKSNGCAATSTGVFASLF 299
L Q L PP + +PQ T + + TSS SL+
Sbjct: 220 -LNYQYLMGTFIPPLQPFVPQPQ----TNPNHHHQHFQPPTSSS------------LSLW 262
Query: 300 ASSTASASASLQPQAP---AFSDLIRSMGCTDPRPT-DFSAPPSSEAISLCLSTSHGSSI 355
A QPQ F + + C D T D ++ A +T S+
Sbjct: 263 MGQDI---APPQPQPDYDWVFGNAKAASACIDNNNTHDEQITQNANASLTTTTTLSAPSL 319
Query: 356 FGTGGQECRQYVPTHQPPAMSATALLQKAAQMGA-----AATN--ASLLRGLGIVSSSAS 408
F + + Q + MSATALLQKAA++GA AATN ++ L+ + S+ +
Sbjct: 320 FSS---DQPQNANANSNVNMSATALLQKAAEIGATSTTTAATNDPSTFLQSFPLKSTDQT 376
Query: 409 TS 410
TS
Sbjct: 377 TS 378
>At1g03840 putative DNA-binding protein
Length = 506
Score = 326 bits (835), Expect = 2e-89
Identities = 152/203 (74%), Positives = 171/203 (83%), Gaps = 8/203 (3%)
Query: 1 MVDLDNVSTASGEASISSSGNNNI--------QSPIPKPTKKKRNLPGMPDPEAEVIALS 52
M D ++SG SSS +++ +S P KKKRNLPG PDPEAEVIALS
Sbjct: 1 MTTEDQTISSSGGYVQSSSTTDHVDHHHHDQHESLNPPLVKKKRNLPGNPDPEAEVIALS 60
Query: 53 PTTLLATNRFVCEICNKGFQRDQNLQLHRRGHNLPWKLRQRSSKEIRKRVYVCPEPTCVH 112
P TL+ATNRF+CEIC KGFQRDQNLQLHRRGHNLPWKL+QR+SKE+RKRVYVCPE +CVH
Sbjct: 61 PKTLMATNRFLCEICGKGFQRDQNLQLHRRGHNLPWKLKQRTSKEVRKRVYVCPEKSCVH 120
Query: 113 HDPSRALGDLTGIKKHFCRKHGEKKWKCEKCSKKYAVQSDWKAHSKVCGSREYKCDCGTV 172
H P+RALGDLTGIKKHFCRKHGEKKWKCEKC+K+YAVQSDWKAHSK CG+REY+CDCGT+
Sbjct: 121 HHPTRALGDLTGIKKHFCRKHGEKKWKCEKCAKRYAVQSDWKAHSKTCGTREYRCDCGTI 180
Query: 173 FSRRDSFITHRAFCDALAEENAK 195
FSRRDSFITHRAFCDALAEE A+
Sbjct: 181 FSRRDSFITHRAFCDALAEETAR 203
>At5g03150 putative protein
Length = 501
Score = 325 bits (832), Expect = 5e-89
Identities = 206/416 (49%), Positives = 241/416 (57%), Gaps = 58/416 (13%)
Query: 18 SSGNNNIQSPIPKPTKKKRNLPGMPDPEAEVIALSPTTLLATNRFVCEICNKGFQRDQNL 77
+S N P KKKRN PG PD A+VIALSPTTL+ATNRFVCEICNKGFQRDQNL
Sbjct: 38 NSNPNPNAKPNSSSAKKKRNQPGTPD--ADVIALSPTTLMATNRFVCEICNKGFQRDQNL 95
Query: 78 QLHRRGHNLPWKLRQRSSKE-IRKRVYVCPEPTCVHHDPSRALGDLTGIKKHFCRKHGEK 136
QLHRRGHNLPWKL+QRS +E I+K+VY+CP TCVHHD SRALGDLTGIKKH+ RKHGEK
Sbjct: 96 QLHRRGHNLPWKLKQRSKQEVIKKKVYICPIKTCVHHDASRALGDLTGIKKHYSRKHGEK 155
Query: 137 KWKCEKCSKKYAVQSDWKAHSKVCGSREYKCDCGTVFSRRDSFITHRAFCDALAEENAK- 195
KWKCEKCSKKYAVQSDWKAH+K CG+REYKCDCGT+FSR+DSFITHRAFCDAL EE A+
Sbjct: 156 KWKCEKCSKKYAVQSDWKAHAKTCGTREYKCDCGTLFSRKDSFITHRAFCDALTEEGARM 215
Query: 196 ---SQNQAV---GKANSESDSKVLTGDSLPAVITTTAAAAATTPQSNSGVSSALETQKLD 249
S N V N ++S V+ +LP P N+ +S D
Sbjct: 216 SSLSNNNPVISTTNLNFGNESNVMNNPNLPHGF---VHRGVHHPDINAAISQFGLGFGHD 272
Query: 250 LPENPPQIIEEPQVVVTTTASALNASCSSSTSSKSN----GCAATSTGVFASLFASSTA- 304
L Q + E + +T L S SSS S TST +L +SST+
Sbjct: 273 LSAMHAQGLSEMVQMASTGNHHLFPSSSSSLPDFSGHHQFQIPMTSTNPSLTLSSSSTSQ 332
Query: 305 SASASLQPQAPAFSDLIRSMGCTDPRPTDFSAPPSSEAISLCLSTSHGSSIFGTGGQECR 364
SASLQ Q L S S +F + E +
Sbjct: 333 QTSASLQHQT--------------------------------LKDSSFSPLF-SSSSENK 359
Query: 365 QYVPTHQPPAMSATALLQKAAQMGAAATNA----SLLRGLGIVSSSASTSSGQQDS 416
Q P MSATALLQKAAQMG+ +N+ S G + SSSA+ S + S
Sbjct: 360 QNKPL---SPMSATALLQKAAQMGSTRSNSSTAPSFFAGPTMTSSSATASPPPRSS 412
>At1g55110 putative zinc finger protein
Length = 455
Score = 321 bits (822), Expect = 7e-88
Identities = 151/208 (72%), Positives = 180/208 (85%), Gaps = 14/208 (6%)
Query: 3 DLDNVSTASGEASISSSGN------NNI------QSPIPKPT-KKKRNLPGMPDPEAEVI 49
++ N+++ASG+ + SSGN +NI Q +P+ + K+KRN PG PDPEAEV+
Sbjct: 20 NMSNLTSASGDQASVSSGNRTETSGSNINQHHQEQCFVPQSSLKRKRNQPGNPDPEAEVM 79
Query: 50 ALSPTTLLATNRFVCEICNKGFQRDQNLQLHRRGHNLPWKLRQRSSKEI-RKRVYVCPEP 108
ALSP TL+ATNRF+CE+CNKGFQRDQNLQLH+RGHNLPWKL+QRS+K++ RK+VYVCPEP
Sbjct: 80 ALSPKTLMATNRFICEVCNKGFQRDQNLQLHKRGHNLPWKLKQRSNKDVVRKKVYVCPEP 139
Query: 109 TCVHHDPSRALGDLTGIKKHFCRKHGEKKWKCEKCSKKYAVQSDWKAHSKVCGSREYKCD 168
CVHH PSRALGDLTGIKKHF RKHGEKKWKCEKCSKKYAVQSDWKAH+K CG++EYKCD
Sbjct: 140 GCVHHHPSRALGDLTGIKKHFFRKHGEKKWKCEKCSKKYAVQSDWKAHAKTCGTKEYKCD 199
Query: 169 CGTVFSRRDSFITHRAFCDALAEENAKS 196
CGT+FSRRDSFITHRAFCDALAEE+A++
Sbjct: 200 CGTLFSRRDSFITHRAFCDALAEESARA 227
>At3g45260 zinc finger like protein
Length = 446
Score = 319 bits (818), Expect = 2e-87
Identities = 183/384 (47%), Positives = 234/384 (60%), Gaps = 52/384 (13%)
Query: 26 SPIPKPT-----KKKRNLPGMPDPEAEVIALSPTTLLATNRFVCEICNKGFQRDQNLQLH 80
+P P PT K+KRNLPG PDP+AEVIALSP +L+ TNRF+CE+CNKGF+RDQNLQLH
Sbjct: 27 NPNPNPTSSNSAKRKRNLPGNPDPDAEVIALSPNSLMTTNRFICEVCNKGFKRDQNLQLH 86
Query: 81 RRGHNLPWKLRQRSSKE-IRKRVYVCPEPTCVHHDPSRALGDLTGIKKHFCRKHGEKKWK 139
RRGHNLPWKL+QR++KE ++K+VY+CPE TCVHHDP+RALGDLTGIKKHF RKHGEKKWK
Sbjct: 87 RRGHNLPWKLKQRTNKEQVKKKVYICPEKTCVHHDPARALGDLTGIKKHFSRKHGEKKWK 146
Query: 140 CEKCSKKYAVQSDWKAHSKVCGSREYKCDCGTVFSRRDSFITHRAFCDALAEENAKSQNQ 199
C+KCSKKYAV SDWKAHSK+CG++EY+CDCGT+FSR+DSFITHRAFCDALAEE+A+
Sbjct: 147 CDKCSKKYAVMSDWKAHSKICGTKEYRCDCGTLFSRKDSFITHRAFCDALAEESAR---- 202
Query: 200 AVGKANSESDSKVLTGDSLPAVITTTAAAAATTPQSNSGVSSALETQKLDLPENPPQIIE 259
+ P + + +++A LD+ N I +
Sbjct: 203 -----------------------------FVSVPPAPAYLNNA-----LDVEVNHGNINQ 228
Query: 260 -EPQVVVTTTASALNASCSSSTSSKSNGCAAT-STGVFASLFASSTASASASLQ-----P 312
Q + TT+S L+ ++ + T T VFAS + S SAS SLQ
Sbjct: 229 NHQQRQLNTTSSQLDQPGFNTNRNNIAFLGQTLPTNVFASSSSPSPRSASDSLQNLWHLQ 288
Query: 313 QAPAFSDLIRSMGCTDPRPTDFSAPPSSEAISLCLSTSHGSSIFGTGGQECRQYVPT-HQ 371
+ L+ + + E + S+GS Y Q
Sbjct: 289 GQSSHQWLLNENNNNNNNILQRGISKNQEEHEMKNVISNGSLFSSEARNNTNNYNQNGGQ 348
Query: 372 PPAMSATALLQKAAQMGAAATNAS 395
+MSATALLQKAAQMG+ +++S
Sbjct: 349 IASMSATALLQKAAQMGSKRSSSS 372
>At3g13810 zinc finger protein, putative
Length = 513
Score = 318 bits (816), Expect = 3e-87
Identities = 155/229 (67%), Positives = 179/229 (77%), Gaps = 26/229 (11%)
Query: 3 DLDNVSTASGEASISSSGN---------------------NNIQSPIPKPTKKKRNLPGM 41
++ N+++ASG+ + SSGN + P + KK+RN PG
Sbjct: 19 NMSNLTSASGDQASVSSGNITEASGSNYFPHHQQQQEQQQQQLVVPDSQTQKKRRNQPGN 78
Query: 42 PDPEAEVIALSPTTLLATNRFVCEICNKGFQRDQNLQLHRRGHNLPWKLRQRSSKE-IRK 100
PDPE+EVIALSP TL+ATNRFVCEICNKGFQRDQNLQLHRRGHNLPWKL+QRS+KE IRK
Sbjct: 79 PDPESEVIALSPKTLMATNRFVCEICNKGFQRDQNLQLHRRGHNLPWKLKQRSNKEVIRK 138
Query: 101 RVYVCPEPTCVHHDPSRALGDLTGIKKHFCRKHGEKKWKCEKCSKKYAVQSDWKAHSKVC 160
+VYVCPE +CVHHDPSRALGDLTGIKKHFCRKHGEKKWKC+KCSKKYAVQSD KAHSK C
Sbjct: 139 KVYVCPEASCVHHDPSRALGDLTGIKKHFCRKHGEKKWKCDKCSKKYAVQSDCKAHSKTC 198
Query: 161 GSREYKCDCGTVFSRRDSFITHRAFCDALAEENAKS----QNQAVGKAN 205
G++EY+CDCGT+FSRRDSFITHRAFC+ALAEE A+ QNQ + N
Sbjct: 199 GTKEYRCDCGTLFSRRDSFITHRAFCEALAEETAREVVIPQNQNNNQPN 247
>At2g02080 C2H2-type zinc finger protein
Length = 516
Score = 313 bits (801), Expect = 2e-85
Identities = 203/495 (41%), Positives = 264/495 (53%), Gaps = 84/495 (16%)
Query: 31 PTKKKRNLPGMPDPEAEVIALSPTTLLATNRFVCEICNKGFQRDQNLQLHRRGHNLPWKL 90
P KK+RN PG P+P+AEV+ALSP TL+ATNRF+C++CNKGFQR+QNLQLHRRGHNLPWKL
Sbjct: 52 PPKKRRNQPGNPNPDAEVVALSPKTLMATNRFICDVCNKGFQREQNLQLHRRGHNLPWKL 111
Query: 91 RQRSSKEIRKRVYVCPEPTCVHHDPSRALGDLTGIKKHFCRKHGEKKWKCEKCSKKYAVQ 150
+Q+S+KE++++VY+CPEPTCVHHDPSRALGDLTGIKKH+ RKHGEKKWKCEKCSK+YAVQ
Sbjct: 112 KQKSTKEVKRKVYLCPEPTCVHHDPSRALGDLTGIKKHYYRKHGEKKWKCEKCSKRYAVQ 171
Query: 151 SDWKAHSKVCGSREYKCDCGTVFSRRDSFITHRAFCDALAEENAKSQNQAVGKANSESDS 210
SDWKAHSK CG++EY+CDCGT+FSRRDS+ITHRAFCDAL +E A++
Sbjct: 172 SDWKAHSKTCGTKEYRCDCGTIFSRRDSYITHRAFCDALIQETARN-------------- 217
Query: 211 KVLTGDSLPAVITTTAAAAATTPQSNS-----GVSSALETQKLDLPENPPQIIEEPQVVV 265
P V T+ AA++ S G SAL L + P P V
Sbjct: 218 --------PTVSFTSMTAASSGVGSGGIYGRLGGGSALSHHHL---SDHPNFGFNPLV-- 264
Query: 266 TTTASALNASCSSSTSSKSNGCAATSTGVFASLFASSTASASASLQPQAPAFSDLIRSMG 325
N + +SS ++ + +S F ASS + + +F + G
Sbjct: 265 -----GYNLNIASS-DNRRDFIPQSSNPNFLIQSASSQGMLNTTPNNNNQSF---MNQHG 315
Query: 326 CTDPRPTDFSAPPSSEAISLCLSTSHGSSIFGTG-GQECRQYVPTHQPPAMSATALLQKA 384
P D I+L S+ +S F G QE + T P S L+
Sbjct: 316 LIQFDPVD--------NINL-KSSGTNNSFFNLGFFQENTKNSETSLPSLYSTDVLVHHR 366
Query: 385 AQMGAAATNASLLRGLGIVSSSASTSSGQQDSLHWGLGPMEPEGSSLVSAGLGLG--LPC 442
+ A +N S L + S +S +L GL SS+++ G G +
Sbjct: 367 EENLNAGSNVSATALLQKATQMGSVTSNDPSALFRGLA-SSSNSSSVIANHFGGGRIMEN 425
Query: 443 DADSGLKELMLG---------------------TPSMFGPKQTTLDFLGL-GMAAGGSAG 480
D + L+ LM +M G + TLDFLG+ GM + G
Sbjct: 426 DNNGNLQGLMNSLAAVNGGGGSGGSIFDVQFGDNGNMSGSDKLTLDFLGVGGMVRNVNRG 485
Query: 481 GGLSALITSIGGGSG 495
GG GGG G
Sbjct: 486 GG--------GGGRG 492
>At4g02670 putative zinc finger protein
Length = 402
Score = 311 bits (798), Expect = 4e-85
Identities = 158/260 (60%), Positives = 192/260 (73%), Gaps = 23/260 (8%)
Query: 8 STASGEASISS----SGNNNIQSPIPKPT-----KKKRNLPGMPDPEAEVIALSPTTLLA 58
S +SG ++S+ SG +N+ S + T KKKR LPG PDP+AEVIALSP TLLA
Sbjct: 19 SASSGNNTLSTIQEFSGFHNVISSVCTHTETHKPKKKRGLPGNPDPDAEVIALSPKTLLA 78
Query: 59 TNRFVCEICNKGFQRDQNLQLHRRGHNLPWKLRQRSSKEI-RKRVYVCPEPTCVHHDPSR 117
TNRFVCEICNKGFQRDQNLQLHRRGHNLPWKL+Q+++KE +K+VYVCPE C HH PSR
Sbjct: 79 TNRFVCEICNKGFQRDQNLQLHRRGHNLPWKLKQKNTKEQQKKKVYVCPETNCAHHHPSR 138
Query: 118 ALGDLTGIKKHFCRKHGEKKWKCEKCSKKYAVQSDWKAHSKVCGSREYKCDCGTVFSRRD 177
ALGDLTGIKKHFCRKHGEKKWKCEKCSK YAVQSDWKAH+K+CG+R+Y+CDCGT+FSR+D
Sbjct: 139 ALGDLTGIKKHFCRKHGEKKWKCEKCSKFYAVQSDWKAHTKICGTRDYRCDCGTLFSRKD 198
Query: 178 SFITHRAFCDALAEENAK-----SQNQAVGKAN--------SESDSKVLTGDSLPAVITT 224
+FITHRAFCDALAEE+A+ S N N ++S S + T L +
Sbjct: 199 TFITHRAFCDALAEESARLHSTSSSNLTNPNPNFQGHHFMFNKSSSLLFTSSPLFIEPSL 258
Query: 225 TAAAAATTPQSNSGVSSALE 244
+ AA +T P + ++ L+
Sbjct: 259 STAALSTPPTAALSATALLQ 278
>At1g14580 putative zinc finger protein
Length = 467
Score = 311 bits (797), Expect = 6e-85
Identities = 131/171 (76%), Positives = 161/171 (93%)
Query: 26 SPIPKPTKKKRNLPGMPDPEAEVIALSPTTLLATNRFVCEICNKGFQRDQNLQLHRRGHN 85
S + P KK+RN PG P+P+AEVIALSP T++ATNRF+CE+CNKGFQR+QNLQLHRRGHN
Sbjct: 46 SSVAPPPKKRRNQPGNPNPDAEVIALSPKTIMATNRFLCEVCNKGFQREQNLQLHRRGHN 105
Query: 86 LPWKLRQRSSKEIRKRVYVCPEPTCVHHDPSRALGDLTGIKKHFCRKHGEKKWKCEKCSK 145
LPWKL+Q+S+KE+R++VY+CPEP+CVHHDP+RALGDLTGIKKH+ RKHGEKKWKC+KCSK
Sbjct: 106 LPWKLKQKSNKEVRRKVYLCPEPSCVHHDPARALGDLTGIKKHYYRKHGEKKWKCDKCSK 165
Query: 146 KYAVQSDWKAHSKVCGSREYKCDCGTVFSRRDSFITHRAFCDALAEENAKS 196
+YAVQSDWKAHSK CG++EY+CDCGT+FSRRDS+ITHRAFCDAL +E+A++
Sbjct: 166 RYAVQSDWKAHSKTCGTKEYRCDCGTIFSRRDSYITHRAFCDALIQESARN 216
>At2g02070 putative C2H2-type zinc finger protein
Length = 602
Score = 311 bits (796), Expect = 7e-85
Identities = 135/175 (77%), Positives = 162/175 (92%), Gaps = 4/175 (2%)
Query: 25 QSPIPK----PTKKKRNLPGMPDPEAEVIALSPTTLLATNRFVCEICNKGFQRDQNLQLH 80
Q+P+P P KKKRN P P+ +AEVIALSP TL+ATNRF+CE+CNKGFQR+QNLQLH
Sbjct: 40 QAPLPPLEAPPQKKKRNQPRTPNSDAEVIALSPKTLMATNRFICEVCNKGFQREQNLQLH 99
Query: 81 RRGHNLPWKLRQRSSKEIRKRVYVCPEPTCVHHDPSRALGDLTGIKKHFCRKHGEKKWKC 140
RRGHNLPWKL+Q+S+KE++++VY+CPEP+CVHHDPSRALGDLTGIKKH+ RKHGEKKWKC
Sbjct: 100 RRGHNLPWKLKQKSTKEVKRKVYLCPEPSCVHHDPSRALGDLTGIKKHYYRKHGEKKWKC 159
Query: 141 EKCSKKYAVQSDWKAHSKVCGSREYKCDCGTVFSRRDSFITHRAFCDALAEENAK 195
+KCSK+YAVQSDWKAHSK CG++EY+CDCGT+FSRRDSFITHRAFCDALA+E+A+
Sbjct: 160 DKCSKRYAVQSDWKAHSKTCGTKEYRCDCGTLFSRRDSFITHRAFCDALAQESAR 214
Score = 37.7 bits (86), Expect = 0.015
Identities = 41/126 (32%), Positives = 55/126 (43%), Gaps = 31/126 (24%)
Query: 373 PAMSATALLQKAAQMGAAATN-------------ASLLR--GLGIVSSSASTSSGQQDSL 417
P MSATALLQKAAQMG+ ++N +S+LR G GI + S +S
Sbjct: 433 PHMSATALLQKAAQMGSTSSNNNNGSNTNNNNNASSILRSFGSGIYGENESNLQDLMNSF 492
Query: 418 HWGLGPMEPEGSSLVSAGLGLGLPCDADSGL-KELMLGTPSMFGPKQTTLDFLGLGMAAG 476
P + V+ G+ P + G+ K L SM T DFLG+G
Sbjct: 493 ------SNPGATGNVN---GVDSPFGSYGGVNKGLSADKQSM------TRDFLGVGQIVK 537
Query: 477 GSAGGG 482
+G G
Sbjct: 538 SMSGSG 543
>At5g60470 putative zinc finger protein
Length = 392
Score = 259 bits (663), Expect = 2e-69
Identities = 109/140 (77%), Positives = 134/140 (94%), Gaps = 1/140 (0%)
Query: 57 LATNRFVCEICNKGFQRDQNLQLHRRGHNLPWKLRQRSSK-EIRKRVYVCPEPTCVHHDP 115
+ATNRF CEICNKGFQR+QNLQLH+RGHNLPWKL+Q+++K +++K+VY+CPE +CVHHDP
Sbjct: 1 MATNRFFCEICNKGFQREQNLQLHKRGHNLPWKLKQKTNKNQVKKKVYICPEKSCVHHDP 60
Query: 116 SRALGDLTGIKKHFCRKHGEKKWKCEKCSKKYAVQSDWKAHSKVCGSREYKCDCGTVFSR 175
+RALGDLTGIKKHF RKHGEKKWKC+KCSKKYAV SDWKAH+K+CGSRE++CDCGT+FSR
Sbjct: 61 ARALGDLTGIKKHFSRKHGEKKWKCDKCSKKYAVISDWKAHNKICGSREFRCDCGTLFSR 120
Query: 176 RDSFITHRAFCDALAEENAK 195
+DSFI+HR+FCD LAEE++K
Sbjct: 121 KDSFISHRSFCDVLAEESSK 140
>At2g01940 putative C2H2-type zinc finger protein
Length = 439
Score = 239 bits (611), Expect = 2e-63
Identities = 135/302 (44%), Positives = 175/302 (57%), Gaps = 27/302 (8%)
Query: 7 VSTASGEASISSSGNNNIQSPIPKPTKKKRNLPGMPDPEAEVIALSPTTLLATNRFVCEI 66
VS++S + +SSS N + + K+KR G PDP+AEV++LSP TLL ++R++CEI
Sbjct: 13 VSSSSSDPFLSSS-ENGVTTTNTSTQKRKRRPAGTPDPDAEVVSLSPRTLLESDRYICEI 71
Query: 67 CNKGFQRDQNLQLHRRGHNLPWKLRQRSSK-EIRKRVYVCPEPTCVHHDPSRALGDLTGI 125
CN+GFQRDQNLQ+HRR H +PWKL +R + E++KRVYVCPEPTC+HH+P ALGDL GI
Sbjct: 72 CNQGFQRDQNLQMHRRRHKVPWKLLKRDNNIEVKKRVYVCPEPTCLHHNPCHALGDLVGI 131
Query: 126 KKHFCRKH-GEKKWKCEKCSKKYAVQSDWKAHSKVCGSREYKCDCGTVFSRRDSFITHRA 184
KKHF RKH K+W CE+CSK YAVQSD+KAH K CG+R + CDCG VFSR +SFI H+
Sbjct: 132 KKHFRRKHSNHKQWVCERCSKGYAVQSDYKAHLKTCGTRGHSCDCGRVFSRVESFIEHQD 191
Query: 185 FCDALAEENAKSQNQAVGKANSESDSKVLTGDSLPAVITT-TAAAAATTPQ--------- 234
C A + S+ + S P+ T A TTPQ
Sbjct: 192 NCSARRVHREPPRPPQTAVTVPACSSRTASTVSTPSSETNYGGTVAVTTPQPLEGRPIHQ 251
Query: 235 -------SNSGVSSALETQKLDL-------PENPPQIIEEPQVVVTTTASALNASCSSST 280
+NS + LE Q L L EN Q ++EP N + S +
Sbjct: 252 RISSSILTNSSNNLNLELQLLPLSSNQNPNQENQQQKVKEPSHHHNHNHDTTNLNLSIAP 311
Query: 281 SS 282
SS
Sbjct: 312 SS 313
>At1g25250 Indeterminate 1 like Zn finger containing protein
Length = 385
Score = 234 bits (596), Expect = 1e-61
Identities = 107/157 (68%), Positives = 126/157 (80%), Gaps = 3/157 (1%)
Query: 33 KKKRNLPGMPDPEAEVIALSPTTLLATNRFVCEICNKGFQRDQNLQLHRRGHNLPWKLRQ 92
K+KR G PDP+AEV++LSP TLL ++R+VCEICN+GFQRDQNLQ+HRR H +PWKL +
Sbjct: 33 KRKRRPAGTPDPDAEVVSLSPRTLLESDRYVCEICNQGFQRDQNLQMHRRRHKVPWKLLK 92
Query: 93 RSSK--EIRKRVYVCPEPTCVHHDPSRALGDLTGIKKHFCRKHG-EKKWKCEKCSKKYAV 149
R K E+RKRVYVCPEPTC+HHDP ALGDL GIKKHF RKH K+W CE+CSK YAV
Sbjct: 93 RDKKDEEVRKRVYVCPEPTCLHHDPCHALGDLVGIKKHFRRKHSVHKQWVCERCSKGYAV 152
Query: 150 QSDWKAHSKVCGSREYKCDCGTVFSRRDSFITHRAFC 186
QSD+KAH K CGSR + CDCG VFSR +SFI H+ C
Sbjct: 153 QSDYKAHLKTCGSRGHSCDCGRVFSRVESFIEHQDTC 189
>At1g68130 putative C2H2-type zinc finger protein
Length = 419
Score = 231 bits (590), Expect = 6e-61
Identities = 114/225 (50%), Positives = 147/225 (64%), Gaps = 16/225 (7%)
Query: 33 KKKRNLPGMPDPEAEVIALSPTTLLATNRFVCEICNKGFQRDQNLQLHRRGHNLPWKLRQ 92
K+KR G PDPEAEV++LSP TLL ++R+VCEICN+GFQRDQNLQ+HRR H +PWKL +
Sbjct: 41 KRKRRPAGTPDPEAEVVSLSPRTLLESDRYVCEICNQGFQRDQNLQMHRRRHKVPWKLLK 100
Query: 93 R-SSKEIRKRVYVCPEPTCVHHDPSRALGDLTGIKKHFCRKH-GEKKWKCEKCSKKYAVQ 150
R +++E+RKRVYVCPEPTC+HH+P ALGDL GIKKHF RKH K+W CE+CSK YAVQ
Sbjct: 101 RETNEEVRKRVYVCPEPTCLHHNPCHALGDLVGIKKHFRRKHSNHKQWICERCSKGYAVQ 160
Query: 151 SDWKAHSKVCGSREYKCDCGTVFSRRDSFITHRAFCDALAEENAKSQNQAVGKANSESDS 210
SD+KAH K CG+R + CDCG VFSR +SFI H+ C + + S+
Sbjct: 161 SDYKAHLKTCGTRGHSCDCGRVFSRVESFIEHQDTCTV--------------RRSQPSNH 206
Query: 211 KVLTGDSLPAVITTTAAAAATTPQSNSGVSSALETQKLDLPENPP 255
++ T TA+ A + + L L ++PP
Sbjct: 207 RLHEQQQHTTNATQTASTAENNENGDLSIGPILPGHPLQRRQSPP 251
>At1g34370 zinc finger protein, putative
Length = 499
Score = 102 bits (253), Expect = 7e-22
Identities = 59/186 (31%), Positives = 89/186 (47%), Gaps = 19/186 (10%)
Query: 41 MPDPEAEVIALSPTTLLATNRFVCEICNKGFQRDQNLQLHRRGHNLPWKLRQRSSKEIRK 100
+P E++ L +LA + C IC KGF+RD NL++H RGH +K +K ++
Sbjct: 223 LPPGSYEILQLEKEEILAPHTHFCTICGKGFKRDANLRMHMRGHGDEYKTAAALAKPNKE 282
Query: 101 RV----------YVCPEPTC---VHHDPSRALGDLTGIKKHFCRKHGEKKWKCEKC-SKK 146
V Y CP C H + L + +K H+ R H +K + C +C +KK
Sbjct: 283 SVPGSEPMLIKRYSCPFLGCKRNKEHKKFQPLKTILCVKNHYKRTHCDKSFTCSRCHTKK 342
Query: 147 YAVQSDWKAHSKVCGSREYKCDCGTVFSRRDSFITHRAFCDALA-----EENAKSQNQAV 201
++V +D K H K CG ++ C CGT FSR+D H A EE S + +
Sbjct: 343 FSVIADLKTHEKHCGKNKWLCSCGTTFSRKDKLFGHIALFQGHTPAIPLEETKPSASTST 402
Query: 202 GKANSE 207
+ +SE
Sbjct: 403 QRGSSE 408
>At5g22890 unknown protein
Length = 235
Score = 101 bits (252), Expect = 9e-22
Identities = 51/150 (34%), Positives = 78/150 (52%), Gaps = 14/150 (9%)
Query: 47 EVIALSPTTLLATNRFVCEICNKGFQRDQNLQLHRRGHNLPWKLRQ----------RSSK 96
+++ L LLA C+IC KGF+RD NL++H R H +K R+ +
Sbjct: 64 DILELDVADLLAKYTHYCQICGKGFKRDANLRMHMRAHGDEYKTREALISPTSQDKKGGY 123
Query: 97 EIRKRVYVCPEPTC---VHHDPSRALGDLTGIKKHFCRKHGEKKWKCEKCS-KKYAVQSD 152
++K Y CP+ C H+ + L + K H+ R H K + C +CS K ++V SD
Sbjct: 124 SLKKHYYSCPQHGCRWNQRHEKFQPLKSVICAKNHYKRSHCPKMYMCRRCSVKHFSVLSD 183
Query: 153 WKAHSKVCGSREYKCDCGTVFSRRDSFITH 182
+ H K CG ++ C CGT FSR+D ++H
Sbjct: 184 LRTHEKHCGDIKWVCSCGTKFSRKDKLMSH 213
>At1g13290 hypothetical protein
Length = 302
Score = 96.7 bits (239), Expect = 3e-20
Identities = 57/167 (34%), Positives = 85/167 (50%), Gaps = 12/167 (7%)
Query: 56 LLATNRFVCEICNKGFQRDQNLQLHRRGHNL-----PWKLR--QRSSKEIRKRVYVCPE- 107
L+ +F C +CNK F R N+Q+H GH P LR + SS +R Y C E
Sbjct: 95 LVGPTQFSCSVCNKTFNRFNNMQMHMWGHGSQYRKGPESLRGTKSSSSILRLPCYCCAEG 154
Query: 108 -PTCVHHDPSRALGDLTGIKKHFCRKHGEKKWKC-EKCSKKYAVQSDWKAHSKVCGSREY 165
+ H S+ L D ++ H+ RKHG K ++C +KC K +AV+ DW+ H K CG + +
Sbjct: 155 CKNNIDHPRSKPLKDFRTLQTHYKRKHGAKPFRCRKKCEKTFAVRGDWRTHEKNCG-KLW 213
Query: 166 KCDCGTVFSRRDSFITH-RAFCDALAEENAKSQNQAVGKANSESDSK 211
C CG+ F + S H RAF D A + +G A+ + + +
Sbjct: 214 FCVCGSDFKHKRSLKDHVRAFGDGHAAHTVSDRVVGIGDADEDDEEE 260
>At1g08290 unknown protein
Length = 337
Score = 88.6 bits (218), Expect = 8e-18
Identities = 47/131 (35%), Positives = 65/131 (48%), Gaps = 10/131 (7%)
Query: 61 RFVCEICNKGFQRDQNLQLHRRGHNLPWKLRQRSSKE-------IRKRVYVCPE--PTCV 111
+F C IC+K F R N+Q+H GH ++ S K +R Y C E +
Sbjct: 179 QFACSICSKTFNRYNNMQMHMWGHGSEFRKGADSLKGTIQPAAILRLPCYCCAEGCKNNI 238
Query: 112 HHDPSRALGDLTGIKKHFCRKHGEKKWKCEKCSKKYAVQSDWKAHSKVCGSREYKCDCGT 171
+H S+ L D ++ H+ RKHG K + C KC K AV+ DW+ H K CG Y C CG+
Sbjct: 239 NHPRSKPLKDFRTLQTHYKRKHGSKPFSCGKCGKALAVKGDWRTHEKNCGKLWY-CTCGS 297
Query: 172 VFSRRDSFITH 182
F + S H
Sbjct: 298 DFKHKRSLKDH 308
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.312 0.127 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,856,500
Number of Sequences: 26719
Number of extensions: 532376
Number of successful extensions: 2593
Number of sequences better than 10.0: 107
Number of HSP's better than 10.0 without gapping: 45
Number of HSP's successfully gapped in prelim test: 63
Number of HSP's that attempted gapping in prelim test: 2072
Number of HSP's gapped (non-prelim): 415
length of query: 519
length of database: 11,318,596
effective HSP length: 104
effective length of query: 415
effective length of database: 8,539,820
effective search space: 3544025300
effective search space used: 3544025300
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC148484.5


