

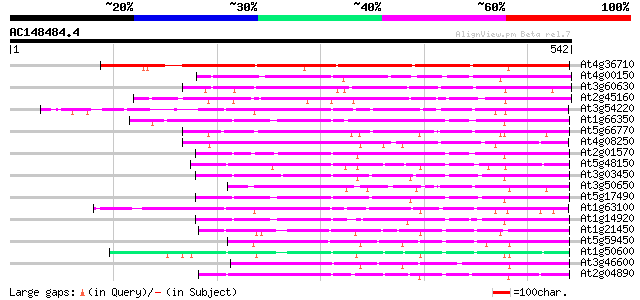
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148484.4 - phase: 0
(542 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g36710 SCARECROW-like protein 459 e-129
At4g00150 scarecrow-like 6 (SCL6) 192 4e-49
At3g60630 scarecrow - like protein 184 8e-47
At2g45160 putative SCARECROW gene regulator 175 5e-44
At3g54220 SCARECROW1 112 7e-25
At1g66350 gibberellin regulatory protein like 97 2e-20
At5g66770 SCARECROW gene regulator 96 7e-20
At4g08250 putative protein 94 3e-19
At2g01570 putative RGA1, giberellin repsonse modulation protein 91 2e-18
At5g48150 SCARECROW gene regulator-like 90 3e-18
At3g03450 RGA1-like protein 89 6e-18
At3g50650 scarecrow-like 7 (SCL7) 89 8e-18
At5g17490 RGA-like protein 88 1e-17
At1g63100 transcription factor SCARECROW, putative 88 1e-17
At1g14920 signal response protein (GAI) 88 1e-17
At1g21450 scarecrow-like 1 (SCL1) 87 3e-17
At5g59450 putative SCARECROW transcriptional regulatory protein ... 85 1e-16
At1g50600 scarecrow-like 5 (SCL5) 84 2e-16
At3g46600 scarecrow-like protein 80 3e-15
At2g04890 putative SCARECROW gene regulator 78 1e-14
>At4g36710 SCARECROW-like protein
Length = 486
Score = 459 bits (1182), Expect = e-129
Identities = 267/470 (56%), Positives = 329/470 (69%), Gaps = 38/470 (8%)
Query: 88 WDSIMKDLGLQDDSSTPIIPLLKNTNNT-SEIYPNPSQDQF-----DQTQ--DFTSLSDI 139
WDSIMK+L L DDS+ + T T S I P + D DQ Q DF S SD+
Sbjct: 38 WDSIMKELELDDDSAPNSLKTGFTTTTTDSTILPLYAVDSNLPGFPDQIQPSDFESSSDV 97
Query: 140 Y-SNNQNLAYNYPNTNTNLDHLVNDFNNNQPNNNTNSNWDFIEELIRAADCFDNNHLQLA 198
Y NQ Y + N ++ N +DFIE+LIR DC +++ LQLA
Sbjct: 98 YPGQNQTTGYGF----------------NSLDSVDNGGFDFIEDLIRVVDCVESDELQLA 141
Query: 199 QAILERLNQRLRSPTGKPLHRAAFHFKDALQSLLSGSNRTNPPRLSSMVEIVQTIRTFKA 258
Q +L RLNQRLRSP G+PL RAAF+FK+AL S L+GSNR NP RLSS EIVQ IR K
Sbjct: 142 QVVLSRLNQRLRSPAGRPLQRAAFYFKEALGSFLTGSNR-NPIRLSSWSEIVQRIRAIKE 200
Query: 259 FSGISPIPMFSIFTTNQALLEALHG---SLYMHVVDFEIGLGIQYASLMKEIAEKAVNGS 315
+SGISPIP+FS FT NQA+L++L S ++HVVDFEIG G QYASLM+EI EK+V+G
Sbjct: 201 YSGISPIPLFSHFTANQAILDSLSSQSSSPFVHVVDFEIGFGGQYASLMREITEKSVSGG 260
Query: 316 PLLRITAVVPEEYAVESRLIRENLNQFAHDLGIRVQVDFVPLRTFETVSFKAVRFVDGEK 375
LR+TAVV EE AVE+RL++ENL QFA ++ IR Q++FV ++TFE +SFKA+RFV+GE+
Sbjct: 261 -FLRVTAVVAEECAVETRLVKENLTQFAAEMKIRFQIEFVLMKTFEMLSFKAIRFVEGER 319
Query: 376 TAILLTPAIFCRLGSEGTAAFLSDVRRITPGVVVFVDGEGWTEAAAAASFRRGVVNSLEF 435
T +L++PAIF RL G F++++RR++P VVVFVD EGWTE A + SFRR V++LEF
Sbjct: 320 TVVLISPAIFRRLS--GITDFVNNLRRVSPKVVVFVDSEGWTEIAGSGSFRREFVSALEF 377
Query: 436 YSMMLESLDASVAAGGGGEWARRIEMLLLRPKIIAAVE-AAGRRTT---PWREAFYGAGM 491
Y+M+LESLDA AA G + +E +LRPKI AAVE AA RR T WREAF AGM
Sbjct: 378 YTMVLESLDA--AAPPGDLVKKIVEAFVLRPKISAAVETAADRRHTGEMTWREAFCAAGM 435
Query: 492 RPVQLSQFADFQAECLLAKVQIRGFHVAKRQAELVLFWHERAMVATSAWR 541
RP+Q SQFADFQAECLL K Q+RGFHVAKRQ ELVL WH RA+VATSAWR
Sbjct: 436 RPIQQSQFADFQAECLLEKAQVRGFHVAKRQGELVLCWHGRALVATSAWR 485
>At4g00150 scarecrow-like 6 (SCL6)
Length = 558
Score = 192 bits (488), Expect = 4e-49
Identities = 131/370 (35%), Positives = 203/370 (54%), Gaps = 29/370 (7%)
Query: 181 EELIRAADCFDNNHLQLAQAILERLNQRLRSPTGKPLHRAAFHFKDALQSLLSGSNRTNP 240
E+L++AA+ +++ LAQ IL RLNQ+L SP GKPL RAAF+FK+AL +LL ++T
Sbjct: 207 EQLVKAAEVIESDTC-LAQGILARLNQQLSSPVGKPLERAAFYFKEALNNLLHNVSQTLN 265
Query: 241 PRLSSMVEIVQTIRTFKAFSGISPIPMFSIFTTNQALLEALHGSLYMHVVDFEIGLGIQY 300
P ++ I +K+FS ISP+ F+ FT+NQALLE+ HG +H++DF+IG G Q+
Sbjct: 266 P-----YSLIFKIAAYKSFSEISPVLQFANFTSNQALLESFHGFHRLHIIDFDIGYGGQW 320
Query: 301 ASLMKEIAEKAVNGSPL-LRIT--AVVPEEYAVESRLIRENLNQFAHDLGIRVQVDFVPL 357
ASLM+E+ + N +PL L+IT A +E ++NL FA ++ I + + + L
Sbjct: 321 ASLMQELVLRD-NAAPLSLKITVFASPANHDQLELGFTQDNLKHFASEINISLDIQVLSL 379
Query: 358 RTFETVSFKAVRFVDGEKTAILLTPAIFCRLGSEGTAAFLSDVRRITPGVVVFVD-GEGW 416
++S+ + E A+ ++ A F L L V+ ++P ++V D G
Sbjct: 380 DLLGSISWP--NSSEKEAVAVNISAASFSHL-----PLVLRFVKHLSPTIIVCSDRGCER 432
Query: 417 TEAAAAASFRRGVVNSLEFYSMMLESLDASVAAGGGGEWARRIEMLLLRPKIIAAV---- 472
T+ F + + +SL ++ + ESLD A + ++IE L++P+I V
Sbjct: 433 TD----LPFSQQLAHSLHSHTALFESLD---AVNANLDAMQKIERFLIQPEIEKLVLDRS 485
Query: 473 EAAGRRTTPWREAFYGAGMRPVQLSQFADFQAECLLAKVQIRGFHVAKRQAELVLFWHER 532
R W+ F G PV S F + QAECL+ + +RGFHV K+ L+L W
Sbjct: 486 RPIERPMMTWQAMFLQMGFSPVTHSNFTESQAECLVQRTPVRGFHVEKKHNSLLLCWQRT 545
Query: 533 AMVATSAWRC 542
+V SAWRC
Sbjct: 546 ELVGVSAWRC 555
>At3g60630 scarecrow - like protein
Length = 623
Score = 184 bits (468), Expect = 8e-47
Identities = 142/401 (35%), Positives = 207/401 (51%), Gaps = 35/401 (8%)
Query: 168 QPNNNTNSNWDFIEELIRAA-----DCFDNNHLQLAQAILERLNQRLRSPTGK------- 215
+ N+ + + I++L AA + DNN + LAQ IL RLN L +
Sbjct: 232 EDQNDQDQSAVIIDQLFSAAAELTTNGGDNNPV-LAQGILARLNHNLNNNNDDTNNNPKP 290
Query: 216 PLHRAAFHFKDALQSLLSGSNRTNPPRLSSMVEIVQTIRTFKAFSGISPIPMFSIFTTNQ 275
P HRAA + +AL SLL S+ +PP LS ++ I ++AFS SP F FT NQ
Sbjct: 291 PFHRAASYITEALHSLLQDSS-LSPPSLSPPQNLIFRIAAYRAFSETSPFLQFVNFTANQ 349
Query: 276 ALLEALHGSLYMHVVDFEIGLGIQYASLMKEIAEKAVNGS--PLLRITA-----VVPEEY 328
+LE+ G +H+VDF+IG G Q+ASL++E+A K S P L+ITA V +E+
Sbjct: 350 TILESFEGFDRIHIVDFDIGYGGQWASLIQELAGKRNRSSSAPSLKITAFASPSTVSDEF 409
Query: 329 AVESRLIRENLNQFAHDLGIRVQVDFVPLRTFETVSFKAVR-FVDGEKTAILLTPAIFCR 387
E R ENL FA + G+ +++ + + ++ + F EK AI + I
Sbjct: 410 --ELRFTEENLRSFAGETGVSFEIELLNMEILLNPTYWPLSLFRSSEKEAIAVNLPI-SS 466
Query: 388 LGSEGTAAFLSDVRRITPGVVVFVDGEGWTEAAAAASFRRGVVNSLEFYSMMLESLDASV 447
+ S L +++I+P VVV D + A F GV+N+L++Y+ +LESLD+
Sbjct: 467 MVSGYLPLILRFLKQISPNVVVCSDRS--CDRNNDAPFPNGVINALQYYTSLLESLDSGN 524
Query: 448 AAGGGGEWARRIEMLLLRPKIIAAVEAAGR---RTTPWREAFYGAGMRPVQLSQFADFQA 504
E A IE ++P I + R R+ PWR F G PV LSQ A+ QA
Sbjct: 525 L--NNAEAATSIERFCVQPSIQKLLTNRYRWMERSPPWRSLFGQCGFTPVTLSQTAETQA 582
Query: 505 ECLLAKVQIRGFHVAKRQA---ELVLFWHERAMVATSAWRC 542
E LL + +RGFH+ KRQ+ LVL W + +V SAW+C
Sbjct: 583 EYLLQRNPMRGFHLEKRQSSSPSLVLCWQRKELVTVSAWKC 623
>At2g45160 putative SCARECROW gene regulator
Length = 640
Score = 175 bits (444), Expect = 5e-44
Identities = 135/444 (30%), Positives = 217/444 (48%), Gaps = 39/444 (8%)
Query: 120 PNPSQDQFDQTQDFTSLSDIYSNNQNLAYNYPNTNTNLDHLVNDFNNNQPNNNTNSNWDF 179
P+P D + F Y NNQ + +++T + + P +
Sbjct: 215 PDPGHDPVRRQHQFQF--PFYHNNQQQQFPSSSSSTAVAMVPVP----SPGMAGDDQSVI 268
Query: 180 IEELIRAADCF------DNNHLQLAQAILERLNQRLRSPTG--KPLHRAAFHFKDALQSL 231
IE+L AA+ + +H LAQ IL RLN L + + P RAA H +AL SL
Sbjct: 269 IEQLFNAAELIGTTGNNNGDHTVLAQGILARLNHHLNTSSNHKSPFQRAASHIAEALLSL 328
Query: 232 LSGSNRTNPPRLSSMVEIVQTIRTFKAFSGISPIPMFSIFTTNQALLEALHGSLY--MHV 289
+ N ++PP L + ++ I +++FS SP F FT NQ++LE+ + S + +H+
Sbjct: 329 IH--NESSPP-LITPENLILRIAAYRSFSETSPFLQFVNFTANQSILESCNESGFDRIHI 385
Query: 290 VDFEIGLGIQYASLMKEIAE----KAVNGSPLLRITAVVPEEYAV----ESRLIRENLNQ 341
+DF++G G Q++SLM+E+A + N + L++T P V E R ENL
Sbjct: 386 IDFDVGYGGQWSSLMQELASGVGGRRRNRASSLKLTVFAPPPSTVSDEFELRFTEENLKT 445
Query: 342 FAHDLGIRVQVDFVPLRTFETVSFKAVRFVDGEKTAILLTPAIFCRLGSEGTAAFLSDVR 401
FA ++ I +++ + + ++ + EK AI + + + S L ++
Sbjct: 446 FAGEVKIPFEIELLSVELLLNPAYWPLSLRSSEKEAIAVNLPVNS-VASGYLPLILRFLK 504
Query: 402 RITPGVVVFVDGEGWTEAAAAASFRRGVVNSLEFYSMMLESLDASVAAGGGGEWARRIEM 461
+++P +VV D G A F V++SL++++ +LESLDA+ IE
Sbjct: 505 QLSPNIVVCSD-RGCDRNDAP--FPNAVIHSLQYHTSLLESLDANQNQDDSS-----IER 556
Query: 462 LLLRPKIIAAVEAAGR---RTTPWREAFYGAGMRPVQLSQFADFQAECLLAKVQIRGFHV 518
++P I + R R+ PWR F G P LSQ A+ QAECLL + +RGFHV
Sbjct: 557 FWVQPSIEKLLMKRHRWIERSPPWRILFTQCGFSPASLSQMAEAQAECLLQRNPVRGFHV 616
Query: 519 AKRQAELVLFWHERAMVATSAWRC 542
KRQ+ LV+ W + +V SAW+C
Sbjct: 617 EKRQSSLVMCWQRKELVTVSAWKC 640
>At3g54220 SCARECROW1
Length = 653
Score = 112 bits (279), Expect = 7e-25
Identities = 127/529 (24%), Positives = 227/529 (42%), Gaps = 90/529 (17%)
Query: 30 PTPNPTPTPTTTTLCYEPRSVLELCRSPSP---EKKPSQQEQEHNPE---VEQEDHASLP 83
P+ + P+P T +EP + ++ +PSP +++ QQ+Q+H P ++Q++
Sbjct: 193 PSSSSDPSPQT----FEP--LYQISNNPSPPQQQQQHQQQQQQHKPPPPPIQQQER---- 242
Query: 84 NLDWWDSIMKDLGLQDDSSTPIIPLLKNTNNTSEIYPNPSQDQFDQTQDFTSLSDIYSNN 143
++ D Q ++ T +P ++ NT+E ++ Q QD L
Sbjct: 243 -----ENSSTDAPPQPETVTATVPAVQT--NTAEALRERKEEIKRQKQDEEGL------- 288
Query: 144 QNLAYNYPNTNTNLDHLVNDFNNNQPNNNTNSNWDFIEELIRAADCFDNNHLQLAQAILE 203
HL+ L++ A+ ++L+ A +L
Sbjct: 289 ---------------HLLT-------------------LLLQCAEAVSADNLEEANKLLL 314
Query: 204 RLNQRLRSPTGKPLHRAAFHFKDALQSLLSGS----NRTNPPRLSSMVEIVQTIRTFKAF 259
++Q L +P G R A +F +A+ + L S P R ++ + F+ F
Sbjct: 315 EISQ-LSTPYGTSAQRVAAYFSEAMSARLLNSCLGIYAALPSRWMPQTHSLKMVSAFQVF 373
Query: 260 SGISPIPMFSIFTTNQALLEALHGSLYMHVVDFEIGLGIQYASLMKEIAEKAVNGSPLLR 319
+GISP+ FS FT NQA+ EA +H++D +I G+Q+ L +A + G P +R
Sbjct: 374 NGISPLVKFSHFTANQAIQEAFEKEDSVHIIDLDIMQGLQWPGLFHILASRP-GGPPHVR 432
Query: 320 ITAVVPEEYAVESRLIRENLNQFAHDLGIRVQVDFVPL-RTFETVSFKAVRFVDGEKTAI 378
+T + A+++ + L+ FA LG+ +F PL + + + E A+
Sbjct: 433 LTGLGTSMEALQA--TGKRLSDFADKLGL--PFEFCPLAEKVGNLDTERLNVRKREAVAV 488
Query: 379 -LLTPAIFCRLGSEGTAAFLSDVRRITPGVVVFVDGEGWTEAAAAASFRRGVVNSLEFYS 437
L +++ GS+ +L ++R+ P VV V+ + + A SF V ++ +YS
Sbjct: 489 HWLQHSLYDVTGSDAHTLWL--LQRLAPKVVTVVE----QDLSHAGSFLGRFVEAIHYYS 542
Query: 438 MMLESLDASVAAGGGGEWARRIEMLLLRPKI--IAAVEAAGR----RTTPWREAFYGAGM 491
+ +SL AS G E +E LL +I + AV R + WRE G
Sbjct: 543 ALFDSLGASY--GEESEERHVVEQQLLSKEIRNVLAVGGPSRSGEVKFESWREKMQQCGF 600
Query: 492 RPVQLSQFADFQAECLLAKVQIRGFHVAKRQAELVLFWHERAMVATSAW 540
+ + L+ A QA LL G+ + L L W + +++ SAW
Sbjct: 601 KGISLAGNAATQATLLLGMFPSDGYTLVDDNGTLKLGWKDLSLLTASAW 649
>At1g66350 gibberellin regulatory protein like
Length = 511
Score = 97.4 bits (241), Expect = 2e-20
Identities = 102/439 (23%), Positives = 190/439 (43%), Gaps = 34/439 (7%)
Query: 116 SEIYPNPSQDQFDQTQDFTSL--SDIYSNNQNLAYNYPNTNTNLDHLVNDFNNNQPNNNT 173
S++ P Q++ D D ++ S +Y ++++ + T ++ ++ + ++
Sbjct: 89 SDLDPTRIQEKPDSEYDLRAIPGSAVYPRDEHVTRR--SKRTRIESELSSTRSVVVLDSQ 146
Query: 174 NSNWDFIEELIRAADCFDNNHLQLAQAILERLNQRLRSPTGKPLHRAAFHFKDALQSLLS 233
+ + L+ A+ N+L+LA A+++ + S G + + A +F + L +
Sbjct: 147 ETGVRLVHALLACAEAVQQNNLKLADALVKHVGLLASSQAGA-MRKVATYFAEGLARRIY 205
Query: 234 GSNRTNPPRLSSMVEIVQTIRTFKAFSGISPIPMFSIFTTNQALLEALHGSLYMHVVDFE 293
+ LSS + +Q F P F+ FT NQA+LE + +HV+D
Sbjct: 206 RIYPRDDVALSSFSDTLQI-----HFYESCPYLKFAHFTANQAILEVFATAEKVHVIDLG 260
Query: 294 IGLGIQYASLMKEIAEKAVNGSPLLRITAVVPEEYAV-ESRLIRENLNQFAHDLGIRVQV 352
+ G+Q+ +L++ +A + NG P R+T + Y++ + + + L Q A +G+ +
Sbjct: 261 LNHGLQWPALIQALALRP-NGPPDFRLTGI---GYSLTDIQEVGWKLGQLASTIGVNFEF 316
Query: 353 DFVPLRTFETVSFKAVRFVDG-EKTAILLTPAIFCRLGSEGTA-AFLSDVRRITPGVVVF 410
+ L + + + G E A+ + L G+ FLS ++ I P ++
Sbjct: 317 KSIALNNLSDLKPEMLDIRPGLESVAVNSVFELHRLLAHPGSIDKFLSTIKSIRPDIMTV 376
Query: 411 VDGEGWTEAAAAASFRRGVVNSLEFYSMMLESLDASVAAGGGGEWARRIEMLLLRPKIIA 470
V+ E F SL +YS + +SL+ G R + L L +I+
Sbjct: 377 VEQEANHNGTV---FLDRFTESLHYYSSLFDSLE------GPPSQDRVMSELFLGRQILN 427
Query: 471 AVEAAG-------RRTTPWREAFYGAGMRPVQLSQFADFQAECLLA-KVQIRGFHVAKRQ 522
V G WR F G +PV + A QA LLA G++V + +
Sbjct: 428 LVACEGEDRVERHETLNQWRNRFGLGGFKPVSIGSNAYKQASMLLALYAGADGYNVEENE 487
Query: 523 AELVLFWHERAMVATSAWR 541
L+L W R ++ATSAWR
Sbjct: 488 GCLLLGWQTRPLIATSAWR 506
>At5g66770 SCARECROW gene regulator
Length = 584
Score = 95.5 bits (236), Expect = 7e-20
Identities = 103/395 (26%), Positives = 175/395 (44%), Gaps = 33/395 (8%)
Query: 168 QPNNNTNSNWDFIEELIRAA-DCF---DNNHLQLAQAILERLNQRLRSPTGKPLHRAAFH 223
+ N++ + ++D L++A DC D++ + ++ +L+ + S G P R AF+
Sbjct: 202 ETNDSEDDDFDLEPPLLKAIYDCARISDSDPNEASKTLLQI--RESVSELGDPTERVAFY 259
Query: 224 FKDALQSLLSGSNRTNPPRLSSMVEIVQTIRTFKAFSGISPIPMFSIFTTNQALLEALHG 283
F +AL + LS ++ P SS I ++K + P F+ T NQA+LEA
Sbjct: 260 FTEALSNRLSPNS---PATSSSSSSTEDLILSYKTLNDACPYSKFAHLTANQAILEATEK 316
Query: 284 SLYMHVVDFEIGLGIQYASLMKEIAEKAVNGSPLLRITAVVPEEY--AVESRLIR--ENL 339
S +H+VDF I GIQ+ +L++ +A + +R++ + + E LI L
Sbjct: 317 SNKIHIVDFGIVQGIQWPALLQALATRTSGKPTQIRVSGIPAPSLGESPEPSLIATGNRL 376
Query: 340 NQFAHDLGIRVQVDFVPLRT-FETVSFKAVRFVDGEKTAILLTPAIFCRLGSEGTA--AF 396
FA L + DF+P+ T ++ + R E A+ ++ L T
Sbjct: 377 RDFAKVLDL--NFDFIPILTPIHLLNGSSFRVDPDEVLAVNFMLQLYKLLDETPTIVDTA 434
Query: 397 LSDVRRITPGVVVFVDGEGWTEAAAAASFRRGVVNSLEFYSMMLESLDASVAAGGGGEWA 456
L + + P VV GE + + F V N+L+FYS + ESL+ ++ G E
Sbjct: 435 LRLAKSLNPRVVTL--GE-YEVSLNRVGFANRVKNALQFYSAVFESLEPNL--GRDSEER 489
Query: 457 RRIEMLLLRPKIIAAV--EAAG------RRTTPWREAFYGAGMRPVQLSQFADFQAECLL 508
R+E L +I + E G WR AG V+LS +A QA+ LL
Sbjct: 490 VRVERELFGRRISGLIGPEKTGIHRERMEEKEQWRVLMENAGFESVKLSNYAVSQAKILL 549
Query: 509 AKVQIRGFH--VAKRQAELVLFWHERAMVATSAWR 541
+ V + + L W++ ++ S+WR
Sbjct: 550 WNYNYSNLYSIVESKPGFISLAWNDLPLLTLSSWR 584
>At4g08250 putative protein
Length = 483
Score = 93.6 bits (231), Expect = 3e-19
Identities = 103/392 (26%), Positives = 164/392 (41%), Gaps = 27/392 (6%)
Query: 168 QPNNNTNSNWDFIEELIRAADCFD--NNHLQLAQAILERLNQRLRSPTGKPLHRAAFHFK 225
+P + + + L+ AAD N +L + IL RL + + R A HF
Sbjct: 93 EPKTDESKGLRLVHLLVAAADASTGANKSRELTRVILARLKDLVSPGDRTNMERLAAHFT 152
Query: 226 DALQSLLSGSNRTNPPR-LSSMVEIVQTIRTFKAFSGISPIPMFSIFTTNQALLEALHGS 284
+ L LL + P + + + I F+ +SP F T QA+LEA+
Sbjct: 153 NGLSKLLERDSVLCPQQHRDDVYDQADVISAFELLQNMSPYVNFGYLTATQAILEAVKYE 212
Query: 285 LYMHVVDFEIGLGIQYASLMKEIAEKAVNGSPL-LRITAVVPEEYAVES-RLIRE---NL 339
+H+VD++I G+Q+ASLM+ + + S LRITA+ +S ++E L
Sbjct: 213 RRIHIVDYDINEGVQWASLMQALVSRNTGPSAQHLRITALSRATNGKKSVAAVQETGRRL 272
Query: 340 NQFAHDLGIRVQVDFVPLRT--FETVSFKAVRFVDGEKTAI--LLTPAIFCRLGSEGTAA 395
FA +G L T F T S K VR GE I +L F +
Sbjct: 273 TAFADSIGQPFSYQHCKLDTNAFSTSSLKLVR---GEAVVINCMLHLPRFSHQTPSSVIS 329
Query: 396 FLSDVRRITPGVVVFVDGEGWTEAAAAASFRRGVVNSLEFYSMMLESLDASVAAGGGGEW 455
FLS+ + + P +V V E F ++ L +S + +SL+A ++
Sbjct: 330 FLSEAKTLNPKLVTLVHEE--VGLMGNQGFLYRFMDLLHQFSAIFDSLEAGLSIANPARG 387
Query: 456 ARRIEMLLLRPKI------IAAVEAAGRRTTPWREAFYGAGMRPVQLSQFADFQAECLLA 509
+E + + P + I A +A W + G +P+++S QA+ LL+
Sbjct: 388 F--VERVFIGPWVANWLTRITANDAEVESFASWPQWLETNGFKPLEVSFTNRCQAKLLLS 445
Query: 510 KVQIRGFHVAK-RQAELVLFWHERAMVATSAW 540
GF V + Q LVL W R +V+ S W
Sbjct: 446 LFN-DGFRVEELGQNGLVLGWKSRRLVSASFW 476
>At2g01570 putative RGA1, giberellin repsonse modulation protein
Length = 587
Score = 90.5 bits (223), Expect = 2e-18
Identities = 96/375 (25%), Positives = 160/375 (42%), Gaps = 28/375 (7%)
Query: 180 IEELIRAADCFDNNHLQLAQAILERLNQRLRSPTGKPLHRAAFHFKDALQSLLSGSNRTN 239
+ L+ A+ N+L LA+A+++++ S G + + A +F +AL + R +
Sbjct: 222 VHALMACAEAIQQNNLTLAEALVKQIGCLAVSQAGA-MRKVATYFAEALARRIY---RLS 277
Query: 240 PPRLSSMVEIVQTIRTFKAFSGISPIPMFSIFTTNQALLEALHGSLYMHVVDFEIGLGIQ 299
PP+ + T++ F P F+ FT NQA+LEA G +HV+DF + G+Q
Sbjct: 278 PPQNQIDHCLSDTLQMH--FYETCPYLKFAHFTANQAILEAFEGKKRVHVIDFSMNQGLQ 335
Query: 300 YASLMKEIAEKAVNGSPLLRITAVVPEEYAVESRL--IRENLNQFAHDLGIRVQV-DFVP 356
+ +LM+ +A + G P R+T + P L + L Q A + + + FV
Sbjct: 336 WPALMQALALRE-GGPPTFRLTGIGPPAPDNSDHLHEVGCKLAQLAEAIHVEFEYRGFVA 394
Query: 357 LRTFE-TVSFKAVRFVDGEKTAILLTPAIFCRLGSE-GTAAFLSDVRRITPGVVVFVDGE 414
+ S +R D E A+ + LG G L V++I P + V+ E
Sbjct: 395 NSLADLDASMLELRPSDTEAVAVNSVFELHKLLGRPGGIEKVLGVVKQIKPVIFTVVEQE 454
Query: 415 GWTEAAAAASFRRGVVNSLEFYSMMLESLDASVAAGGGGEWARRIEMLLLRPKIIAAVEA 474
F SL +YS + +SL+ G + + + L +I V
Sbjct: 455 SNHNGPV---FLDRFTESLHYYSTLFDSLE-----GVPNSQDKVMSEVYLGKQICNLVAC 506
Query: 475 AG-------RRTTPWREAFYGAGMRPVQLSQFADFQAECLLAKVQI-RGFHVAKRQAELV 526
G + W F +G+ P L A QA LL+ +G+ V + L+
Sbjct: 507 EGPDRVERHETLSQWGNRFGSSGLAPAHLGSNAFKQASMLLSVFNSGQGYRVEESNGCLM 566
Query: 527 LFWHERAMVATSAWR 541
L WH R ++ TSAW+
Sbjct: 567 LGWHTRPLITTSAWK 581
>At5g48150 SCARECROW gene regulator-like
Length = 490
Score = 90.1 bits (222), Expect = 3e-18
Identities = 97/388 (25%), Positives = 167/388 (43%), Gaps = 33/388 (8%)
Query: 175 SNWDFIEELIRAADCFDNNHLQLAQAILERLNQRLRSPTGKPLHRAAFHFKDALQSLLSG 234
S D +L+ A N L +A +++E+L Q + S +G+P+ R + + L + L+
Sbjct: 115 SRRDLRADLVSCAKAMSENDLMMAHSMMEKLRQMV-SVSGEPIQRLGAYLLEGLVAQLAS 173
Query: 235 SNRTNPPRLSSMVEIVQT--IRTFKAFSGISPIPMFSIFTTNQALLEALHGSLYMHVVDF 292
S + L+ E T + + P F + N A+ EA+ +H++DF
Sbjct: 174 SGSSIYKALNRCPEPASTELLSYMHILYEVCPYFKFGYMSANGAIAEAMKEENRVHIIDF 233
Query: 293 EIGLGIQYASLMKEIAEKAVNGSPLLRITAV--VPEEYAVESRL--IRENLNQFAHDLGI 348
+IG G Q+ +L++ A + G P +RIT + + YA L + L + A +
Sbjct: 234 QIGQGSQWVTLIQAFAARP-GGPPRIRITGIDDMTSAYARGGGLSIVGNRLAKLAKQFNV 292
Query: 349 RVQVDFVPLRTFETVSFKAVRFVDGEKTAILLTPAIFCRLGSEGTAA------FLSDVRR 402
+ + V + E V K + GE A+ + + E + L V+
Sbjct: 293 PFEFNSVSVSVSE-VKPKNLGVRPGEALAVNFA-FVLHHMPDESVSTENHRDRLLRMVKS 350
Query: 403 ITPGVVVFVDGEGWTEAAAAASFRRGVVNSLEFYSMMLESLDASVAAGGGGEWARRIEM- 461
++P VV V+ E T AA F + ++ +Y+ M ES+D ++ + +RI +
Sbjct: 351 LSPKVVTLVEQESNTNTAA---FFPRFMETMNYYAAMFESIDVTLPR----DHKQRINVE 403
Query: 462 --LLLRPKI-IAAVEAAGR-----RTTPWREAFYGAGMRPVQLSQFADFQAECLLAKVQI 513
L R + I A E A R WR F AG P LS + + LL
Sbjct: 404 QHCLARDVVNIIACEGADRVERHELLGKWRSRFGMAGFTPYPLSPLVNSTIKSLLRNYSD 463
Query: 514 RGFHVAKRQAELVLFWHERAMVATSAWR 541
+ + + +R L L W R +VA+ AW+
Sbjct: 464 K-YRLEERDGALYLGWMHRDLVASCAWK 490
>At3g03450 RGA1-like protein
Length = 547
Score = 89.0 bits (219), Expect = 6e-18
Identities = 91/378 (24%), Positives = 159/378 (41%), Gaps = 29/378 (7%)
Query: 180 IEELIRAADCFDNNHLQLAQAILERLNQRLRSPTGKPLHRAAFHFKDALQSLLSGSNRTN 239
+ L+ A+ +L LA A+++R+ S G + + A +F AL +
Sbjct: 181 VHALVACAEAIHQENLNLADALVKRVGTLAGSQAGA-MGKVATYFAQALARRIYRDYTAE 239
Query: 240 PPRLSSMVEIVQTIRTFKAFSGISPIPMFSIFTTNQALLEALHGSLYMHVVDFEIGLGIQ 299
+++ + + + P F+ FT NQA+LEA+ + +HV+D + G+Q
Sbjct: 240 TDVCAAVNPSFEEVLEMHFYESC-PYLKFAHFTANQAILEAVTTARRVHVIDLGLNQGMQ 298
Query: 300 YASLMKEIAEKAVNGSPLLRITAVVPEEYAVESRL--IRENLNQFAHDLGIRVQVDFVPL 357
+ +LM+ +A + G P R+T + P + L + L QFA ++G V+ +F L
Sbjct: 299 WPALMQALALRP-GGPPSFRLTGIGPPQTENSDSLQQLGWKLAQFAQNMG--VEFEFKGL 355
Query: 358 RTFETVSFKAVRFVDGEKTAILLTPAIF------CRLGSEGTAAFLSDVRRITPGVVVFV 411
+ F ++ L+ ++F R GS L+ V+ I P +V V
Sbjct: 356 AAESLSDLEPEMFETRPESETLVVNSVFELHRLLARSGS--IEKLLNTVKAIKPSIVTVV 413
Query: 412 DGEGWTEAAAAASFRRGVVNSLEFYSMMLESLDASVAAGGGGEWARRIEMLLLRPKIIAA 471
+ E F +L +YS + +SL+ S + R + + L +I+
Sbjct: 414 EQEANHNGIV---FLDRFNEALHYYSSLFDSLEDSYSLPSQD---RVMSEVYLGRQILNV 467
Query: 472 VEAAG-------RRTTPWREAFYGAGMRPVQLSQFADFQAECLLAKVQI-RGFHVAKRQA 523
V A G WR AG P+ L A QA LL+ G+ V +
Sbjct: 468 VAAEGSDRVERHETAAQWRIRMKSAGFDPIHLGSSAFKQASMLLSLYATGDGYRVEENDG 527
Query: 524 ELVLFWHERAMVATSAWR 541
L++ W R ++ TSAW+
Sbjct: 528 CLMIGWQTRPLITTSAWK 545
>At3g50650 scarecrow-like 7 (SCL7)
Length = 542
Score = 88.6 bits (218), Expect = 8e-18
Identities = 94/361 (26%), Positives = 152/361 (42%), Gaps = 51/361 (14%)
Query: 211 SPTGKPLHRAAFHFKDALQSLLSGSNRTNPPRLSSMVEIVQTIRTFKAFSGISPIPMFSI 270
S +G P+ R ++F +AL T P SS + I ++K + P F+
Sbjct: 203 SESGDPIQRVGYYFAEALSH-----KETESPSSSSSSSLEDFILSYKTLNDACPYSKFAH 257
Query: 271 FTTNQALLEALHGSLYMHVVDFEIGLGIQYASLMKEIAEKAVNGSPLLRITAVV------ 324
T NQA+LEA + S +H+VDF I GIQ+++L++ +A ++ +RI+ +
Sbjct: 258 LTANQAILEATNQSNNIHIVDFGIFQGIQWSALLQALATRSSGKPTRIRISGIPAPSLGD 317
Query: 325 ---PEEYAVESRLIRENLNQFAH--DLGIRVQVDFVPLRTFETVSFKAVRFVDGEKTAIL 379
P A +R L FA DL P++ SF+ VD ++ ++
Sbjct: 318 SPGPSLIATGNR-----LRDFAAILDLNFEFYPVLTPIQLLNGSSFR----VDPDEVLVV 368
Query: 380 LTPAIFCRLGSE-----GTAAFLSDVRRITPGVVVFVDGEGWTEAAAAASFRRGVVNSLE 434
+L E GTA L+ R + P +V GE + + F V NSL
Sbjct: 369 NFMLELYKLLDETATTVGTALRLA--RSLNPRIVTL--GE-YEVSLNRVEFANRVKNSLR 423
Query: 435 FYSMMLESLDASVAAGGGGEWARRIEMLLLRPKIIAAVEAAGRRTTP------------W 482
FYS + ESL+ ++ + R+E +L +I+ V + P W
Sbjct: 424 FYSAVFESLEPNL--DRDSKERLRVERVLFGRRIMDLVRSDDDNNKPGTRFGLMEEKEQW 481
Query: 483 REAFYGAGMRPVQLSQFADFQAECLLAKVQIRGFH--VAKRQAELVLFWHERAMVATSAW 540
R AG PV+ S +A QA+ LL + V + L W+ ++ S+W
Sbjct: 482 RVLMEKAGFEPVKPSNYAVSQAKLLLWNYNYSTLYSLVESEPGFISLAWNNVPLLTVSSW 541
Query: 541 R 541
R
Sbjct: 542 R 542
>At5g17490 RGA-like protein
Length = 523
Score = 87.8 bits (216), Expect = 1e-17
Identities = 96/374 (25%), Positives = 159/374 (41%), Gaps = 27/374 (7%)
Query: 180 IEELIRAADCFDNNHLQLAQAILERLNQRLRSPTGKPLHRAAFHFKDALQSLLSGSNRTN 239
++ L+ A+ +L LA A+++R+ S G + + A +F +AL + + +
Sbjct: 158 VQALVACAEAVQLENLSLADALVKRVGLLAASQAGA-MGKVATYFAEALARRIYRIHPSA 216
Query: 240 PPRLSSMVEIVQTIRTFKAFSGISPIPMFSIFTTNQALLEALHGSLYMHVVDFEIGLGIQ 299
S EI+Q F P F+ FT NQA+LEA+ S +HV+D + G+Q
Sbjct: 217 AAIDPSFEEILQM-----NFYDSCPYLKFAHFTANQAILEAVTTSRVVHVIDLGLNQGMQ 271
Query: 300 YASLMKEIAEKAVNGSPLLRITAVVPEEYAVESRLIRENLNQFAHDLGIRVQVDFVPLRT 359
+ +LM+ +A + G P R+T V + + L Q A +G V+ F L T
Sbjct: 272 WPALMQALALRP-GGPPSFRLTGVGNPSNREGIQELGWKLAQLAQAIG--VEFKFNGLTT 328
Query: 360 FETVSFKAVRFVDGEKTAILLTPAIF---CRLGSEGT-AAFLSDVRRITPGVVVFVDGEG 415
+ F ++ L+ ++F L G+ L+ V+ + PG+V V+ E
Sbjct: 329 ERLSDLEPDMFETRTESETLVVNSVFELHPVLSQPGSIEKLLATVKAVKPGLVTVVEQEA 388
Query: 416 WTEAAAAASFRRGVVNSLEFYSMMLESLDASVAAGGGGEWARRIEMLLLRPKIIAAVEAA 475
F +L +YS + +SL+ V R + + L +I+ V
Sbjct: 389 NHNGDV---FLDRFNEALHYYSSLFDSLEDGVVIPSQD---RVMSEVYLGRQILNLVATE 442
Query: 476 G-------RRTTPWREAFYGAGMRPVQLSQFADFQAECLLA-KVQIRGFHVAKRQAELVL 527
G WR+ AG PV L A QA LLA G+ V + L+L
Sbjct: 443 GSDRIERHETLAQWRKRMGSAGFDPVNLGSDAFKQASLLLALSGGGDGYRVEENDGSLML 502
Query: 528 FWHERAMVATSAWR 541
W + ++A SAW+
Sbjct: 503 AWQTKPLIAASAWK 516
>At1g63100 transcription factor SCARECROW, putative
Length = 658
Score = 87.8 bits (216), Expect = 1e-17
Identities = 115/492 (23%), Positives = 201/492 (40%), Gaps = 56/492 (11%)
Query: 82 LPNLDWWDSIMKDL-GLQDDSSTPIIPL-LKNTNNTSEIYPNPSQDQFDQTQDFTSLSDI 139
LPN W DS++ +L G+ D +P +K + S + + SLS
Sbjct: 185 LPN--WVDSVITELAGIGDKDVESSLPAAVKEASGGSST---------SASSESRSLSHR 233
Query: 140 YSNNQNLAYN-YPNTNTNLDHLVNDFNNNQPNNNTNSNWDFIEELIRAADCFDNNHLQLA 198
N + N Y + + + NNN N+ +++ + L D + ++
Sbjct: 234 VPEPTNGSRNPYSHRGATEERTTGNINNNNNRNDLQRDFELVNLLTGCLDAIRSRNIAAI 293
Query: 199 QAILERLNQRLRSPTGK-PLHRAAFHFKDALQSLLSGS----NRTNPPRLSSMVEIVQTI 253
+ R L SP G+ P+ R ++ +AL ++ PPR ++
Sbjct: 294 NHFIARTGD-LASPRGRTPMTRLIAYYIEALALRVARMWPHIFHIAPPREFDRTVEDESG 352
Query: 254 RTFKAFSGISPIPMFSIFTTNQALLEALHGSLYMHVVDFEIGLGIQYASLMKEIAEKAVN 313
+ + ++PIP F FT N+ LL A G +H++DF+I G+Q+ S + +A + +N
Sbjct: 353 NALRFLNQVTPIPKFIHFTANEMLLRAFEGKERVHIIDFDIKQGLQWPSFFQSLASR-IN 411
Query: 314 GSPLLRITAVVPEEYAVESRLIRENLNQFAHDLGIRVQVDFVP-LRTFETVSFKAVRFVD 372
+RIT + E +E + L+ FA + +Q +F P + E V + +
Sbjct: 412 PPHHVRITGI--GESKLELNETGDRLHGFAE--AMNLQFEFHPVVDRLEDVRLWMLHVKE 467
Query: 373 GEKTAILLTPAIFCRLGSEGTAA----FLSDVRRITPGVVVFVDGEGWTEAAAAASFRRG 428
GE A+ + L +GT A FL +R P +V + E +
Sbjct: 468 GESVAVNCVMQMHKTL-YDGTGAAIRDFLGLIRSTNPIALVLAEQEA---EHNSEQLETR 523
Query: 429 VVNSLEFYSMMLESLDASVAAGGGGEWARRIEMLLLRPKI--IAAVEAAGRRT-----TP 481
V NSL++YS M +++ ++A ++E +L +I I A E + R+
Sbjct: 524 VCNSLKYYSAMFDAIHTNLAT--DSLMRVKVEEMLFGREIRNIVACEGSHRQERHVGFRH 581
Query: 482 WREAFYGAGMRPVQLSQFADFQAECLLAKV--QIRGFHVAKRQAE-----------LVLF 528
WR G R + +S+ Q++ LL GF +R E + L
Sbjct: 582 WRRMLEQLGFRSLGVSEREVLQSKMLLRMYGSDNEGFFNVERSDEDNGGEGGRGGGVTLR 641
Query: 529 WHERAMVATSAW 540
W E+ + SAW
Sbjct: 642 WSEQPLYTISAW 653
>At1g14920 signal response protein (GAI)
Length = 533
Score = 87.8 bits (216), Expect = 1e-17
Identities = 95/384 (24%), Positives = 164/384 (41%), Gaps = 46/384 (11%)
Query: 180 IEELIRAADCFDNNHLQLAQAILERLNQRLRSPTGKPLHRAAFHFKDALQSLLSGSNRTN 239
+ L+ A+ +L +A+A+++++ S G + + A +F +AL + + +
Sbjct: 170 VHALLACAEAVQKENLTVAEALVKQIGFLAVSQIGA-MRKVATYFAEALARRIYRLSPSQ 228
Query: 240 PPRLSSMVEIVQTIRTFKAFSGISPIPMFSIFTTNQALLEALHGSLYMHVVDFEIGLGIQ 299
P S+ + +Q F P F+ FT NQA+LEA G +HV+DF + G+Q
Sbjct: 229 SPIDHSLSDTLQM-----HFYETCPYLKFAHFTANQAILEAFQGKKRVHVIDFSMSQGLQ 283
Query: 300 YASLMKEIAEKAVNGSPLLRITAVVPEEYAVESRLIRENLNQFAHDLGIRVQVDFVPLRT 359
+ +LM+ +A + G P+ R+T + P +N + + H++G ++ +
Sbjct: 284 WPALMQALALRP-GGPPVFRLTGIGPP--------APDNFD-YLHEVGCKLAHLAEAIHV 333
Query: 360 -FETVSFKAVRFVDGEKTAILLTPA------------IFCRLGSEGTA-AFLSDVRRITP 405
FE F A D + + + L P+ + LG G L V +I P
Sbjct: 334 EFEYRGFVANTLADLDASMLELRPSEIESVAVNSVFELHKLLGRPGAIDKVLGVVNQIKP 393
Query: 406 GVVVFVDGEGWTEAAAAASFRRGVVNSLEFYSMMLESLDASVAAGGGGEWARRIEMLLLR 465
+ V+ E + F SL +YS + +SL+ V +G + + + L
Sbjct: 394 EIFTVVEQESNHNSPI---FLDRFTESLHYYSTLFDSLE-GVPSGQD----KVMSEVYLG 445
Query: 466 PKIIAAVEAAG-------RRTTPWREAFYGAGMRPVQLSQFADFQAECLLAKVQ-IRGFH 517
+I V G + WR F AG + A QA LLA G+
Sbjct: 446 KQICNVVACDGPDRVERHETLSQWRNRFGSAGFAAAHIGSNAFKQASMLLALFNGGEGYR 505
Query: 518 VAKRQAELVLFWHERAMVATSAWR 541
V + L+L WH R ++ATSAW+
Sbjct: 506 VEESDGCLMLGWHTRPLIATSAWK 529
>At1g21450 scarecrow-like 1 (SCL1)
Length = 593
Score = 86.7 bits (213), Expect = 3e-17
Identities = 98/391 (25%), Positives = 159/391 (40%), Gaps = 55/391 (14%)
Query: 183 LIRAADCFDNNHLQLAQAILERLNQRLRSPTGKPLHRAAFHFKDALQSLLSGSNR----- 237
LI A L+ A +++ L Q + S G P R A + + L + ++ S +
Sbjct: 226 LISCARALSEGKLEEALSMVNELRQ-IVSIQGDPSQRIAAYMVEGLAARMAASGKFIYRA 284
Query: 238 ---TNPP---RLSSMVEIVQTIRTFKAFSGISPIPMFSIFTTNQALLEALHGSLYMHVVD 291
PP RL++M + + FK F N A+LEA+ G +H++D
Sbjct: 285 LKCKEPPSDERLAAMQVLFEVCPCFK----------FGFLAANGAILEAIKGEEEVHIID 334
Query: 292 FEIGLGIQYASLMKEIAEKAVNGSPLLRITAVVPEEYAVES----RLIRENLNQFAHDLG 347
F+I G QY +L++ IAE P LR+T + E S R+I L Q A D G
Sbjct: 335 FDINQGNQYMTLIRSIAE-LPGKRPRLRLTGIDDPESVQRSIGGLRIIGLRLEQLAEDNG 393
Query: 348 IRVQVDFVPLRTFETVSFKAVRFVDGEKTAILLTPAIFCRLGSEGTAA------FLSDVR 401
+ + +P +T VS + GE T I+ + E L V+
Sbjct: 394 VSFKFKAMPSKT-SIVSPSTLGCKPGE-TLIVNFAFQLHHMPDESVTTVNQRDELLHMVK 451
Query: 402 RITPGVVVFVDGEGWTEAAAAASFRRGVVNSLEFYSMMLESLDASVAAGGGGEWARRIEM 461
+ P +V V+ + T + F + + E+YS + ESLD ++ + +E
Sbjct: 452 SLNPKLVTVVEQDVNTN---TSPFFPRFIEAYEYYSAVFESLDMTLPR--ESQERMNVER 506
Query: 462 LLLRPKIIAAV-----------EAAGRRTTPWREAFYGAGMRPVQLSQFADFQAECLLAK 510
L I+ V EAAG+ WR AG P +S + L+ +
Sbjct: 507 QCLARDIVNIVACEGEERIERYEAAGK----WRARMMMAGFNPKPMSAKVTNNIQNLIKQ 562
Query: 511 VQIRGFHVAKRQAELVLFWHERAMVATSAWR 541
+ + + EL W E++++ SAWR
Sbjct: 563 QYCNKYKLKEEMGELHFCWEEKSLIVASAWR 593
>At5g59450 putative SCARECROW transcriptional regulatory protein
(F2O15.13)
Length = 610
Score = 84.7 bits (208), Expect = 1e-16
Identities = 85/352 (24%), Positives = 151/352 (42%), Gaps = 29/352 (8%)
Query: 211 SPTGKPLHRAAFHFKDALQSLLSG--SNRTNPPRLSSMVEIVQTIRTFKAFSGISPIPMF 268
S G R AF+F +AL++ ++G S + P SS +V ++ +K F PI +
Sbjct: 255 SSNGDGTQRLAFYFAEALEARITGNISPPVSNPFPSSTTSMVDILKAYKLFVHTCPIYVT 314
Query: 269 SIFTTNQALLEALHGSLYMHVVDFEIGLGIQYASLMKEIAEKAVNGSPLLRITAV-VPEE 327
F N+++ E + +H+VDF + G Q+ L++ ++ K G P+LR+T + +P+
Sbjct: 315 DYFAANKSIYELAMKATKLHIVDFGVLYGFQWPCLLRALS-KRPGGPPMLRVTGIELPQA 373
Query: 328 YAVESRLIRE---NLNQFAHDLGIRVQVDFVPLRTFETVSFKAVRFVDGEKTAI------ 378
S + E L +F + + +F+ + +ET++ + GE T +
Sbjct: 374 GFRPSDRVEETGRRLKRFCDQFNVPFEFNFI-AKKWETITLDELMINPGETTVVNCIHRL 432
Query: 379 LLTPAIFCRLGSEGTAAFLSDVRRITPGVVVFVDGEGWTEAAAAASFRRGVVNSLEFYSM 438
TP L S L R I P + VF + G + F +L YS
Sbjct: 433 QYTPDETVSLDSPRDTV-LKLFRDINPDLFVFAEINGMYN---SPFFMTRFREALFHYSS 488
Query: 439 MLESLDASVAAGGGGEWARRI----EMLLLRPKIIAAVEAAGRRTTP-----WREAFYGA 489
+ + D ++ A E+ R E+L+ + + E A R P WR A
Sbjct: 489 LFDMFDTTIHA--EDEYKNRSLLERELLVRDAMSVISCEGAERFARPETYKQWRVRILRA 546
Query: 490 GMRPVQLSQFADFQAECLLAKVQIRGFHVAKRQAELVLFWHERAMVATSAWR 541
G +P +S+ +A+ ++ K R F + ++ W R + A S W+
Sbjct: 547 GFKPATISKQIMKEAKEIVRKRYHRDFVIDSDNNWMLQGWKGRVIYAFSCWK 598
>At1g50600 scarecrow-like 5 (SCL5)
Length = 597
Score = 84.0 bits (206), Expect = 2e-16
Identities = 112/508 (22%), Positives = 196/508 (38%), Gaps = 83/508 (16%)
Query: 97 LQDDSSTPIIPLLKNTNNTSEIYPNPSQDQFDQTQDFTSLSDIYSNNQNLAYNYP----- 151
L+ S T P L N NN+S S + + +LS +++ N P
Sbjct: 110 LESSSGTKSHPCLNNKNNSSSTTSFSSNESPISQANNNNLSRFNNHSPEENNNSPLSGSS 169
Query: 152 --NTN-TNLDHLVNDFN--------NNQPNNNTN-------------------SNWDFIE 181
NTN T L ++ D +N NN S D
Sbjct: 170 ATNTNETELSLMLKDLETAMMEPDVDNSYNNQGGFGQQHGVVSSAMYRSMEMISRGDLKG 229
Query: 182 ELIRAADCFDNNHLQLAQAILERLNQRLRSPTGKPLHRAAFHFKDALQSLLSGSNR---- 237
L A +N L++ ++ +L Q + S +G+P+ R + + L + L+ S
Sbjct: 230 VLYECAKAVENYDLEMTDWLISQLQQMV-SVSGEPVQRLGAYMLEGLVARLASSGSSIYK 288
Query: 238 -------TNPPRLSSMVEIVQTIRTFKAFSGISPIPMFSIFTTNQALLEALHGSLYMHVV 290
T P L+ M + + FK F + N A+ EA+ ++H++
Sbjct: 289 ALRCKDPTGPELLTYMHILYEACPYFK----------FGYESANGAIAEAVKNESFVHII 338
Query: 291 DFEIGLGIQYASLMKEIAEKAVNGSPLLRITAVVPEEYAVESR----LIRENLNQFAHDL 346
DF+I G Q+ SL++ + + G P +RIT + + + L+ + L + A
Sbjct: 339 DFQISQGGQWVSLIRALGARP-GGPPNVRITGIDDPRSSFARQGGLELVGQRLGKLAEMC 397
Query: 347 GIRVQVDFVPLRTFETVSFKAVRFVDGEKTAILLTPAIFCRLGSEGTAA------FLSDV 400
G+ + L E V + + +GE A+ P + + E L V
Sbjct: 398 GVPFEFHGAALCCTE-VEIEKLGVRNGEALAVNF-PLVLHHMPDESVTVENHRDRLLRLV 455
Query: 401 RRITPGVVVFVDGEGWTEAAAAASFRRGVVNSLEFYSMMLESLDASVAAGGGGEWARRIE 460
+ ++P VV V+ E T A F V ++ Y + ES+D +A + +E
Sbjct: 456 KHLSPNVVTLVEQEANTN---TAPFLPRFVETMNHYLAVFESIDVKLAR--DHKERINVE 510
Query: 461 MLLLRPKIIAAVEAAG----RRTTP---WREAFYGAGMRPVQLSQFADFQAECLLAKVQI 513
L +++ + G R P WR F+ AG +P LS + + + LL
Sbjct: 511 QHCLAREVVNLIACEGVEREERHEPLGKWRSRFHMAGFKPYPLSSYVNATIKGLLESYSE 570
Query: 514 RGFHVAKRQAELVLFWHERAMVATSAWR 541
+ + + +R L L W + ++ + AWR
Sbjct: 571 K-YTLEERDGALYLGWKNQPLITSCAWR 597
>At3g46600 scarecrow-like protein
Length = 583
Score = 80.1 bits (196), Expect = 3e-15
Identities = 74/344 (21%), Positives = 143/344 (41%), Gaps = 23/344 (6%)
Query: 214 GKPLHRAAFHFKDALQSLLSGSNRTNPPRLSSMVEIVQTIRTFKAFSGISPIPMFSIFTT 273
G R +HF +AL++ ++G+ T SS +V ++ +K F P + FT
Sbjct: 243 GDATQRLGYHFAEALEARITGTMTTPISATSSRTSMVDILKAYKGFVQACPTLIMCYFTA 302
Query: 274 NQALLEALHGSLYMHVVDFEIGLGIQYASLMKEIAEKAVNGSPLLRITAV-VPEEYAVES 332
N+ + E + +H++DF I G Q+ L++ ++++ + G PLLR+T + +P+ S
Sbjct: 303 NRTINELASKATTLHIIDFGILYGFQWPCLIQALSKRDI-GPPLLRVTGIELPQSGFRPS 361
Query: 333 RLIRE---NLNQFAHDLGIRVQVDFVPLRTFETVSFKAVRFVDGEKTAI------LLTPA 383
+ E L +F + + F+ + +E ++ + GE T + TP
Sbjct: 362 ERVEETGRRLKRFCDKFNVPFEYSFI-AKNWENITLDDLVINSGETTVVNCILRLQYTPD 420
Query: 384 IFCRLGSEGTAAFLSDVRRITPGVVVFVDGEG-WTEAAAAASFRRGVVNSLEFYSMMLES 442
L S A L R I P + VF + G + FR + + S + +
Sbjct: 421 ETVSLNSPRDTA-LKLFRDINPDLFVFAEINGTYNSPFFLTRFREALFHC----SSLFDM 475
Query: 443 LDASVAAGGGGEWARRIEMLLLRPKIIAAVEAAGRRTTP-----WREAFYGAGMRPVQLS 497
+ +++ E+++ + A E + R P W+ AG RP +LS
Sbjct: 476 YETTLSEDDNCRTLVERELIIRDAMSVIACEGSERFARPETYKQWQVRILRAGFRPAKLS 535
Query: 498 QFADFQAECLLAKVQIRGFHVAKRQAELVLFWHERAMVATSAWR 541
+ + ++ + + F + + W R + A S W+
Sbjct: 536 KQIVKDGKEIVKERYHKDFVIDNDNHWMFQGWKGRVLYAVSCWK 579
>At2g04890 putative SCARECROW gene regulator
Length = 413
Score = 77.8 bits (190), Expect = 1e-14
Identities = 85/373 (22%), Positives = 153/373 (40%), Gaps = 27/373 (7%)
Query: 183 LIRAADCFDNNHLQLAQAILERLNQRLRSPTGKPLHRAAFHFKDALQSLLSGSNRTNPPR 242
L+ A N+L +A+ + L + S +G+P+ R + + L + L+ S +
Sbjct: 54 LVACAKAVSENNLLMARWCMGELRGMV-SISGEPIQRLGAYMLEGLVARLAASGSSIYKS 112
Query: 243 LSSMV-EIVQTIRTFKAFSGISPIPMFSIFTTNQALLEALHGSLYMHVVDFEIGLGIQYA 301
L S E + + + P F + N A+ EA+ +H++DF+IG G Q+
Sbjct: 113 LQSREPESYEFLSYVYVLHEVCPYFKFGYMSANGAIAEAMKDEERIHIIDFQIGQGSQWI 172
Query: 302 SLMKEIAEKAVNGSPLLRITAVVPEEYAVESRLIRENLNQFAHDLGIRVQVDFVPLRTFE 361
+L++ A + G+P +RIT V V +++ L + A + + + V + E
Sbjct: 173 ALIQAFAARP-GGAPNIRITGVGDGSVLV---TVKKRLEKLAKKFDVPFRFNAVSRPSCE 228
Query: 362 TVSFKAVRFVDGEKTAILLTPAIFCRLGSEGTA------AFLSDVRRITPGVVVFVDGEG 415
V + + DGE + + L E + L V+ ++P VV V+ E
Sbjct: 229 -VEVENLDVRDGEALGVNFAYMLH-HLPDESVSMENHRDRLLRMVKSLSPKVVTLVEQEC 286
Query: 416 WTEAAAAASFRRGVVNSLEFYSMMLESLDASVAAGGGGEWARRIEMLLLRPKIIAAVEAA 475
T + F + +L +Y+ M ES+D V + IE + ++ +
Sbjct: 287 NTN---TSPFLPRFLETLSYYTAMFESID--VMLPRNHKERINIEQHCMARDVVNIIACE 341
Query: 476 GRRTT-------PWREAFYGAGMRPVQLSQFADFQAECLLAKVQIRGFHVAKRQAELVLF 528
G W+ F AG P LS LL G+ + +R L L
Sbjct: 342 GAERIERHELLGKWKSRFSMAGFEPYPLSSIISATIRALLRDYS-NGYAIEERDGALYLG 400
Query: 529 WHERAMVATSAWR 541
W +R +V++ AW+
Sbjct: 401 WMDRILVSSCAWK 413
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.133 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,497,711
Number of Sequences: 26719
Number of extensions: 590329
Number of successful extensions: 3199
Number of sequences better than 10.0: 125
Number of HSP's better than 10.0 without gapping: 53
Number of HSP's successfully gapped in prelim test: 77
Number of HSP's that attempted gapping in prelim test: 2707
Number of HSP's gapped (non-prelim): 343
length of query: 542
length of database: 11,318,596
effective HSP length: 104
effective length of query: 438
effective length of database: 8,539,820
effective search space: 3740441160
effective search space used: 3740441160
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC148484.4


