

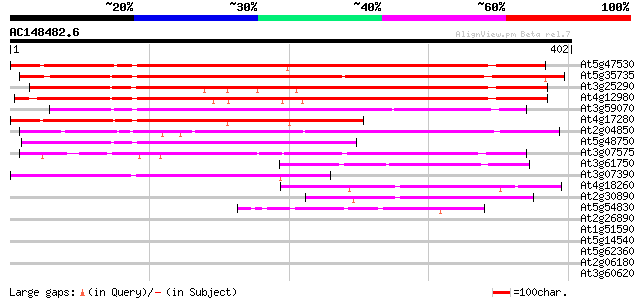
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148482.6 - phase: 0
(402 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g47530 unknown protein 405 e-113
At5g35735 unknown protein 364 e-101
At3g25290 unknown protein 319 1e-87
At4g12980 unknown protein 310 1e-84
At3g59070 putative protein 268 5e-72
At4g17280 unknown protein 231 7e-61
At2g04850 unknown protein 226 2e-59
At5g48750 putative protein 169 3e-42
At3g07575 unknown protein 123 2e-28
At3g61750 putative protein 107 9e-24
At3g07390 auxin-induced protein (AIR12) 97 1e-20
At4g18260 unknown protein 67 2e-11
At2g30890 hypothetical protein 65 7e-11
At5g54830 unknown protein 60 2e-09
At2g26890 unknown protein 31 1.1
At1g51590 mannosyl-oligosaccharide 1,2-alpha-mannosidase, putative 31 1.1
At5g14540 unknown protein 30 1.8
At5g62360 DC1.2 homologue - like protein 29 4.0
At2g06180 hypothetical protein 29 4.0
At3g60620 phosphatidate cytidylyltransferase - like protein 29 5.2
>At5g47530 unknown protein
Length = 395
Score = 405 bits (1040), Expect = e-113
Identities = 197/388 (50%), Positives = 273/388 (69%), Gaps = 15/388 (3%)
Query: 1 MVKNLRFMFLISILMFVTTTLAQTTTQTCKSQNFTNNAIFTSCRDLPQLTSYLHWTYDQT 60
M + + +S+ +F+ T A Q C + F+ N +F SC DLP L S+LH+TYD +
Sbjct: 1 MAISSNLLLCLSLFIFIITKSA--LAQKCSNYKFSTNRLFESCNDLPVLDSFLHYTYDSS 58
Query: 61 TGKLDIAFRHKGITDTNRWVAWAINPNNDLASSMNGAQALVAILQSSGTPKAYTSSIANS 120
+G L IA+RH +T +WVAWA+NP + + M GAQA+VA QS GT +AYTS I++
Sbjct: 59 SGNLQIAYRHTKLTP-GKWVAWAVNPTS---TGMVGAQAIVAYPQSDGTVRAYTSPISSY 114
Query: 121 RTQLAESNISYPHSGLIATHENNEVTIYASITLPVGTPSLVH-LWQDGAMSGSTPQMHDM 179
+T L E+ +S+ S L AT++NNE+ IYA + LP+ +++ +WQDG++SG+ P H
Sbjct: 115 QTSLLEAELSFNVSQLSATYQNNEMVIYAILNLPLANGGIINTVWQDGSLSGNNPLPHPT 174
Query: 180 TSANTQSKESLDLRSGAS---EQGSGGGSLSRRRNTHGVLNAISWGILMPLGAVIARYLK 236
+ N +S +L+L SGAS G+GG S R+RN HG+LN +SWGI+MP+GA+IARYLK
Sbjct: 175 SGNNVRSVSTLNLVSGASGSTSTGAGGASKLRKRNIHGILNGVSWGIMMPIGAIIARYLK 234
Query: 237 VFKSADPAWFYLHVTCQSAAYIVGVAGWGTGLKLGSDSAGVTYSTHRTLGIVIFCLGTLQ 296
V KSADPAWFYLHV CQS+AYI+GVAGW TGLKLG++SAG+ ++ HR +GI +FCL T+Q
Sbjct: 235 VSKSADPAWFYLHVFCQSSAYIIGVAGWATGLKLGNESAGIQFTFHRAVGIALFCLATIQ 294
Query: 297 VFALLLRPKKDHKIRFYWNLYHWGVGYATIIISIINIFKGFEALEVSAADRYDNWKHAYT 356
VFA+ LRPK +HK R YWN+YH VGY+ II++++N+FKG + L W++AYT
Sbjct: 295 VFAMFLRPKPEHKYRVYWNIYHHTVGYSVIILAVVNVFKGLDILSPE-----KQWRNAYT 349
Query: 357 GIIAALGGVAVLLEAYTWIIVIKRKKSE 384
II LG VAV+LE +TW +VIKR K+E
Sbjct: 350 AIIVVLGIVAVVLEGFTWYVVIKRGKAE 377
>At5g35735 unknown protein
Length = 404
Score = 364 bits (934), Expect = e-101
Identities = 189/402 (47%), Positives = 252/402 (62%), Gaps = 26/402 (6%)
Query: 8 MFLISILMFVTTTLAQTTTQTCKSQNFTNNAIFTSCRDLPQLTSYLHWTYDQTTGKLDIA 67
+F + + V T Q+ C + FTNN F C DL L S+LHWTY++ G + IA
Sbjct: 11 LFAVLATLLVLTVNGQSL---CNTHRFTNNLAFADCSDLSALGSFLHWTYNEQNGTVSIA 67
Query: 68 FRHKGITDTNRWVAWAINPNNDLASSMNGAQALVAILQSSGTP-KAYTSSIANSRTQLAE 126
+RH G T + WVAW +NP++ + M G QALVA ++ +AYTSS+++ T+L
Sbjct: 68 YRHPG-TSASSWVAWGLNPSS---TQMVGTQALVAFTNTTTNQFQAYTSSVSSYGTRLER 123
Query: 127 SNISYPHSGLIATHENNEVTIYASITLPVGTPSLVHLWQDGAMSGSTPQMHDMTSANTQS 186
S++S+ SGL AT + EVTI+A++ L + LWQ G + P H + N +S
Sbjct: 124 SSLSFGVSGLSATLVSGEVTIFATLELSPNLITANQLWQVGPVVNGVPASHQTSGDNMRS 183
Query: 187 KESLDLRSGASEQGSGG-GSLSRRRNTHGVLNAISWGILMPLGAVIARYLKVFKSADPAW 245
+D R+G + G GG G R+RNTHGVLNA+SWG+LMP+GA++ARY+KVF ADP W
Sbjct: 184 SGRIDFRTGQASAGGGGSGDRLRKRNTHGVLNAVSWGVLMPMGAMMARYMKVF--ADPTW 241
Query: 246 FYLHVTCQSAAYIVGVAGWGTGLKLGSDSAGVTYSTHRTLGIVIFCLGTLQVFALLLRPK 305
FYLH+ Q + Y++GVAGW TG+KLG+DS G +YSTHR LGI +F TLQVFALL+RPK
Sbjct: 242 FYLHIAFQVSGYVIGVAGWATGIKLGNDSPGTSYSTHRNLGIALFTFATLQVFALLVRPK 301
Query: 306 KDHKIRFYWNLYHWGVGYATIIISIINIFKGFEALEVSAADRYDNWKHAYTGIIAALGGV 365
DHK R YWN+YH VGY TII+SI+NIFKGF+ L D D W+ AY GI+ LG
Sbjct: 302 PDHKYRTYWNVYHHTVGYTTIILSIVNIFKGFDIL-----DPEDKWRWAYIGILIFLGAC 356
Query: 366 AVLLEAYTWIIVIKRKK----------SENKLQGMNGTNGNG 397
++LE TW IV++RK S G+NGT G
Sbjct: 357 VLILEPLTWFIVLRRKSRGGNTVAAPTSSKYSNGVNGTTTTG 398
>At3g25290 unknown protein
Length = 393
Score = 319 bits (818), Expect = 1e-87
Identities = 166/383 (43%), Positives = 234/383 (60%), Gaps = 21/383 (5%)
Query: 15 MFVTTTLAQTTTQTCKSQNFTNNAIFTSCRDLPQLTSYLHWTYDQTTGKLDIAFRHKGIT 74
+F+ ++ +QTCKSQ F+ + + C DLPQL ++LH++YD + L + F
Sbjct: 14 IFLALLISPAVSQTCKSQTFSGDKTYPHCLDLPQLKAFLHYSYDASNTTLAVVFSAPP-A 72
Query: 75 DTNRWVAWAINPNNDLASSMNGAQALVAILQSSGTPKAYTSSIANSRTQLAESNISYPHS 134
W+AWAINP A+ M G+Q LVA + +S + L S +++
Sbjct: 73 KPGGWIAWAINPK---ATGMVGSQTLVAYKDPGNGVAVVKTLNISSYSSLIPSKLAFDVW 129
Query: 135 GLIA---THENNEVTIYASITLP---VGTPSLVHLWQDGAMSGSTPQM--HDMTSANTQS 186
+ A + + I+A + +P V + +WQ G G + H SAN S
Sbjct: 130 DMKAEEAARDGGSLRIFARVKVPADLVAKGKVNQVWQVGPELGPGGMIGRHAFDSANLAS 189
Query: 187 KESLDLRSGASEQGSGGG----SLSRRRNTHGVLNAISWGILMPLGAVIARYLKVFKSAD 242
SLDL+ S GG + + RN HG+LNA+SWGIL P+GA+IARY++VF SAD
Sbjct: 190 MSSLDLKGDNSGGTISGGDEVNAKIKNRNIHGILNAVSWGILFPIGAIIARYMRVFDSAD 249
Query: 243 PAWFYLHVTCQSAAYIVGVAGWGTGLKLGSDSAGVTYSTHRTLGIVIFCLGTLQVFALLL 302
PAWFYLHV+CQ +AY++GVAGW TGLKLG++S G+ +S HR +GI +F L T+Q+FA+LL
Sbjct: 250 PAWFYLHVSCQFSAYVIGVAGWATGLKLGNESEGIRFSAHRNIGIALFTLATIQMFAMLL 309
Query: 303 RPKKDHKIRFYWNLYHWGVGYATIIISIINIFKGFEALEVSAADRYDNWKHAYTGIIAAL 362
RPKKDHK RFYWN+YH GVGYA + + IIN+FKG L+ D +K AY +IA L
Sbjct: 310 RPKKDHKYRFYWNIYHHGVGYAILTLGIINVFKGLNILKPQ-----DTYKTAYIAVIAVL 364
Query: 363 GGVAVLLEAYTWIIVIKRKKSEN 385
GG+A+LLEA TW++V+KRK + +
Sbjct: 365 GGIALLLEAITWVVVLKRKSNNS 387
>At4g12980 unknown protein
Length = 394
Score = 310 bits (793), Expect = 1e-84
Identities = 167/394 (42%), Positives = 239/394 (60%), Gaps = 26/394 (6%)
Query: 4 NLRFMFLISILMFVTTTLAQTTTQTCKSQNFTNNAIFTSCRDLPQLTSYLHWTYDQTTGK 63
+L F+F +L + A + + +C SQ F+ + C DLP L + LH++YD +
Sbjct: 9 SLSFLFWALLL-----SPAVSQSSSCSSQTFSGVKSYPHCLDLPDLKAILHYSYDASNTT 63
Query: 64 LDIAFRHKGITDTNRWVAWAINPNNDLASSMNGAQALVAILQSSGTPKAYTSSIANSRTQ 123
L + F + W+AWAINP + + M G+QALVA S + T+ S +
Sbjct: 64 LAVVFSAPP-SKPGGWIAWAINPKS---TGMAGSQALVASKDPSTGVASVTTLNIVSYSS 119
Query: 124 LAESNISYPHSGLIATHENNE---VTIYASITLPV---GTPSLVHLWQDG-AMSGSTPQM 176
L S +S+ + A N+ + I+A + +P + + +WQ G +S Q
Sbjct: 120 LVPSKLSFDVWDVKAEEAANDGGALRIFAKVKVPADLAASGKVNQVWQVGPGVSNGRIQA 179
Query: 177 HDMTSANTQSKESLDLRS---GASEQGSGGGSLSR--RRNTHGVLNAISWGILMPLGAVI 231
HD + N S SLDL G G GG SR +RN HG+LNA+SWG+L P+GA+I
Sbjct: 180 HDFSGPNLNSVGSLDLTGTTPGVPVSGGGGAGNSRIHKRNIHGILNAVSWGLLFPIGAMI 239
Query: 232 ARYLKVFKSADPAWFYLHVTCQSAAYIVGVAGWGTGLKLGSDSAGVTYSTHRTLGIVIFC 291
ARY+++F+SADPAWFYLHV+CQ +AY++GVAGW TGLKLGS+S G+ Y+THR +GI +F
Sbjct: 240 ARYMRIFESADPAWFYLHVSCQFSAYVIGVAGWATGLKLGSESKGIQYNTHRNIGICLFS 299
Query: 292 LGTLQVFALLLRPKKDHKIRFYWNLYHWGVGYATIIISIINIFKGFEALEVSAADRYDNW 351
+ TLQ+FA+LLRP+KDHK RF WN+YH GVGY+ +I+ IIN+FKG L +
Sbjct: 300 IATLQMFAMLLRPRKDHKFRFVWNIYHHGVGYSILILGIINVFKGLSILNPK-----HTY 354
Query: 352 KHAYTGIIAALGGVAVLLEAYTWIIVIKRKKSEN 385
K AY +I LGG+ +LLE TW+IV+KRK +++
Sbjct: 355 KTAYIAVIGTLGGITLLLEVVTWVIVLKRKSAKS 388
>At3g59070 putative protein
Length = 466
Score = 268 bits (684), Expect = 5e-72
Identities = 146/346 (42%), Positives = 207/346 (59%), Gaps = 14/346 (4%)
Query: 29 CKSQNFTNNAIFTSCRDLPQLTSYLHWTYDQTTGKLDIAFRHKGITDTNRWVAWAINPNN 88
C+S +F N F SC DL L SYLH+ Y Q TG L+IA+ H + +++ W++WAINP +
Sbjct: 37 CESYSFNNGKSFRSCTDLLVLNSYLHFNYAQETGVLEIAYHHSNL-ESSSWISWAINPTS 95
Query: 89 DLASSMNGAQALVAILQS-SGTPKAYTSSIANSRTQLAESNISYPHSGLIATHENNEVTI 147
M GAQALVA S SG +AYTSSI + L ES +S + + A + N E+ I
Sbjct: 96 ---KGMVGAQALVAYRNSTSGVMRAYTSSINSYSPMLQESPLSLRVTQVSAEYSNGEMMI 152
Query: 148 YASITLPVGTPSLVHLWQDGAMS-GSTPQMHDMTSANTQSKESLDLRSG-ASEQGSGGGS 205
+A++ LP T + HLWQDG + G MH M+ N +S SLDL SG + S +
Sbjct: 153 FATLVLPPNTTVVNHLWQDGPLKEGDRLGMHAMSGDNLKSMASLDLLSGQVTTTKSVNRN 212
Query: 206 LSRRRNTHGVLNAISWGILMPLGAVIARYLKVFKSADPAWFYLHVTCQSAAYIVG-VAGW 264
+ + H ++NA+SWGILMP+G + ARY+K ++ DP WFY+HV CQ+ Y G + G
Sbjct: 213 MLLVKQIHAIVNALSWGILMPIGVMAARYMKNYEVLDPTWFYIHVVCQTTGYFSGLIGGL 272
Query: 265 GTGLKLGSDSAGVTYSTHRTLGIVIFCLGTLQVFALLLRPKKDHKIRFYWNLYHWGVGYA 324
GT + + + G+ + H +G+++F LG LQ+ +L RP KDHK R YWN YH +GY
Sbjct: 273 GTAIYMARHT-GMRTTLHTVIGLLLFALGFLQILSLKARPNKDHKYRKYWNWYHHTMGYI 331
Query: 325 TIIISIINIFKGFEALEVSAADRYDNWKHAYTGIIAALGGVAVLLE 370
I++SI NI+KG L+ + WK AYT II + AV++E
Sbjct: 332 VIVLSIYNIYKGLSILQPGSI-----WKIAYTTIICCIAAFAVVME 372
>At4g17280 unknown protein
Length = 297
Score = 231 bits (588), Expect = 7e-61
Identities = 119/258 (46%), Positives = 176/258 (68%), Gaps = 10/258 (3%)
Query: 1 MVKNLRFMFLISILMFVTTTLAQTTTQTCKSQNFTNNAIFTSCRDLPQLTSYLHWTYDQT 60
++K L + +S+++ ++ T + QTC F++N +F SC DLP L S+LH+TY+ +
Sbjct: 7 IMKFLNQILCLSLILSISMTTL-SFAQTCSKYKFSSNNVFDSCNDLPFLDSFLHYTYESS 65
Query: 61 TGKLDIAFRHKGITDTNRWVAWAINPNNDLASSMNGAQALVAILQSSGTPKAYTSSIANS 120
TG L IA+RH +T + +WVAWA+NP + + M GAQA+VA QS GT + YTS I +
Sbjct: 66 TGSLHIAYRHTKLT-SGKWVAWAVNPTS---TGMVGAQAIVAYPQSDGTVRVYTSPIRSY 121
Query: 121 RTQLAESNISYPHSGLIATHENNEVTIYASITLP--VGTPSLVH-LWQDGAMSGSTPQMH 177
+T L E ++S+ SGL AT++NNE+ + AS+ L +G ++ +WQDG+MSG++ H
Sbjct: 122 QTSLLEGDLSFNVSGLSATYQNNEIVVLASLKLAQDLGNGGTINTVWQDGSMSGNSLLPH 181
Query: 178 DMTSANTQSKESLDLRSGASEQ--GSGGGSLSRRRNTHGVLNAISWGILMPLGAVIARYL 235
+ N +S +L+L SG S G+GG S R+RN HG+LN +SWGI+MPLGA+IARYL
Sbjct: 182 PTSGNNVRSVSTLNLVSGVSAAAGGAGGSSKLRKRNIHGILNGVSWGIMMPLGAIIARYL 241
Query: 236 KVFKSADPAWFYLHVTCQ 253
+V KSADPAWFY+HV+ +
Sbjct: 242 RVAKSADPAWFYIHVSAR 259
>At2g04850 unknown protein
Length = 404
Score = 226 bits (576), Expect = 2e-59
Identities = 137/405 (33%), Positives = 205/405 (49%), Gaps = 29/405 (7%)
Query: 8 MFLISILMFVTTTLAQTTTQTCKSQNFTNNAIFTSCRDLPQLTSYLHWTYDQTTGKLDIA 67
+ L +L+ + T L ++ C + T + F C LP + + WTY LD+
Sbjct: 4 LILSFLLLLLATKLPESLAGHCTTTTATKS--FEKCISLPTQQASIAWTYHPHNATLDLC 61
Query: 68 FRHKGITDTNRWVAWAINPNNDLASSMNGAQALVAILQSSG-----TPKAYTSSIANSR- 121
F I+ + WV W INP D + M G++ L+A + P SS+ +
Sbjct: 62 FFGTFISPSG-WVGWGINP--DSPAQMTGSRVLIAFPDPNSGQLILLPYVLDSSVKLQKG 118
Query: 122 ---------TQLAESNISYPHSGLIATHENN-EVTIYASITLPVGTPSLVHLWQDGA-MS 170
+L+ S+ S + G +AT N V IYAS+ L + H+W G +
Sbjct: 119 PLLSRPLDLVRLSSSSASL-YGGKMATIRNGASVQIYASVKLSSNNTKIHHVWNRGLYVQ 177
Query: 171 GSTPQMHDMTSANTQSKESLDLRSGASEQGSGGGSLSRRRNTHGVLNAISWGILMPLGAV 230
G +P +H TS + S + D+ SG + GS + + THGV+NAISWG L+P GAV
Sbjct: 178 GYSPTIHPTTSTDLSSFSTFDVTSGFATVNQNSGSRALKV-THGVVNAISWGFLLPAGAV 236
Query: 231 IARYLKVFKSADPAWFYLHVTCQSAAYIVGVAGWGTGLKLGSDSAGVTYSTHRTLGIVIF 290
ARYL+ +S P WFY+H Q +++G G+ G+ LG +S GVTY HR+LGI F
Sbjct: 237 TARYLRQMQSIGPTWFYIHAAIQLTGFLLGTIGFSIGIVLGHNSPGVTYGLHRSLGIATF 296
Query: 291 CLGTLQVFALLLRPKKDHKIRFYWNLYHWGVGYATIIISIINIFKGFEALEVSAADRYDN 350
LQ ALL RPK +K R YW YH VGYA +++ ++N+F+GFE L +
Sbjct: 297 TAAALQTLALLFRPKTTNKFRRYWKSYHHFVGYACVVMGVVNVFQGFEVLREGRSYA--- 353
Query: 351 WKHAYTGIIAALGGVAVLLEAYTWIIVIKRKKSEN-KLQGMNGTN 394
K Y ++ L GV V +E +W++ ++ K E K G+ G +
Sbjct: 354 -KLGYCLCLSTLVGVCVAMEVNSWVVFCRKAKEEKMKRDGLTGVD 397
>At5g48750 putative protein
Length = 255
Score = 169 bits (427), Expect = 3e-42
Identities = 93/243 (38%), Positives = 138/243 (56%), Gaps = 7/243 (2%)
Query: 9 FLISILMFVTTTLAQTTTQTCKSQNFTNNAIFTSCRDLPQLTSYLHWTYDQTTGKLDIAF 68
FL + T +C S NF N F SC DLP L S+LH++Y + TG L++A+
Sbjct: 14 FLFVLAPCFTRATTNEVQVSCDSHNFNNGKHFRSCVDLPVLDSFLHYSYVRETGVLEVAY 73
Query: 69 RHKGITDTNRWVAWAINPNNDLASSMNGAQALVAILQS-SGTPKAYTSSIANSRTQLAES 127
RH I +++ W+AW INP + M GAQ L+A S SG +AYTSSI L E
Sbjct: 74 RHTNI-ESSSWIAWGINPTS---KGMIGAQTLLAYRNSTSGFMRAYTSSINGYTPMLQEG 129
Query: 128 NISYPHSGLIATHENNEVTIYASITLPVGTPSLVHLWQDGAMS-GSTPQMHDMTSANTQS 186
+S+ + L A + N E+TI+A++ P T + HLWQDG + G MH M+ + +S
Sbjct: 130 PLSFRVTQLSAEYLNREMTIFATMVWPSNTTVVNHLWQDGPLKEGDRLGMHAMSGNHLKS 189
Query: 187 KESLDLRSG-ASEQGSGGGSLSRRRNTHGVLNAISWGILMPLGAVIARYLKVFKSADPAW 245
+LDL SG + ++ ++ HG++NA+ WGI +P+G + ARY++ +K DP W
Sbjct: 190 MANLDLLSGQVMTTKAANDNMLLVKSIHGLVNAVCWGIFIPIGVMAARYMRTYKGLDPTW 249
Query: 246 FYL 248
FY+
Sbjct: 250 FYI 252
>At3g07575 unknown protein
Length = 369
Score = 123 bits (308), Expect = 2e-28
Identities = 102/381 (26%), Positives = 167/381 (43%), Gaps = 39/381 (10%)
Query: 8 MFLISILMFVTTTLA------QTTTQTCKSQNFTNNAIFTSCRDLPQLTSYLHWTYDQTT 61
++ +SI++FV L+ Q T +C S N+ F TS L T T
Sbjct: 3 LYSVSIIIFVLIALSTIVNAQQAATDSCNSTLPLNDLTFN--------TSLLQCTEAWTP 54
Query: 62 GKLDIAFRHKGITDTNRWVAWAINPNNDLA--------SSMNGAQALVAILQS---SGTP 110
+ + T N W P++ M G+ A+V + S SGT
Sbjct: 55 QNFILRYAR---TAENTWSFILSAPDSSAFIGIGFSTNGQMIGSSAIVGWIPSDGGSGTV 111
Query: 111 KAYT-SSIANSRTQLAESNISYPHSGLIATHENNEVTIYASITLPVGTPSLVHLWQDGAM 169
K Y + + +++ + L ++ + + +T + SL++
Sbjct: 112 KPYLLGGKSPGEVNPDQGDLTIVNGSLKIESVSSRLYMRFQLTATLPRQSLLYAVGPAGF 171
Query: 170 SGSTPQMHDMTSANTQSKESLDLRSGASEQGSGGGSLSRRRNTHGVLNAISWGILMPLGA 229
S+P + + +++ +G S+ S+ + THG++N WGIL+ +GA
Sbjct: 172 FPSSPDFR-LREHRFVTTTTINYNTG-SQSVVKVSPHSKLKKTHGLMNMFGWGILIIVGA 229
Query: 230 VIARYLKVFKSADPAWFYLHVTCQSAAYIVGVAGWGTGLKLGSDSAGVTYSTHRTLGIVI 289
++AR++K + DP WFY H+ Q+ +++G+ G GL L + S H+ LGI I
Sbjct: 230 IVARHMKQW---DPTWFYAHIALQTTGFLLGLTGVICGLVLENRLKANNVSKHKGLGITI 286
Query: 290 FCLGTLQVFALLLRPKKDHKIRFYWNLYHWGVGYATIIISIINIFKGFEALEVSAADRYD 349
+G LQ+ ALL RP K K R YWN YH +G II++I NIF G + A
Sbjct: 287 LVMGVLQMLALLARPDKQSKYRKYWNWYHHNIGRLLIILAISNIFYG-----IHLAKAGT 341
Query: 350 NWKHAYTGIIAALGGVAVLLE 370
+W Y +A L A+ LE
Sbjct: 342 SWNGGYGFAVAVLALTAIGLE 362
>At3g61750 putative protein
Length = 398
Score = 107 bits (268), Expect = 9e-24
Identities = 62/179 (34%), Positives = 93/179 (51%), Gaps = 8/179 (4%)
Query: 194 SGASEQGSGGGSLSRRRNTHGVLNAISWGILMPLGAVIARYLKVFKSADPAWFYLHVTCQ 253
S AS S + S + HGV+ + WG L+P+GA++ARYL+ DP W+YLH+ Q
Sbjct: 201 SKASGATSIKTTTSTEKTKHGVMAILGWGFLLPVGAILARYLR---HKDPLWYYLHIGFQ 257
Query: 254 SAAYIVGVAGWGTGLKLGSDSAGVTYSTHRTLGIVIFCLGTLQVFALLLRPKKDHKIRFY 313
+I G+A G++L + HR +GI + L TLQV A RP+K+ K+R Y
Sbjct: 258 FTGFIFGLAAVILGIQL-YNRIQPDIPAHRGIGIFLLVLSTLQVLAFFARPQKETKMRRY 316
Query: 314 WNLYHWGVGYATIIISIINIFKGFEALEVSAADRYDNWKHAYTGIIAALGGVAVLLEAY 372
WN YH +G ++ +NI G A + D WK Y +++ V+LE +
Sbjct: 317 WNWYHHWIGRISLFFGAVNIVLGIR----MADNGGDGWKIGYGFVLSVTLLAFVVLEIF 371
>At3g07390 auxin-induced protein (AIR12)
Length = 273
Score = 97.4 bits (241), Expect = 1e-20
Identities = 68/252 (26%), Positives = 110/252 (42%), Gaps = 25/252 (9%)
Query: 1 MVKNLRFMFLISILMFVTTTLAQTTTQTCKSQNFTNNAIFTSCRDLPQLTSYLHWTYDQT 60
M + + ++++ FV+ + Q CKSQN + F SC DLP L SYLH+TY+ +
Sbjct: 22 MASSSSSLLILAVACFVSLISPAISQQACKSQNLNSAGPFDSCEDLPVLNSYLHYTYNSS 81
Query: 61 TGKLDIAFRHKGITDTNRWVAWAINPNNDLASSMNGAQALVAILQSSGTPKAYTSSIANS 120
L +AF WVAWAINP + M G+QA +A G + +S
Sbjct: 82 NSSLSVAFVATPSQANGGWVAWAINPT---GTKMAGSQAFLAYRSGGGAAPVVKTYNISS 138
Query: 121 RTQLAESNISYPHSGLIA-THENNEVTIYASITLPVGTPSLVHLWQ-DGAMSGSTPQMHD 178
+ L E +++ L A + + I+ ++ +P G S+ +WQ G ++ P +H
Sbjct: 139 YSSLVEGKLAFDFWNLRAESLSGGRIAIFTTVKVPAGADSVNQVWQIGGNVTNGRPGVHP 198
Query: 179 MTSANTQSKESLDL-------------------RSGASEQG-SGGGSLSRRRNTHGVLNA 218
N S L SG++ G + GG + T V
Sbjct: 199 FGPDNLGSHRVLSFTEDAAPGSAPSPGSAPAPGTSGSTTPGTAAGGPGNAGSLTRNVNFG 258
Query: 219 ISWGILMPLGAV 230
++ GIL+ LG++
Sbjct: 259 VNLGILVLLGSI 270
>At4g18260 unknown protein
Length = 545
Score = 67.0 bits (162), Expect = 2e-11
Identities = 55/207 (26%), Positives = 88/207 (41%), Gaps = 11/207 (5%)
Query: 195 GASEQGSGGGSLSRRRNTHGVLNAISWGILMPLGAVIARYLKVFKSAD---PAWFYLHVT 251
G+ EQ + HG+L +S G LMP+G + R +FYLHV
Sbjct: 46 GSLEQDKLSHQMINSIKLHGILLWVSMGFLMPVGILFIRMANKAHENGIKVKVFFYLHVI 105
Query: 252 CQSAAYIVGVAGWGTGLKLGSDSAGVTYSTHRTLGIVIFCLGTLQVFALLLRPKKDHKIR 311
Q A ++ G L+ +S + H+ LG+ ++ LQ + +P + K R
Sbjct: 106 FQILAVVLATIGAILSLRTLENSFD---NNHQRLGLALYAAMWLQFLTGVFKPSRGSKRR 162
Query: 312 FYWNLYHWGVGYATIIISIINIFKGFEALEVSAADRYDN--WKHAYTGIIAALGGVAVLL 369
W L HW +G I+ I+NI+ G +A + + D+ W +T + L V L
Sbjct: 163 LRWFLLHWILGTIVSIVGIVNIYTGIQAYQKKTSLSRDSSLWTILFTVQVTCL--VFFYL 220
Query: 370 EAYTWIIVIKRKKSENKLQGM-NGTNG 395
W K++ ++L N TNG
Sbjct: 221 YQDKWEHFQKQRVVLDELDHQNNNTNG 247
>At2g30890 hypothetical protein
Length = 257
Score = 65.1 bits (157), Expect = 7e-11
Identities = 43/166 (25%), Positives = 78/166 (46%), Gaps = 6/166 (3%)
Query: 213 HGVLNAISWGILMPLGAVIARYLKVFKSADPAW---FYLHVTCQSAAYIVGVAGWGTGLK 269
HG + + G+LMP+G + R + + F+LHVT Q A I+ G +
Sbjct: 56 HGFMLWAAMGVLMPIGIISIRLMSIKDQPIITLRRLFFLHVTSQMVAVILVTIGAVMSVI 115
Query: 270 LGSDSAGVTYSTHRTLGIVIFCLGTLQVFALLLRPKKDHKIRFYWNLYHWGVGYATIIIS 329
++S + H+ LGI ++ + Q LRP ++ K R W + HW +G + I+
Sbjct: 116 NFNNSFS---NHHQQLGIGLYVIVWFQALLGFLRPPREEKARRKWFVGHWILGTSIAILG 172
Query: 330 IINIFKGFEALEVSAADRYDNWKHAYTGIIAALGGVAVLLEAYTWI 375
IINI+ G A + + W +T ++ + V + + +++I
Sbjct: 173 IINIYTGLHAYAKKTSKSANLWTILFTAQLSCIALVYLFQDKWSYI 218
>At5g54830 unknown protein
Length = 907
Score = 60.1 bits (144), Expect = 2e-09
Identities = 51/184 (27%), Positives = 78/184 (41%), Gaps = 20/184 (10%)
Query: 164 WQDGAMSGSTPQMHDMTSANTQSKESLDLRSGASEQGSGGGSLSRRRNTHGVLNAISWGI 223
W DG ++ MH +TS Q + L G++E L HG + ++WGI
Sbjct: 648 WTDGQLTERN--MHSVTS---QRPVRVMLTRGSAEADQ---DLRPVLGVHGFMMFLAWGI 699
Query: 224 LMPLGAVIARYLKVFKSADPAWFYLHVTCQSAAYIVGVAGWGTGLKLGSDSAGVTY-STH 282
L+P G + ARYLK K WF +H+ Q + + G L ++ G ++ STH
Sbjct: 700 LLPGGILSARYLKHIKG--DGWFKIHMYLQCSGLAIVFLGL---LFAVAELNGFSFSSTH 754
Query: 283 RTLGIVIFCLGTLQVFALLLRPKKD------HKIRFYWNLYHWGVGYATIIISIINIFKG 336
G L Q LRP K R W H VG + +++ ++ +F G
Sbjct: 755 VKFGFTAIVLACAQPVNAWLRPAKPAQGELISSKRLIWEYSHSIVGQSAVVVGVVALFTG 814
Query: 337 FEAL 340
+ L
Sbjct: 815 MKHL 818
>At2g26890 unknown protein
Length = 2535
Score = 31.2 bits (69), Expect = 1.1
Identities = 25/92 (27%), Positives = 41/92 (44%), Gaps = 15/92 (16%)
Query: 84 INPNNDLASSMNGAQALVAILQSSGTPKAYTSSIANSRT---------------QLAESN 128
I+P S ++G Q LVA L++ + ++ A ++ E N
Sbjct: 375 IDPPCGRVSLISGPQHLVADLETCSLHLKHLAAAAKDAVAEGGSVPGCRARLWRRIREFN 434
Query: 129 ISYPHSGLIATHENNEVTIYASITLPVGTPSL 160
P++G+ A E EVT+ A IT+ TP+L
Sbjct: 435 ACIPYTGVPANSEVPEVTLMALITMLPSTPNL 466
>At1g51590 mannosyl-oligosaccharide 1,2-alpha-mannosidase, putative
Length = 560
Score = 31.2 bits (69), Expect = 1.1
Identities = 31/123 (25%), Positives = 52/123 (42%), Gaps = 10/123 (8%)
Query: 81 AWAINPNNDLASSMNGAQALVAILQSSGTPKAYTSSIANSRTQLAESNISYPHSGLIATH 140
+WA ++ LA S +A+ Q +G PK Y + T+L N ++P GL+ +
Sbjct: 237 SWAAGGDSILADSGTEQLEFIALSQRTGDPK-YQQKVEKVITEL---NKNFPADGLLPIY 292
Query: 141 EN--NEVTIYASITLPVGTPS----LVHLWQDGAMSGSTPQMHDMTSANTQSKESLDLRS 194
N N Y++ T S L+ +W G + + DM + + SL +S
Sbjct: 293 INPDNANPSYSTTTFGAMGDSFYEYLLKVWVQGNKTSAVKPYRDMWEKSMKGLLSLVKKS 352
Query: 195 GAS 197
S
Sbjct: 353 TPS 355
>At5g14540 unknown protein
Length = 530
Score = 30.4 bits (67), Expect = 1.8
Identities = 28/92 (30%), Positives = 39/92 (41%), Gaps = 18/92 (19%)
Query: 118 ANSRTQLAESNISYPHSGLIATHENNEVTIYASITLPVGTPSLVHLWQDGAMSGSTPQMH 177
+NS S SYP++G + + N TPS+ Q G+ SG+ PQ+
Sbjct: 386 SNSGFPSGYSPESYPYTGPPSQYGN--------------TPSVKPTHQSGSGSGAYPQL- 430
Query: 178 DMTSANTQSKESLDLRSGASEQGSGGGSLSRR 209
+ L + S S GSGGGS S R
Sbjct: 431 ---PMARPLPQGLPMASAISSGGSGGGSDSPR 459
>At5g62360 DC1.2 homologue - like protein
Length = 203
Score = 29.3 bits (64), Expect = 4.0
Identities = 18/58 (31%), Positives = 30/58 (51%)
Query: 7 FMFLISILMFVTTTLAQTTTQTCKSQNFTNNAIFTSCRDLPQLTSYLHWTYDQTTGKL 64
F+ +I++L V TT TTT T ++ ++ FT+ L + H + QT+ KL
Sbjct: 17 FLIIIAMLKLVHTTTTTTTTTTTNTEFVKSSCTFTTYPRLCFSSLSTHASLIQTSPKL 74
>At2g06180 hypothetical protein
Length = 793
Score = 29.3 bits (64), Expect = 4.0
Identities = 29/97 (29%), Positives = 44/97 (44%), Gaps = 3/97 (3%)
Query: 117 IANSRTQLAESNI--SYPHSGLIATHENNEVTIYASITLPVGTPSLVHLWQDGAMSGSTP 174
+A +RT LA S+ S H L A + + A + LPV + + D ++ S
Sbjct: 376 VAAARTSLAASHTQASTSHPSLPAANARPLLRRRARL-LPVVRLTAELIVIDQEVAPSRE 434
Query: 175 QMHDMTSANTQSKESLDLRSGASEQGSGGGSLSRRRN 211
+ +AN ++ S+D RS QG G GS R N
Sbjct: 435 ERVKPLAANPEAAVSVDRRSSGRSQGRGRGSRGRGPN 471
>At3g60620 phosphatidate cytidylyltransferase - like protein
Length = 399
Score = 28.9 bits (63), Expect = 5.2
Identities = 15/67 (22%), Positives = 27/67 (39%), Gaps = 10/67 (14%)
Query: 318 HWGVGYATIIISIINIF----------KGFEALEVSAADRYDNWKHAYTGIIAALGGVAV 367
HW VG I+IS I K F + + W+ A+ G++ + +
Sbjct: 263 HWTVGLVAILISFCGIIASDTFAFLGGKAFGRTPLISISPKKTWEGAFAGLVGCISITIL 322
Query: 368 LLEAYTW 374
L ++ +W
Sbjct: 323 LSKSLSW 329
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.133 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,706,816
Number of Sequences: 26719
Number of extensions: 356783
Number of successful extensions: 1050
Number of sequences better than 10.0: 23
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 982
Number of HSP's gapped (non-prelim): 24
length of query: 402
length of database: 11,318,596
effective HSP length: 102
effective length of query: 300
effective length of database: 8,593,258
effective search space: 2577977400
effective search space used: 2577977400
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC148482.6


