

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148482.4 + phase: 0
(337 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
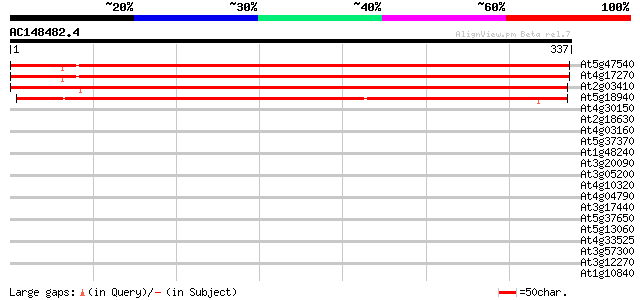
Sequences producing significant alignments: (bits) Value
At5g47540 unknown protein 551 e-157
At4g17270 unknown protein 516 e-147
At2g03410 unknown protein 434 e-122
At5g18940 unknown protein 270 6e-73
At4g30150 hypothetical protein 34 0.099
At2g18630 unknown protein 34 0.099
At4g03160 hypothetical protein 32 0.38
At5g37370 unknown protein 30 1.9
At1g48240 NPSN12 like protein 30 1.9
At3g20090 cytochrome P450 like protein 30 2.4
At3g05200 RING-H2 zinc finger protein ATL6 (ATL6) 29 3.2
At4g10320 isoleucine-tRNA ligase - like protein 29 4.2
At4g04790 hypothetical protein 29 4.2
At3g17440 unknown protein 29 4.2
At5g37650 putative protein 28 5.4
At5g13060 unknown protein 28 5.4
At4g33525 metal-transporting P-type ATPase 28 5.4
At3g57300 helicase-like protein 28 7.1
At3g12270 hypothetical protein 28 7.1
At1g10840 ranslation initiation factor like protein 28 7.1
>At5g47540 unknown protein
Length = 343
Score = 551 bits (1421), Expect = e-157
Identities = 282/339 (83%), Positives = 313/339 (92%), Gaps = 4/339 (1%)
Query: 1 MKGLFKPKPRTPTDIVRQTRDLLLFFDRNT---ESRDSKREEKQMTELCKNIRELKSILY 57
MKGLFK KPRTP D+VRQTRDLLLF DR+T + RDSKR+EK M EL +NIR++KSILY
Sbjct: 1 MKGLFKSKPRTPADLVRQTRDLLLFSDRSTSLPDLRDSKRDEK-MAELSRNIRDMKSILY 59
Query: 58 GNSESEPVSEACAQLTQEFFKENTLRLLIKCIPKLNLEARKDATQVVANLQRQPVQSKLI 117
GNSE+EPV+EACAQLTQEFFKE+TLRLLI C+PKLNLE RKDATQVVANLQRQ V S+LI
Sbjct: 60 GNSEAEPVAEACAQLTQEFFKEDTLRLLITCLPKLNLETRKDATQVVANLQRQQVNSRLI 119
Query: 118 ASDYLENNMDLMDILIVGYENTDMALHYGAMLRECIRHQIVAKYVLNSPHMKKFFDYIQL 177
ASDYLE N+DLMD+LI G+ENTDMALHYGAM RECIRHQIVAKYVL S H+KKFFDYIQL
Sbjct: 120 ASDYLEANIDLMDVLIEGFENTDMALHYGAMFRECIRHQIVAKYVLESDHVKKFFDYIQL 179
Query: 178 PNFDIAADAAATFKELMTRHKSTVAEFLSKNYEWFFADYNSKLLESSNYITRRLAVKLLG 237
PNFDIAADAAATFKEL+TRHKSTVAEFL+KN +WFFADYNSKLLESSNYITRR A+KLLG
Sbjct: 180 PNFDIAADAAATFKELLTRHKSTVAEFLTKNEDWFFADYNSKLLESSNYITRRQAIKLLG 239
Query: 238 DMLLDRSNSAVMTQYVSSRENLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPADIVSI 297
D+LLDRSNSAVMT+YVSSR+NLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPADIV+I
Sbjct: 240 DILLDRSNSAVMTKYVSSRDNLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPADIVNI 299
Query: 298 FVANKSKMLRLLDEFKIDKEDEQFEADKAQVMEEIASLE 336
VAN+SK+LRLL + K DKEDE+FEADK+QV+ EIA+LE
Sbjct: 300 LVANRSKLLRLLADLKPDKEDERFEADKSQVLREIAALE 338
>At4g17270 unknown protein
Length = 343
Score = 516 bits (1329), Expect = e-147
Identities = 264/339 (77%), Positives = 303/339 (88%), Gaps = 4/339 (1%)
Query: 1 MKGLFKPKPRTPTDIVRQTRDLLLFFDRNT---ESRDSKREEKQMTELCKNIRELKSILY 57
M+GLFK KPRTP DIVRQTRDLLL+ DR+ + R+SKREEK M EL K+IR+LK ILY
Sbjct: 1 MRGLFKSKPRTPADIVRQTRDLLLYADRSNSFPDLRESKREEK-MVELSKSIRDLKLILY 59
Query: 58 GNSESEPVSEACAQLTQEFFKENTLRLLIKCIPKLNLEARKDATQVVANLQRQPVQSKLI 117
GNSE+EPV+EACAQLTQEFFK +TLR L+ +P LNLEARKDATQVVANLQRQ V S+LI
Sbjct: 60 GNSEAEPVAEACAQLTQEFFKADTLRRLLTSLPNLNLEARKDATQVVANLQRQQVNSRLI 119
Query: 118 ASDYLENNMDLMDILIVGYENTDMALHYGAMLRECIRHQIVAKYVLNSPHMKKFFDYIQL 177
A+DYLE+N+DLMD L+ G+ENTDMALHYG M RECIRHQIVAKYVL+S H+KKFF YIQL
Sbjct: 120 AADYLESNIDLMDFLVDGFENTDMALHYGTMFRECIRHQIVAKYVLDSEHVKKFFYYIQL 179
Query: 178 PNFDIAADAAATFKELMTRHKSTVAEFLSKNYEWFFADYNSKLLESSNYITRRLAVKLLG 237
PNFDIAADAAATFKEL+TRHKSTVAEFL KN +WFFADYNSKLLES+NYITRR A+KLLG
Sbjct: 180 PNFDIAADAAATFKELLTRHKSTVAEFLIKNEDWFFADYNSKLLESTNYITRRQAIKLLG 239
Query: 238 DMLLDRSNSAVMTQYVSSRENLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPADIVSI 297
D+LLDRSNSAVMT+YVSS +NLRILMNLLRESSK+IQIEAFHVFKLF ANQNKP+DI +I
Sbjct: 240 DILLDRSNSAVMTKYVSSMDNLRILMNLLRESSKTIQIEAFHVFKLFVANQNKPSDIANI 299
Query: 298 FVANKSKMLRLLDEFKIDKEDEQFEADKAQVMEEIASLE 336
VAN++K+LRLL + K DKEDE+F+ADKAQV+ EIA+L+
Sbjct: 300 LVANRNKLLRLLADIKPDKEDERFDADKAQVVREIANLK 338
>At2g03410 unknown protein
Length = 348
Score = 434 bits (1116), Expect = e-122
Identities = 213/338 (63%), Positives = 276/338 (81%), Gaps = 3/338 (0%)
Query: 1 MKGLFKPKPRTPTDIVRQTRDLLLFFDRNTESRDSKREEKQ--MTELCKNIRELKSILYG 58
MKGLFK K R P +IVRQTRDL+ + E D++ ++ ELC+NIR+LKSILYG
Sbjct: 1 MKGLFKNKSRLPGEIVRQTRDLIALAESEEEETDARNSKRLGICAELCRNIRDLKSILYG 60
Query: 59 NSESEPVSEACAQLTQEFFKENTLRLLIKCIPKLNLEARKDATQVVANLQRQPVQSKLIA 118
N E+EPV EAC LTQEFF+ +TLR LIK IPKL+LEARKDATQ+VANLQ+Q V+ +L+A
Sbjct: 61 NGEAEPVPEACLLLTQEFFRADTLRPLIKSIPKLDLEARKDATQIVANLQKQQVEFRLVA 120
Query: 119 SDYLENNMDLMDILIVGYENT-DMALHYGAMLRECIRHQIVAKYVLNSPHMKKFFDYIQL 177
S+YLE+N+D++D L+ G ++ ++ALHY ML+EC+RHQ+VAKY+L S +++KFFDY+QL
Sbjct: 121 SEYLESNLDVIDSLVEGIDHDHELALHYTGMLKECVRHQVVAKYILESKNLEKFFDYVQL 180
Query: 178 PNFDIAADAAATFKELMTRHKSTVAEFLSKNYEWFFADYNSKLLESSNYITRRLAVKLLG 237
P FD+A DA+ F+EL+TRHKSTVAE+L+KNYEWFFA+YN+KLLE +Y T+R A KLLG
Sbjct: 181 PYFDVATDASKIFRELLTRHKSTVAEYLAKNYEWFFAEYNTKLLEKGSYFTKRQASKLLG 240
Query: 238 DMLLDRSNSAVMTQYVSSRENLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPADIVSI 297
D+L+DRSNS VM +YVSS +NLRI+MNLLRE +K+IQ+EAFH+FKLF AN+NKP DIV+I
Sbjct: 241 DVLMDRSNSGVMVKYVSSLDNLRIMMNLLREPTKNIQLEAFHIFKLFVANENKPEDIVAI 300
Query: 298 FVANKSKMLRLLDEFKIDKEDEQFEADKAQVMEEIASL 335
VAN++K+LRL + K +KED FE DKA VM EIA+L
Sbjct: 301 LVANRTKILRLFADLKPEKEDVGFETDKALVMNEIATL 338
>At5g18940 unknown protein
Length = 345
Score = 270 bits (691), Expect = 6e-73
Identities = 145/334 (43%), Positives = 214/334 (63%), Gaps = 5/334 (1%)
Query: 5 FKP-KPRTPTDIVRQTRDLLLFFDRNTESRDSKREEKQMTELCKNIRELKSILYGNSESE 63
FKP +P+TP ++V+ RD L+ D T + K EK + E+ KN L+ IL G+ E+E
Sbjct: 6 FKPSRPKTPQEVVKAIRDSLMALDTKTVV-EVKALEKALEEVEKNFSSLRGILSGDGETE 64
Query: 64 PVSEACAQLTQEFFKENTLRLLIKCIPKLNLEARKDATQVVANLQRQPVQSKLIASDYLE 123
P ++ QL EF KE+ + L+I + L E RKD + L +Q V Y E
Sbjct: 65 PNADQAVQLALEFCKEDVVSLVIHKLHILGWETRKDLLHCWSILLKQKVGDTYCCVQYFE 124
Query: 124 NNMDLMDILIVGYENTDMALHYGAMLRECIRHQIVAKYVLNSPHMKKFFDYIQLPNFDIA 183
+ +L+D L+V Y+N ++ALH G+MLRECI+ +AKY+L S + FF +++LPNFD+A
Sbjct: 125 EHFELLDSLVVCYDNKEIALHCGSMLRECIKFPSLAKYILESACFELFFKFVELPNFDVA 184
Query: 184 ADAAATFKELMTRHKSTVAEFLSKNYEWFFADYNSKLLESSNYITRRLAVKLLGDMLLDR 243
+DA +TFK+L+T+H S V+EFL+ +Y FF D +LL SSNY+TRR ++KLL D LL+
Sbjct: 185 SDAFSTFKDLLTKHDSVVSEFLTSHYTEFF-DVYERLLTSSNYVTRRQSLKLLSDFLLEP 243
Query: 244 SNSAVMTQYVSSRENLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPADIVSIFVANKS 303
NS +M +++ L+++M LL++SSK+IQI AFH+FK+F AN NKP ++ I N
Sbjct: 244 PNSHIMKKFILEVRYLKVIMTLLKDSSKNIQISAFHIFKIFVANPNKPQEVKIILARNHE 303
Query: 304 KMLRLLDEFKIDK--EDEQFEADKAQVMEEIASL 335
K+L LL + K ED+QFE +K ++EEI L
Sbjct: 304 KLLELLHDLSPGKGSEDDQFEEEKELIIEEIQKL 337
>At4g30150 hypothetical protein
Length = 1966
Score = 34.3 bits (77), Expect = 0.099
Identities = 43/173 (24%), Positives = 78/173 (44%), Gaps = 17/173 (9%)
Query: 123 ENNMDL-MDILIVGYENTDMALHYGAMLRECIRHQIVAKYVLNS-PHMKK------FFDY 174
ENN+D +G E L + ++L E H+ V +Y+ S H+ K F D
Sbjct: 1081 ENNVDKGSQKKKIGLEQFSFGLLFDSVLYE---HEFVRRYLAPSFSHVLKMTAETFFKDI 1137
Query: 175 IQLPNFDIAADAAATFKELMTRHKSTVAEFLSKNYEWFFADYNSKLLESSNYITRRLAVK 234
+ NFD +D + E++ +S++A K F + + LL++ + + +
Sbjct: 1138 TEEVNFDSPSD----WSEVLILLESSIANLSGKLKSEAFLEAHVSLLDNRKFTACQNLLN 1193
Query: 235 LLGDMLLDRSNSAVMTQYVSSRENLR--ILMNLLRESSKSIQIEAFHVFKLFA 285
LLG M + +N Y S +L I+ ++LR +K + ++F LF+
Sbjct: 1194 LLGVMPKEYTNKKSFQLYASYVLDLERFIVFSMLRCLNKLSCGDMQNLFSLFS 1246
>At2g18630 unknown protein
Length = 392
Score = 34.3 bits (77), Expect = 0.099
Identities = 17/80 (21%), Positives = 43/80 (53%), Gaps = 4/80 (5%)
Query: 89 IPKLNLEARKDATQVVANLQRQPVQSKLIASDYLENNMDLMDILIVGYENTDMALHYGAM 148
I L+ ++ ++ TQ + ++ + V+ L + + NN DL ++ + +E+T + + +
Sbjct: 70 IKSLSFDSLREVTQCLLDMNQDVVKVILQDKEDIWNNQDLFSLVNLYFESTAKTMDFCSE 129
Query: 149 LRECI----RHQIVAKYVLN 164
L C+ R Q++ ++ +N
Sbjct: 130 LENCLNRARRSQVIIQFAVN 149
>At4g03160 hypothetical protein
Length = 192
Score = 32.3 bits (72), Expect = 0.38
Identities = 18/63 (28%), Positives = 30/63 (47%), Gaps = 1/63 (1%)
Query: 24 LFFDRNTESRDSKREEKQMTELCKNIRELKSILYGNSESEPVSEACAQLTQEFFKENTLR 83
+FFD+ E D + EE +C++ E+KS + N + QL K+N R
Sbjct: 22 VFFDQEEEEEDEE-EEYDEESVCEDDLEVKSCMQTNENKGKKRKVAEQLMDSDVKDNQYR 80
Query: 84 LLI 86
L++
Sbjct: 81 LML 83
>At5g37370 unknown protein
Length = 393
Score = 30.0 bits (66), Expect = 1.9
Identities = 22/69 (31%), Positives = 33/69 (46%), Gaps = 3/69 (4%)
Query: 267 RESSKSIQIEAFHVFKLFAANQNKPADIVSIFVANKSKMLRLLDEFKIDKEDEQFEADKA 326
R S+S+QIE K ++N+ K A V+ +N +K+ L + K DE F K
Sbjct: 322 RSRSRSVQIEREPTPKRDSSNKEKSAVTVN---SNLAKLKDLYGDASSQKRDEGFGTRKD 378
Query: 327 QVMEEIASL 335
EE+ L
Sbjct: 379 SSSEEVIKL 387
>At1g48240 NPSN12 like protein
Length = 265
Score = 30.0 bits (66), Expect = 1.9
Identities = 28/128 (21%), Positives = 54/128 (41%), Gaps = 10/128 (7%)
Query: 26 FDRNTESRDSKREEKQMTELCKNIRELKSILY--------GNSESEP-VSEACAQLTQEF 76
F R + +DS R+ KQ+ EL + +R+ K ++ G + + P V++ Q
Sbjct: 29 FQRLDKIKDSSRQSKQLEELAEKMRDCKRLVKEFDRELKDGEARNSPQVNKQLNDEKQSM 88
Query: 77 FKE-NTLRLLIKCIPKLNLEARKDATQVVANLQRQPVQSKLIASDYLENNMDLMDILIVG 135
KE N+ L K + + A + +P + + +N +L+D +
Sbjct: 89 IKELNSYVALRKTYLNTLGNKKVELFDTGAGVSGEPTAEENVQMASTMSNQELVDAGMKR 148
Query: 136 YENTDMAL 143
+ TD A+
Sbjct: 149 MDETDQAI 156
>At3g20090 cytochrome P450 like protein
Length = 386
Score = 29.6 bits (65), Expect = 2.4
Identities = 39/153 (25%), Positives = 59/153 (38%), Gaps = 30/153 (19%)
Query: 28 RNTESRDSKREEKQMTELCKNIRELKSILYGNSESEPV----SEACAQLTQEFFKENTLR 83
+N SK M + + +S N E+E V E+ A + + FF R
Sbjct: 39 KNENIEISKESAMLMNNILCRMSMGRSFSEENGEAERVRGLVGESYALVKKIFFAATLRR 98
Query: 84 LLIKC-IPKLNLEARKDATQVVANLQRQPVQSKLIASDYLENN---MDLMDILIVGYENT 139
LL K IP E + + L+R V+ K D LE MD+MD+L+ Y +
Sbjct: 99 LLEKLRIPLFRKEIMGVSDRFDELLERIIVEHK----DKLEKEHQVMDMMDVLLAAYRDK 154
Query: 140 DMALHYGAMLRECIRHQIVAKYVLNSPHMKKFF 172
+ A+Y + H+K FF
Sbjct: 155 N------------------AEYKITRNHIKSFF 169
>At3g05200 RING-H2 zinc finger protein ATL6 (ATL6)
Length = 398
Score = 29.3 bits (64), Expect = 3.2
Identities = 22/95 (23%), Positives = 41/95 (43%), Gaps = 10/95 (10%)
Query: 49 IRELKSILYGNSESEPVSEA---CAQLTQEFFKENTLRLLIKCIPKLN-------LEARK 98
+ + LY + +++ + + CA EF + TLRLL KC + LEA
Sbjct: 105 VETFPTFLYSDVKTQKLGKGELECAICLNEFEDDETLRLLPKCDHVFHPHCIDAWLEAHV 164
Query: 99 DATQVVANLQRQPVQSKLIASDYLENNMDLMDILI 133
ANL Q + + + E +++L +++
Sbjct: 165 TCPVCRANLAEQVAEGESVEPGGTEPDLELQQVVV 199
>At4g10320 isoleucine-tRNA ligase - like protein
Length = 1190
Score = 28.9 bits (63), Expect = 4.2
Identities = 18/79 (22%), Positives = 42/79 (52%), Gaps = 11/79 (13%)
Query: 187 AATFKELMTRHKSTVAEFLSKNYEWFFADYNSKLLESSNYITRRLAVKLLGDML---LDR 243
A T K+++TR+++ +++ + W + L N I R+L +K +++ +D+
Sbjct: 62 AGTIKDIVTRYQTMTGHHVTRRFGW-----DCHGLPVENEIDRKLNIKRRDEVIKMGIDK 116
Query: 244 SNS---AVMTQYVSSRENL 259
N +++T+YV+ E +
Sbjct: 117 YNEECRSIVTRYVAEWEKV 135
>At4g04790 hypothetical protein
Length = 731
Score = 28.9 bits (63), Expect = 4.2
Identities = 18/68 (26%), Positives = 31/68 (45%), Gaps = 1/68 (1%)
Query: 241 LDRSNSAVMTQYVSSRENLRILMNLLRESSKSIQ-IEAFHVFKLFAANQNKPADIVSIFV 299
+D + + +Y S+ L L+ L + I+ F LFA K +DIV +
Sbjct: 504 VDPKSIISLIEYSDSKGELSTLVQLADDLQDDTSWIDGFFRMILFAVRNKKSSDIVDLLK 563
Query: 300 ANKSKMLR 307
NK ++L+
Sbjct: 564 RNKVRLLK 571
>At3g17440 unknown protein
Length = 269
Score = 28.9 bits (63), Expect = 4.2
Identities = 30/128 (23%), Positives = 53/128 (40%), Gaps = 10/128 (7%)
Query: 26 FDRNTESRDSKREEKQMTELCKNIRELKSIL------YGNSESEPVSEACAQLT---QEF 76
F R + +DS R+ KQ+ EL +RE K ++ + E+ E QL Q
Sbjct: 29 FQRLDKIKDSTRQSKQLEELTDKMRECKRLVKEFDRELKDEEARNSPEVNKQLNDEKQSM 88
Query: 77 FKE-NTLRLLIKCIPKLNLEARKDATQVVANLQRQPVQSKLIASDYLENNMDLMDILIVG 135
KE N+ L K + + + A + +P + + +N +L+D +
Sbjct: 89 IKELNSYVALRKTYMSTLGNKKVELFDMGAGVSGEPTAEENVQVASSMSNQELVDAGMKR 148
Query: 136 YENTDMAL 143
+ TD A+
Sbjct: 149 MDETDQAI 156
>At5g37650 putative protein
Length = 607
Score = 28.5 bits (62), Expect = 5.4
Identities = 31/109 (28%), Positives = 49/109 (44%), Gaps = 16/109 (14%)
Query: 160 KYVLNSPHMKKFFDYIQLPNFDIAADAAATFKELMTRHKSTVAEFLSKNYEWFFADYNSK 219
KYVLN P K ++ ++ N D+ D+ + + T E LS FFA
Sbjct: 296 KYVLN-PAFK--LEHFRVDNQDLGVDSDRELYYMF--RQCTPMEVLS-----FFA----- 340
Query: 220 LLESSNYITRRLAVKLLGDMLLDRSNSAVMTQYVSSRENLRILMNLLRE 268
+ S+Y +R +A+K L D L D ++S R +L+ L+E
Sbjct: 341 -IPGSDYRSREIAIKRLYDSLCDHTSSQWEIDVSEIRGLQPLLITCLKE 388
>At5g13060 unknown protein
Length = 709
Score = 28.5 bits (62), Expect = 5.4
Identities = 22/77 (28%), Positives = 35/77 (44%), Gaps = 4/77 (5%)
Query: 219 KLLESSNYITRRLAVKLLGDMLLDRSNSAVMTQYVSSRENLRILMNLLRESSKSIQIEAF 278
K+LESS+ ++ LG + D N A ++ R + L+NLL + S+Q A
Sbjct: 330 KMLESSDEQVVEMSAFALGRLAQDAHNQAG----IAHRGGIISLLNLLDVKTGSVQHNAA 385
Query: 279 HVFKLFAANQNKPADIV 295
A N+ AD +
Sbjct: 386 FALYGLADNEENVADFI 402
>At4g33525 metal-transporting P-type ATPase
Length = 632
Score = 28.5 bits (62), Expect = 5.4
Identities = 16/42 (38%), Positives = 25/42 (59%), Gaps = 4/42 (9%)
Query: 79 ENTLRLLIKCIPKLNLEARKDATQVVANLQRQPVQSKLIASD 120
+NTL +I+ K+ R+DA QVV NL RQ + +++ D
Sbjct: 407 DNTLAAVIRFEDKV----REDAAQVVENLTRQGIDVYMLSGD 444
>At3g57300 helicase-like protein
Length = 1507
Score = 28.1 bits (61), Expect = 7.1
Identities = 15/52 (28%), Positives = 27/52 (51%), Gaps = 1/52 (1%)
Query: 17 RQTRDLLLFFDRNTESRDSKREEKQMTELCKNIRELKSILYGNSESEPVSEA 68
+Q ++ F R E R+SKR+++++ L K + ++S P SEA
Sbjct: 430 KQEKEAAEAFKREQEQRESKRQQQRLNFLIKQTELYSHFMQNKTDSNP-SEA 480
>At3g12270 hypothetical protein
Length = 590
Score = 28.1 bits (61), Expect = 7.1
Identities = 13/28 (46%), Positives = 18/28 (63%)
Query: 308 LLDEFKIDKEDEQFEADKAQVMEEIASL 335
LL F D+EDE+ D+ +VMEE+ L
Sbjct: 140 LLYSFADDEEDEEVTFDREEVMEELQKL 167
>At1g10840 ranslation initiation factor like protein
Length = 337
Score = 28.1 bits (61), Expect = 7.1
Identities = 15/39 (38%), Positives = 21/39 (53%), Gaps = 1/39 (2%)
Query: 77 FKENTLRLLIKCIPKLNLEARKDATQVVANLQRQPVQSK 115
F EN + LIKC+ L++E +K NL RQ Q +
Sbjct: 224 FLENNMEFLIKCMDDLSMEQQK-FQYYYRNLSRQQAQQQ 261
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.133 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,604,910
Number of Sequences: 26719
Number of extensions: 254094
Number of successful extensions: 853
Number of sequences better than 10.0: 25
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 16
Number of HSP's that attempted gapping in prelim test: 835
Number of HSP's gapped (non-prelim): 28
length of query: 337
length of database: 11,318,596
effective HSP length: 100
effective length of query: 237
effective length of database: 8,646,696
effective search space: 2049266952
effective search space used: 2049266952
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC148482.4


