

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148469.4 - phase: 0 /pseudo
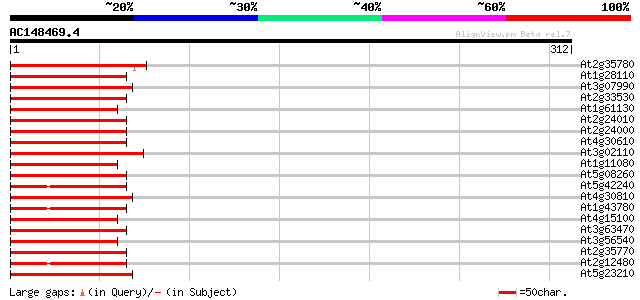
(312 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g35780 serine carboxypeptidase II like protein 78 7e-15
At1g28110 serine carboxypeptidase II (At1g28110) 78 7e-15
At3g07990 putative serine carboxypeptidase II 76 2e-14
At2g33530 putative serine carboxypeptidase II 76 3e-14
At1g61130 76 3e-14
At2g24010 putative serine carboxypeptidase II 73 2e-13
At2g24000 putative serine carboxypeptidase II 73 2e-13
At4g30610 SERINE CARBOXYPEPTIDASE II-like protein 72 3e-13
At3g02110 serine carboxypeptidase II like protein 72 3e-13
At1g11080 Serine carboxypeptidase isolog 71 9e-13
At5g08260 serine-type carboxypeptidase II-like protein 70 2e-12
At5g42240 serine carboxypeptidase II-like 69 3e-12
At4g30810 serine carboxypeptidase II like protein 68 7e-12
At1g43780 serine carboxypeptidase II, putative 68 7e-12
At4g15100 hydroxynitrile lyase like protein 67 1e-11
At3g63470 serin carboxypeptidase - like protein 67 1e-11
At3g56540 serine carboxypeptidase-like protein 67 1e-11
At2g35770 serine carboxypeptidase II like protein 67 2e-11
At2g12480 serine carboxypeptidase II like protein 67 2e-11
At5g23210 serine carboxypeptidase II-like protein 65 4e-11
>At2g35780 serine carboxypeptidase II like protein
Length = 452
Score = 77.8 bits (190), Expect = 7e-15
Identities = 38/83 (45%), Positives = 53/83 (63%), Gaps = 7/83 (8%)
Query: 1 MLYLESPAGVGFSYSANTSDYFMVTDERTARDVLIFLQGWVTKFQKYQNSDFFITGESYA 60
+L+LESPAGVGFSYS TSD + D+RTA D +FL W +F +Y++ +F+I GESYA
Sbjct: 123 LLFLESPAGVGFSYSNTTSDLYTAGDQRTAEDAYVFLVKWFERFPQYKHREFYIAGESYA 182
Query: 61 VWNLPLIT-------IPGLNFYG 76
+P ++ P +NF G
Sbjct: 183 GHYVPQLSQIVYEKRNPAINFKG 205
>At1g28110 serine carboxypeptidase II (At1g28110)
Length = 461
Score = 77.8 bits (190), Expect = 7e-15
Identities = 39/65 (60%), Positives = 44/65 (67%)
Query: 1 MLYLESPAGVGFSYSANTSDYFMVTDERTARDVLIFLQGWVTKFQKYQNSDFFITGESYA 60
MLYLE+P GVGFSYS +S Y V D+ TARD L+FLQ W KF Y N FITGESYA
Sbjct: 120 MLYLETPVGVGFSYSTQSSHYEGVNDKITARDNLVFLQRWFLKFPHYLNRSLFITGESYA 179
Query: 61 VWNLP 65
+P
Sbjct: 180 GHYVP 184
>At3g07990 putative serine carboxypeptidase II
Length = 459
Score = 76.3 bits (186), Expect = 2e-14
Identities = 35/68 (51%), Positives = 48/68 (70%)
Query: 1 MLYLESPAGVGFSYSANTSDYFMVTDERTARDVLIFLQGWVTKFQKYQNSDFFITGESYA 60
+L+LESPAGVGFSYS TSD + D+RTA D IFL W +F +Y++ +F+I GESYA
Sbjct: 127 LLFLESPAGVGFSYSNTTSDLYTTGDQRTAEDSYIFLVNWFERFPQYKHREFYIVGESYA 186
Query: 61 VWNLPLIT 68
+P ++
Sbjct: 187 GHFVPQLS 194
>At2g33530 putative serine carboxypeptidase II
Length = 465
Score = 75.9 bits (185), Expect = 3e-14
Identities = 37/65 (56%), Positives = 45/65 (68%)
Query: 1 MLYLESPAGVGFSYSANTSDYFMVTDERTARDVLIFLQGWVTKFQKYQNSDFFITGESYA 60
MLYLE+P GVGFSY+ +S Y V D+ TA+D L+FLQ W KF +Y N FITGESYA
Sbjct: 122 MLYLETPVGVGFSYANESSSYEGVNDKITAKDNLVFLQKWFLKFPQYLNRSLFITGESYA 181
Query: 61 VWNLP 65
+P
Sbjct: 182 GHYVP 186
>At1g61130
Length = 476
Score = 75.9 bits (185), Expect = 3e-14
Identities = 36/60 (60%), Positives = 43/60 (71%)
Query: 1 MLYLESPAGVGFSYSANTSDYFMVTDERTARDVLIFLQGWVTKFQKYQNSDFFITGESYA 60
+L+LESPAGVGFSYS +SDY + D+ TARD FLQ W +F Y+ DFFI GESYA
Sbjct: 122 ILFLESPAGVGFSYSNTSSDYRKLGDDFTARDSYTFLQKWFLRFPAYKEKDFFIAGESYA 181
>At2g24010 putative serine carboxypeptidase II
Length = 425
Score = 73.2 bits (178), Expect = 2e-13
Identities = 36/65 (55%), Positives = 48/65 (73%)
Query: 1 MLYLESPAGVGFSYSANTSDYFMVTDERTARDVLIFLQGWVTKFQKYQNSDFFITGESYA 60
+L+LESPAGVGFSY+ +SD DERTA++ LIFL W+++F +YQ DF+I GESYA
Sbjct: 92 ILFLESPAGVGFSYTNTSSDLKDSGDERTAQENLIFLIKWMSRFPQYQYRDFYIVGESYA 151
Query: 61 VWNLP 65
+P
Sbjct: 152 GHYVP 156
>At2g24000 putative serine carboxypeptidase II
Length = 474
Score = 73.2 bits (178), Expect = 2e-13
Identities = 34/65 (52%), Positives = 48/65 (73%)
Query: 1 MLYLESPAGVGFSYSANTSDYFMVTDERTARDVLIFLQGWVTKFQKYQNSDFFITGESYA 60
+L+LESP GVGFSY+ +SD+ DERTA++ LIFL W+++F +Y+ DF+I GESYA
Sbjct: 132 LLFLESPVGVGFSYTNTSSDFEESGDERTAQENLIFLISWMSRFPQYRYRDFYIVGESYA 191
Query: 61 VWNLP 65
+P
Sbjct: 192 GHYVP 196
>At4g30610 SERINE CARBOXYPEPTIDASE II-like protein
Length = 465
Score = 72.4 bits (176), Expect = 3e-13
Identities = 35/65 (53%), Positives = 48/65 (73%)
Query: 1 MLYLESPAGVGFSYSANTSDYFMVTDERTARDVLIFLQGWVTKFQKYQNSDFFITGESYA 60
+L+LESPAGVG+SY+ +SD DERTA+D LIFL W+++F +Y+ DF+I GESYA
Sbjct: 124 LLFLESPAGVGYSYTNTSSDLKDSGDERTAQDNLIFLIKWLSRFPQYKYRDFYIAGESYA 183
Query: 61 VWNLP 65
+P
Sbjct: 184 GHYVP 188
>At3g02110 serine carboxypeptidase II like protein
Length = 473
Score = 72.4 bits (176), Expect = 3e-13
Identities = 34/74 (45%), Positives = 50/74 (66%)
Query: 1 MLYLESPAGVGFSYSANTSDYFMVTDERTARDVLIFLQGWVTKFQKYQNSDFFITGESYA 60
+L+LE+PAGVGFSY+ +SD F D RTA+D L FL W+ +F +Y + + +ITGESYA
Sbjct: 128 LLFLEAPAGVGFSYTNRSSDLFNTGDRRTAKDSLQFLIQWLHRFPRYNHREIYITGESYA 187
Query: 61 VWNLPLITIPGLNF 74
+P + +N+
Sbjct: 188 GHYVPQLAKEIMNY 201
>At1g11080 Serine carboxypeptidase isolog
Length = 465
Score = 70.9 bits (172), Expect = 9e-13
Identities = 33/60 (55%), Positives = 42/60 (70%)
Query: 1 MLYLESPAGVGFSYSANTSDYFMVTDERTARDVLIFLQGWVTKFQKYQNSDFFITGESYA 60
ML+LESP GVGFSYS +SDY + D+ TARD FL W KF +++ + F+I GESYA
Sbjct: 141 MLFLESPVGVGFSYSNTSSDYQKLGDDFTARDAYTFLCNWFEKFPEHKENTFYIAGESYA 200
>At5g08260 serine-type carboxypeptidase II-like protein
Length = 480
Score = 69.7 bits (169), Expect = 2e-12
Identities = 33/65 (50%), Positives = 46/65 (70%)
Query: 1 MLYLESPAGVGFSYSANTSDYFMVTDERTARDVLIFLQGWVTKFQKYQNSDFFITGESYA 60
ML+LE+P GVGFSY+ N+ D + DE TA D L FL W KF ++++S+F+I+GESYA
Sbjct: 131 MLFLEAPVGVGFSYTNNSMDLQKLGDEVTASDSLAFLINWFMKFPEFRSSEFYISGESYA 190
Query: 61 VWNLP 65
+P
Sbjct: 191 GHYVP 195
>At5g42240 serine carboxypeptidase II-like
Length = 473
Score = 69.3 bits (168), Expect = 3e-12
Identities = 34/65 (52%), Positives = 46/65 (70%), Gaps = 1/65 (1%)
Query: 1 MLYLESPAGVGFSYSANTSDYFMVTDERTARDVLIFLQGWVTKFQKYQNSDFFITGESYA 60
+L++ESPAGVG+SYS +SDY D+ TA D+L+FL W KF K ++ D F+TGESYA
Sbjct: 123 LLFVESPAGVGWSYSNKSSDY-NTGDKSTANDMLVFLLRWFEKFPKLKSRDLFLTGESYA 181
Query: 61 VWNLP 65
+P
Sbjct: 182 GHYIP 186
>At4g30810 serine carboxypeptidase II like protein
Length = 479
Score = 67.8 bits (164), Expect = 7e-12
Identities = 32/68 (47%), Positives = 46/68 (67%)
Query: 1 MLYLESPAGVGFSYSANTSDYFMVTDERTARDVLIFLQGWVTKFQKYQNSDFFITGESYA 60
+L+L++P GVG+SYS +SD D+RTA D L FL WV +F +Y+ DF+I GESYA
Sbjct: 129 ILFLDAPVGVGYSYSNTSSDLKSNGDKRTAEDSLKFLLKWVERFPEYKGRDFYIVGESYA 188
Query: 61 VWNLPLIT 68
+P ++
Sbjct: 189 GHYIPQLS 196
>At1g43780 serine carboxypeptidase II, putative
Length = 479
Score = 67.8 bits (164), Expect = 7e-12
Identities = 31/65 (47%), Positives = 47/65 (71%), Gaps = 1/65 (1%)
Query: 1 MLYLESPAGVGFSYSANTSDYFMVTDERTARDVLIFLQGWVTKFQKYQNSDFFITGESYA 60
+L+++SPAGVG+SYS TSDY DE TA+D+L+F+ W+ KF +++ + F+ GESYA
Sbjct: 128 LLFVDSPAGVGWSYSNTTSDY-TTGDESTAKDMLVFMLRWLEKFPQFKTRNLFLAGESYA 186
Query: 61 VWNLP 65
+P
Sbjct: 187 GHYVP 191
>At4g15100 hydroxynitrile lyase like protein
Length = 407
Score = 67.4 bits (163), Expect = 1e-11
Identities = 30/60 (50%), Positives = 40/60 (66%)
Query: 1 MLYLESPAGVGFSYSANTSDYFMVTDERTARDVLIFLQGWVTKFQKYQNSDFFITGESYA 60
ML+LESP GVGFSYS +SDY + D +D FL W KF +++ ++F+I GESYA
Sbjct: 54 MLFLESPVGVGFSYSNTSSDYLNLDDHFAKKDAYTFLCNWFEKFPEHKGNEFYIAGESYA 113
>At3g63470 serin carboxypeptidase - like protein
Length = 502
Score = 67.4 bits (163), Expect = 1e-11
Identities = 33/65 (50%), Positives = 42/65 (63%)
Query: 1 MLYLESPAGVGFSYSANTSDYFMVTDERTARDVLIFLQGWVTKFQKYQNSDFFITGESYA 60
+L+LESPAGVGFSY+ TSD D TA D IFL W+ +F +Y+ D +I GESYA
Sbjct: 172 VLFLESPAGVGFSYTNTTSDLEKHGDRNTAADNYIFLVNWLERFPEYKGRDLYIAGESYA 231
Query: 61 VWNLP 65
+P
Sbjct: 232 GHYVP 236
>At3g56540 serine carboxypeptidase-like protein
Length = 264
Score = 67.0 bits (162), Expect = 1e-11
Identities = 31/60 (51%), Positives = 41/60 (67%)
Query: 1 MLYLESPAGVGFSYSANTSDYFMVTDERTARDVLIFLQGWVTKFQKYQNSDFFITGESYA 60
ML+LESPAG GFSY+ T+D D +TA D +FL W+ +F +Y+ DF+I GESYA
Sbjct: 169 MLFLESPAGTGFSYTNTTTDMENPGDMKTAADNYVFLVKWLERFPEYKGRDFYIAGESYA 228
>At2g35770 serine carboxypeptidase II like protein
Length = 462
Score = 66.6 bits (161), Expect = 2e-11
Identities = 31/65 (47%), Positives = 43/65 (65%)
Query: 1 MLYLESPAGVGFSYSANTSDYFMVTDERTARDVLIFLQGWVTKFQKYQNSDFFITGESYA 60
+L+L+SPAGVGFSY+ +SD V D+RT D FL W+ +F +Y+ F+I GESYA
Sbjct: 129 VLFLDSPAGVGFSYTNTSSDELTVGDKRTGEDAYRFLVRWLERFPEYKERAFYIAGESYA 188
Query: 61 VWNLP 65
+P
Sbjct: 189 GHYIP 193
>At2g12480 serine carboxypeptidase II like protein
Length = 442
Score = 66.6 bits (161), Expect = 2e-11
Identities = 32/65 (49%), Positives = 45/65 (69%), Gaps = 1/65 (1%)
Query: 1 MLYLESPAGVGFSYSANTSDYFMVTDERTARDVLIFLQGWVTKFQKYQNSDFFITGESYA 60
+L++ESPAGVG+SYS +SDY D+ T D+L+FL W KF + ++ D F+TGESYA
Sbjct: 122 LLFVESPAGVGWSYSNRSSDY-NTGDKSTVNDMLVFLLRWFNKFPELKSRDLFLTGESYA 180
Query: 61 VWNLP 65
+P
Sbjct: 181 GHYIP 185
>At5g23210 serine carboxypeptidase II-like protein
Length = 499
Score = 65.5 bits (158), Expect = 4e-11
Identities = 31/68 (45%), Positives = 44/68 (64%)
Query: 1 MLYLESPAGVGFSYSANTSDYFMVTDERTARDVLIFLQGWVTKFQKYQNSDFFITGESYA 60
+L+LESP GVGFSY+ + D + D TARD FL W +F +Y++ DF+I GESYA
Sbjct: 143 LLFLESPVGVGFSYTNTSRDIKQLGDTVTARDSYNFLVNWFKRFPQYKSHDFYIAGESYA 202
Query: 61 VWNLPLIT 68
+P ++
Sbjct: 203 GHYVPQLS 210
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.340 0.150 0.525
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,830,775
Number of Sequences: 26719
Number of extensions: 269667
Number of successful extensions: 703
Number of sequences better than 10.0: 56
Number of HSP's better than 10.0 without gapping: 54
Number of HSP's successfully gapped in prelim test: 2
Number of HSP's that attempted gapping in prelim test: 612
Number of HSP's gapped (non-prelim): 59
length of query: 312
length of database: 11,318,596
effective HSP length: 99
effective length of query: 213
effective length of database: 8,673,415
effective search space: 1847437395
effective search space used: 1847437395
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC148469.4


