

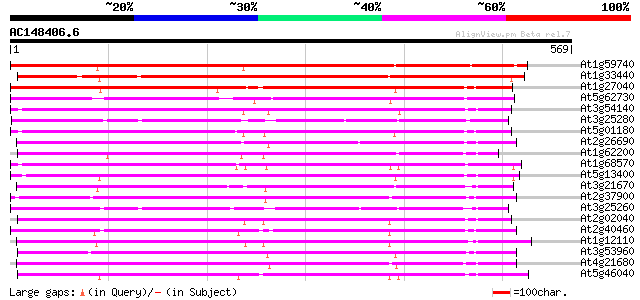
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148406.6 + phase: 0
(569 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g59740 nitrate transporter NTL1, putative 421 e-118
At1g33440 nitrate transporter NTL1, putative 416 e-116
At1g27040 nitrate transporter like protein 408 e-114
At5g62730 nitrate transporter NTL1 - like protein 368 e-102
At3g54140 peptide transport - like protein 365 e-101
At3g25280 nitrate transporter, putative 359 2e-99
At5g01180 oligopeptide transporter - like protein 353 1e-97
At2g26690 putative nitrate transporter 348 4e-96
At1g62200 unknown protein (At1g62200) 348 4e-96
At1g68570 peptide transporter like 342 3e-94
At5g13400 peptide transporter - like protein 340 1e-93
At3g21670 nitrate transporter 339 3e-93
At2g37900 putative peptide/amino acid transporter 339 3e-93
At3g25260 nitrate transporter, putative 338 5e-93
At2g02040 histidine transport protein (PTR2-B) 338 5e-93
At2g40460 putative PTR2 family peptide transporter 328 5e-90
At1g12110 nitrate/chlorate transporter CHL1 328 6e-90
At3g53960 transporter like protein 315 6e-86
At4g21680 peptide transporter - like protein 311 5e-85
At5g46040 peptide transporter 306 3e-83
>At1g59740 nitrate transporter NTL1, putative
Length = 591
Score = 421 bits (1082), Expect = e-118
Identities = 225/536 (41%), Positives = 333/536 (61%), Gaps = 15/536 (2%)
Query: 1 MGFVTNMVSLVLYFIGVMHFDLSSSANTLTNFMGSTFLLSLVGAFISDTYLNRFTTCLVF 60
MG +L+ Y I MHF LS +AN +TNF+G+ F+ +L+G ++SD +L F T ++F
Sbjct: 60 MGIAAVGNNLITYVINEMHFPLSKAANIVTNFVGTIFIFALLGGYLSDAFLGSFWTIIIF 119
Query: 61 GSLEVMALVMITVQAALDNLHPKACGK---SSCVE--GGIAVMFYTSLCLYALGMGGVRG 115
G +E+ ++++VQA L L P C +C E G A++F+ +L L ALG G V+
Sbjct: 120 GFVELSGFILLSVQAHLPQLKPPKCNPLIDQTCEEAKGFKAMIFFMALYLVALGSGCVKP 179
Query: 116 SLTAFGANQFDEKDPNEAKALATFFNWLLLSSTLGSVIGVTGIVWVSTQKAWHWGFFIIT 175
++ A GA+QF + P ++K L+++FN + ++G +I +T +VWV T GF +
Sbjct: 180 NMIAHGADQFSQSHPKQSKRLSSYFNAAYFAFSMGELIALTLLVWVQTHSGMDIGFGVSA 239
Query: 176 VASSIGFVTLAIGKPFYRIKTPGESPILRIIQVIVVAFKNRKLQLPESNEQLYEVYKDAT 235
A ++G ++L G ++R K P S I VIV A RKL P L+ + A
Sbjct: 240 AAMTMGIISLVSGTMYFRNKRPRRSIFTPIAHVIVAAILKRKLASPSDPRMLHGDHHVAN 299
Query: 236 ----IEKIVHTNQMRFLDKAAI-LQENSESQQPWKVCTVTQVEEVKILTRMLPILASTIV 290
+ HT + RFLDKA I +Q+ + + PW++CTVTQVE+VK L ++PI ASTIV
Sbjct: 300 DVVPSSTLPHTPRFRFLDKACIKIQDTNTKESPWRLCTVTQVEQVKTLISLVPIFASTIV 359
Query: 291 MNTCLAQLQTFSVQQGNSMNLKL-GSFTVPASSIPVIPLIFLCTLIPIYELFFVPFIRKI 349
NT LAQLQTFSVQQG+SMN +L SF +P +S+ IP I L L+P+Y+ F VPF RK+
Sbjct: 360 FNTILAQLQTFSVQQGSSMNTRLSNSFHIPPASLQAIPYIMLIFLVPLYDSFLVPFARKL 419
Query: 350 THHPSGVTQLQRVGVGLVLSAISMTIAGFIEVKRRDQGRKDPSKPISLFWLSFQYAIFGI 409
T H SG+ L R+G+GL LS SM A +E KRRD D + +S+FW++ Q+ IFGI
Sbjct: 420 TGHNSGIPPLTRIGIGLFLSTFSMVSAAMLEKKRRDSSVLD-GRILSIFWITPQFLIFGI 478
Query: 410 ADMFTLVGLLEFFYREAPSTMKSLSTSFTFLSMSLGYFLSTVFVNVINTVTKRITPSKQG 469
++MFT VGL+EFFY+++ M+S + T+ S S G++ S+V V+V+N +T SK G
Sbjct: 479 SEMFTAVGLIEFFYKQSAKGMESFLMALTYCSYSFGFYFSSVLVSVVNKITSTSVDSK-G 537
Query: 470 WLQGFDLNQNNLNLFYWFLAILSCLNFFNFLYWASWYKYKSEEDSNNSKELGETHL 525
WL DLN++ L+LFYW LA+LS LNF ++L+W+ W S NN+ +G+ ++
Sbjct: 538 WLGENDLNKDRLDLFYWLLAVLSLLNFLSYLFWSRWNIKSSRR--NNTNVVGDENI 591
>At1g33440 nitrate transporter NTL1, putative
Length = 596
Score = 416 bits (1068), Expect = e-116
Identities = 224/530 (42%), Positives = 325/530 (61%), Gaps = 25/530 (4%)
Query: 9 SLVLYFIGVMHFDLSSSANTLTNFMGSTFLLSLVGAFISDTYLNRFTTCLVFGSLEVMAL 68
+L+ Y MHF LS SAN +TNF+G+ FLLSL+G F+SD+YL F T LVFG +E+
Sbjct: 65 NLITYVFNEMHFPLSKSANLVTNFIGTVFLLSLLGGFLSDSYLGSFRTMLVFGVIEISVF 124
Query: 69 VMITVQAALDNLHPKACGKSS----CVE--GGIAVMFYTSLCLYALGMGGVRGSLTAFGA 122
QA L L P C S CVE G A YT+LCL ALG G ++ ++ + GA
Sbjct: 125 -----QAHLPELRPPECNMKSTTIHCVEANGYKAATLYTALCLVALGSGCLKPNIISHGA 179
Query: 123 NQFDEKDPNEAKALATFFNWLLLSSTLGSVIGVTGIVWVSTQKAWHWGFFIITVASSIGF 182
NQF KD + L++FFN + ++G +I +T +VWV T GF + + G
Sbjct: 180 NQFQRKD---LRKLSSFFNAAYFAFSMGQLIALTLLVWVQTHSGMDVGFGVSAAVMAAGM 236
Query: 183 VTLAIGKPFYRIKTPGESPILRIIQVIVVAFKNRKLQLPESNEQLYEVYKDAT-IEKIVH 241
++L G FYR K P S I QV V A RK P + +++ D ++ ++H
Sbjct: 237 ISLVAGTSFYRNKPPSGSIFTPIAQVFVAAITKRKQICPSNPNMVHQPSTDLVRVKPLLH 296
Query: 242 TNQMRFLDKAAILQENSESQQPWKVCTVTQVEEVKILTRMLPILASTIVMNTCLAQLQTF 301
+N+ RFLDKA I + + PW++CT+ QV +VKIL ++PI A TI+ NT LAQLQTF
Sbjct: 297 SNKFRFLDKACIKTQGKAMESPWRLCTIEQVHQVKILLSVIPIFACTIIFNTILAQLQTF 356
Query: 302 SVQQGNSMNLKL-GSFTVPASSIPVIPLIFLCTLIPIYELFFVPFIRKITHHPSGVTQLQ 360
SVQQG+SMN + +F +P +S+ IP I L +P+YE FFVP RK+T + SG++ LQ
Sbjct: 357 SVQQGSSMNTHITKTFQIPPASLQAIPYIILIFFVPLYETFFVPLARKLTGNDSGISPLQ 416
Query: 361 RVGVGLVLSAISMTIAGFIEVKRRDQGRKDPSKPISLFWLSFQYAIFGIADMFTLVGLLE 420
R+G GL L+ SM A +E KRR + + + +S+FW++ Q+ IFG+++MFT VGL+E
Sbjct: 417 RIGTGLFLATFSMVAAALVEKKRR-ESFLEQNVMLSIFWIAPQFLIFGLSEMFTAVGLVE 475
Query: 421 FFYREAPSTMKSLSTSFTFLSMSLGYFLSTVFVNVINTVT-KRITPSKQGWLQGFDLNQN 479
FFY+++ +M+S T+ T+ S S G++LS+V V+ +N VT + +K+GWL DLN++
Sbjct: 476 FFYKQSSQSMQSFLTAMTYCSYSFGFYLSSVLVSTVNRVTSSNGSGTKEGWLGDNDLNKD 535
Query: 480 NLNLFYWFLAILSCLNFFNFLYWASWYK-------YKSEEDSNNSKELGE 522
L+ FYW LA LS +NFFN+L+W+ WY + +E +S + E GE
Sbjct: 536 RLDHFYWLLASLSFINFFNYLFWSRWYSCDPSATHHSAEVNSLEALENGE 585
>At1g27040 nitrate transporter like protein
Length = 563
Score = 408 bits (1048), Expect = e-114
Identities = 206/525 (39%), Positives = 326/525 (61%), Gaps = 23/525 (4%)
Query: 1 MGFVTNMVSLVLYFIGVMHFDLSSSANTLTNFMGSTFLLSLVGAFISDTYLNRFTTCLVF 60
+ F+ N +LVLY MH L+ S++ +T FM + FLL+L+G F++D + + F L+
Sbjct: 42 LAFLANASNLVLYLKNFMHMSLARSSSEVTTFMATAFLLALLGGFLADAFFSTFVIFLIS 101
Query: 61 GSLEVMALVMITVQAALDNLHPKACGKSSC-----VEGGIAVMFYTSLCLYALGMGGVRG 115
S+E + L+++T+QA +L P C S+ V G A + L L +LG+GG++G
Sbjct: 102 ASIEFLGLILLTIQARRPSLMPPPCKSSAALRCEVVGGSKAAFLFVGLYLVSLGIGGIKG 161
Query: 116 SLTAFGANQFDEKDPNEAKALATFFNWLLLSSTLGSVIGVTGIVWVSTQKAWHWGFFIIT 175
SL + GA QFDE P K +TFFN+ + + G+++ VT +VW+ K W WGF + T
Sbjct: 162 SLPSHGAEQFDEGTPKGRKQRSTFFNYYVFCLSCGALVAVTFVVWIEDNKGWEWGFGVST 221
Query: 176 VASSIGFVTLAIGKPFYRIKTPGESPILRIIQVI----VVAFKNRKLQLPESNEQLYEVY 231
++ + + +G FY+ K P SP+ I +V+ +V+ ++ ++ ++ +
Sbjct: 222 ISIFLSILVFLLGSRFYKNKIPRGSPLTTIFKVLLAASIVSCSSKTSSNHFTSREVQSEH 281
Query: 232 KDATIEKIVHTNQMRFLDKAAILQENSESQQPWKVCTVTQVEEVKILTRMLPILASTIVM 291
++ T + + TN + L+KA ++ W CTV QVE+VKI+ +MLPI TI++
Sbjct: 282 EEKTPSQSL-TNSLTCLNKAI----EGKTHHIWLECTVQQVEDVKIVLKMLPIFGCTIML 336
Query: 292 NTCLAQLQTFSVQQGNSMNLKLGSFTVPASSIPVIPLIFLCTLIPIYELFFVPFIRKITH 351
N CLAQL T+SV Q +MN K+ +F VP++S+PV P++F+ L P Y+ +PF RK+T
Sbjct: 337 NCCLAQLSTYSVHQAATMNRKIVNFNVPSASLPVFPVVFMLILAPTYDHLIIPFARKVTK 396
Query: 352 HPSGVTQLQRVGVGLVLSAISMTIAGFIEVKRRDQGRK------DPSKPISLFWLSFQYA 405
G+T LQR+GVGLVLS ++M +A +E+KR+ R+ + + PI+ W++ QY
Sbjct: 397 SEIGITHLQRIGVGLVLSIVAMAVAALVELKRKQVAREAGLLDSEETLPITFLWIALQYL 456
Query: 406 IFGIADMFTLVGLLEFFYREAPSTMKSLSTSFTFLSMSLGYFLSTVFVNVINTVTKRITP 465
G AD+FTL GLLEFF+ EAPS+M+SL+TS ++ S++LGY+LS+V V ++N VTK +
Sbjct: 457 FLGSADLFTLAGLLEFFFTEAPSSMRSLATSLSWASLALGYYLSSVMVPIVNRVTK--SA 514
Query: 466 SKQGWLQGFDLNQNNLNLFYWFLAILSCLNFFNFLYWASWYKYKS 510
+ WL G LN+N L+LFYW + +LS +NF ++L+WA YKY S
Sbjct: 515 GQSPWL-GEKLNRNRLDLFYWLMCVLSVVNFLHYLFWAKRYKYIS 558
>At5g62730 nitrate transporter NTL1 - like protein
Length = 589
Score = 368 bits (944), Expect = e-102
Identities = 204/542 (37%), Positives = 302/542 (55%), Gaps = 58/542 (10%)
Query: 1 MGFVTNMVSLVLYFIGVMHFDLSSSANTLTNFMGSTFLLSLVGAFISDTYLNRFTTCLVF 60
+ F+ N +LVLY M F S +AN +T FMG+ F L+L+G F++D + F LV
Sbjct: 75 LAFLANASNLVLYLSTKMGFSPSGAANAVTAFMGTAFFLALLGGFLADAFFTTFHIYLVS 134
Query: 61 GSLEVMALVMITVQAALDNLHPKACGKSSCVEGGIAVMFYTSLCLYALGMGGVRGSLTAF 120
++E + L+++TVQA + P + V + L L ALG+GG++GSL
Sbjct: 135 AAIEFLGLMVLTVQAHEHSTEPWS-----------RVFLFVGLYLVALGVGGIKGSLPPH 183
Query: 121 GANQFDEKDPNEAKALATFFNWLLLSSTLGSVIGVTGIVWVSTQKAWHWGFFIITVASSI 180
GA QFDE+ + + + FFN+ + S + G++I VT +VW+ K W +GF + T A I
Sbjct: 184 GAEQFDEETSSGRRQRSFFFNYFIFSLSCGALIAVTVVVWLEDNKGWSYGFGVSTAAILI 243
Query: 181 GFVTLAIGKPFYRIKTPGESPILRIIQVIVVAFKNRKLQLPESNEQLYEVYKDATIEKIV 240
G YR+K P SPI + +V+ A LY YK +IV
Sbjct: 244 SVPVFLAGSRVYRLKVPSGSPITTLFKVLTAA--------------LYAKYKKRRTSRIV 289
Query: 241 HTNQMR-----------------FLDK--AAILQENSESQQPWKVCTVTQVEEVKILTRM 281
T R FL +++E +P + CT QV++VKI+ ++
Sbjct: 290 VTCHTRNDCDDSVTKQNCDGDDGFLGSFLGEVVRERESLPRPLR-CTEEQVKDVKIVIKI 348
Query: 282 LPILASTIVMNTCLAQLQTFSVQQGNSMNLKLGSFTVPASSIPVIPLIFLCTLIPIYELF 341
LPI STI++N CLAQL TFSVQQ ++MN KLGSFTVP +++PV P++F+ L P Y
Sbjct: 349 LPIFMSTIMLNCCLAQLSTFSVQQASTMNTKLGSFTVPPAALPVFPVVFMMILAPTYNHL 408
Query: 342 FVPFIRKITHHPSGVTQLQRVGVGLVLSAISMTIAGFIEVKRR-----------DQGRKD 390
+P RK T +G+T LQR+G GLVLS ++M +A +E KR+
Sbjct: 409 LLPLARKSTKTETGITHLQRIGTGLVLSIVAMAVAALVETKRKHVVVSCCSNNNSSSYSS 468
Query: 391 PSKPISLFWLSFQYAIFGIADMFTLVGLLEFFYREAPSTMKSLSTSFTFLSMSLGYFLST 450
PI+ W++ QY G AD+FTL G++EFF+ EAPSTM+SL+TS ++ S+++GY+ S+
Sbjct: 469 SPLPITFLWVAIQYVFLGSADLFTLAGMMEFFFTEAPSTMRSLATSLSWASLAMGYYFSS 528
Query: 451 VFVNVINTVTKRITPSKQGWLQGFDLNQNNLNLFYWFLAILSCLNFFNFLYWASWYKYKS 510
V V+ +N VT WL G +LNQ +L FYW + +LS +NF ++L+WAS Y Y+S
Sbjct: 529 VLVSAVNFVTG--LNHHNPWLLGENLNQYHLERFYWLMCVLSGINFLHYLFWASRYVYRS 586
Query: 511 EE 512
+
Sbjct: 587 NQ 588
>At3g54140 peptide transport - like protein
Length = 570
Score = 365 bits (938), Expect = e-101
Identities = 194/523 (37%), Positives = 312/523 (59%), Gaps = 21/523 (4%)
Query: 2 GFVTNMVSLVLYFIGVMHFDLSSSANTLTNFMGSTFLLSLVGAFISDTYLNRFTTCLVFG 61
G TN+V+ Y ++ +++AN +TN+ G+ ++ L+GAFI+D YL R+ T F
Sbjct: 47 GMGTNLVN---YLESRLNQGNATAANNVTNWSGTCYITPLIGAFIADAYLGRYWTIATFV 103
Query: 62 SLEVMALVMITVQAALDNLHPKACGKSSC-VEGGIAVMFYTSLCLYALGMGGVRGSLTAF 120
+ V + ++T+ A++ L P C +C +F+ +L + ALG GG++ +++F
Sbjct: 104 FIYVSGMTLLTLSASVPGLKPGNCNADTCHPNSSQTAVFFVALYMIALGTGGIKPCVSSF 163
Query: 121 GANQFDEKDPNEAKALATFFNWLLLSSTLGSVIGVTGIVWVSTQKAWHWGFFIITVASSI 180
GA+QFDE D NE ++FFNW S +G++I T +VW+ W WGF + TVA I
Sbjct: 164 GADQFDENDENEKIKKSSFFNWFYFSINVGALIAATVLVWIQMNVGWGWGFGVPTVAMVI 223
Query: 181 GFVTLAIGKPFYRIKTPGESPILRIIQVIVVAFKNRKLQLPESNEQLYEVYKDAT----I 236
G FYR++ PG SP+ RI QVIV AF+ +++PE L+E D +
Sbjct: 224 AVCFFFFGSRFYRLQRPGGSPLTRIFQVIVAAFRKISVKVPEDKSLLFETADDESNIKGS 283
Query: 237 EKIVHTNQMRFLDKAAILQENSESQ----QPWKVCTVTQVEEVKILTRMLPILASTIVMN 292
K+VHT+ ++F DKAA+ ++ + PW++C+VTQVEE+K + +LP+ A+ IV
Sbjct: 284 RKLVHTDNLKFFDKAAVESQSDSIKDGEVNPWRLCSVTQVEELKSIITLLPVWATGIVFA 343
Query: 293 TCLAQLQTFSVQQGNSMNLKLG-SFTVPASSIPVIPLIFLCTLIPIYELFFVPFIRKITH 351
T +Q+ T V QGN+M+ +G +F +P++S+ + + + P+Y+ F +P RK T
Sbjct: 344 TVYSQMSTMFVLQGNTMDQHMGKNFEIPSASLSLFDTVSVLFWTPVYDQFIIPLARKFTR 403
Query: 352 HPSGVTQLQRVGVGLVLSAISMTIAGFIEVKRRDQGRKDPSKP-----ISLFWLSFQYAI 406
+ G TQLQR+G+GLV+S +M AG +EV R D + + +S+FW QY +
Sbjct: 404 NERGFTQLQRMGIGLVVSIFAMITAGVLEVVRLDYVKTHNAYDQKQIHMSIFWQIPQYLL 463
Query: 407 FGIADMFTLVGLLEFFYREAPSTMKSLSTSFTFLSMSLGYFLSTVFVNVINTVTKRITPS 466
G A++FT +G LEFFY +AP M+SL ++ + +++LG +LSTV V V+ +TK+
Sbjct: 464 IGCAEVFTFIGQLEFFYDQAPDAMRSLCSALSLTTVALGNYLSTVLVTVVMKITKK--NG 521
Query: 467 KQGWLQGFDLNQNNLNLFYWFLAILSCLNFFNFLYWASWYKYK 509
K GW+ +LN+ +L+ F++ LA LS LNF +L+ + YKYK
Sbjct: 522 KPGWIPD-NLNRGHLDYFFYLLATLSFLNFLVYLWISKRYKYK 563
>At3g25280 nitrate transporter, putative
Length = 521
Score = 359 bits (922), Expect = 2e-99
Identities = 204/507 (40%), Positives = 303/507 (59%), Gaps = 30/507 (5%)
Query: 3 FVTNMVSLVLYFIGVMHFDLSSSANTLTNFMGSTFLLSLVGAFISDTYLNRFTTCLVFGS 62
F+ N + V YF+G MH+ +++AN +TNFMG++FLL+L G FI+D+++ FTT +VF
Sbjct: 42 FIANGFNFVKYFMGSMHYTPATAANMVTNFMGTSFLLTLFGGFIADSFVTHFTTFIVFCC 101
Query: 63 LEVMALVMITVQAALDNLHPKACGKSSCVEGGIAVMFYTSLCLYALGMGGVRGSLTAFGA 122
+E+M L+++T QA L P+ S ++ I +T L A+G GG++ SL + G
Sbjct: 102 IELMGLILLTFQAHNPKLLPEKDKTPSTLQSAI---LFTGLYAMAIGTGGLKASLPSHGG 158
Query: 123 NQFDEKDPNEAKALATFFNWLLLSSTLGSVIGVTGIVWVSTQKAWHWGFFIITVASSIGF 182
+Q D ++P + ++ FF+WL S G ++ VT ++W+ +K W W F I +
Sbjct: 159 DQIDRRNP---RLISRFFDWLYFSICSGCLLAVTVVLWIEEKKGWIWSFNISVGILATAL 215
Query: 183 VTLAIGKPFYRIKTPGESPILRIIQVIVVAFKNR-KLQLPESNEQ-LYEVYKDATIEKIV 240
+G PFYR K P SP+ +I VI+ A +NR K L E + L +YK+ +
Sbjct: 216 CIFTVGLPFYRFKRPNGSPLKKIAIVIISAARNRNKSDLDEEMMRGLISIYKNNS----- 270
Query: 241 HTNQMRFLDKAAILQENSESQQPWKVCTVTQVEEVKILTRMLPILASTIVMNTCLAQLQT 300
N+++++DKA + + SE T+VEE + +LPI STIVM+ C+AQL T
Sbjct: 271 -HNKLKWIDKATLNKNISE----------TEVEETRTFLGLLPIFGSTIVMSCCVAQLST 319
Query: 301 FSVQQGNSMNLKL-GSFTVPASSIPVIPLIFLCTLIPIYELFFVPFIRKITHHPSGVTQL 359
FS QQG MN KL SF +P S+ IPLIF+ IP+YE FF I ++ S L
Sbjct: 320 FSAQQGMLMNKKLFHSFEIPVPSLTAIPLIFMLLSIPLYE-FFGKKISSGNNNRSSSFNL 378
Query: 360 QRVGVGLVLSAISMTIAGFIEVKRRDQGRKDPSKPISLFWLSFQYAIFGIADMFTLVGLL 419
+R+G+GL LS++SM ++ +E KR+ + + + IS+ WL FQY + ++DM TL G+L
Sbjct: 379 KRIGLGLALSSVSMAVSAIVEAKRKHEVVHNNFR-ISVLWLVFQYLMLSVSDMLTLGGML 437
Query: 420 EFFYREAPSTMKSLSTSFTFLSMSLGYFLSTVFVNVINTVTKRITPSKQGWLQGFDLNQN 479
EFFYREAPS MKS+ST+ + S +LG+FLST V V N VT R+ WL G DLN+
Sbjct: 438 EFFYREAPSNMKSISTALGWCSTALGFFLSTTLVEVTNAVTGRL---GHQWLGGEDLNKT 494
Query: 480 NLNLFYWFLAILSCLNFFNFLYWASWY 506
L LFY L +L+ LN N+++WA Y
Sbjct: 495 RLELFYVLLCVLNTLNLLNYIFWAKRY 521
>At5g01180 oligopeptide transporter - like protein
Length = 570
Score = 353 bits (907), Expect = 1e-97
Identities = 189/523 (36%), Positives = 310/523 (59%), Gaps = 22/523 (4%)
Query: 2 GFVTNMVSLVLYFIGVMHFDLSSSANTLTNFMGSTFLLSLVGAFISDTYLNRFTTCLVFG 61
G TN+++ Y M+ + S++ +++N+ G+ + L+GAFI+D YL R+ T F
Sbjct: 48 GMSTNLIN---YLEKQMNMENVSASKSVSNWSGTCYATPLIGAFIADAYLGRYWTIASFV 104
Query: 62 SLEVMALVMITVQAALDNLHPKACGKSSCVEGGIAVMFYTSLCLYALGMGGVRGSLTAFG 121
+ + + ++T+ A++ L P G++ G + + +L L ALG GG++ +++FG
Sbjct: 105 VIYIAGMTLLTISASVPGLTPTCSGETCHATAGQTAITFIALYLIALGTGGIKPCVSSFG 164
Query: 122 ANQFDEKDPNEAKALATFFNWLLLSSTLGSVIGVTGIVWVSTQKAWHWGFFIITVASSIG 181
A+QFD+ D E ++ ++FFNW +G++I + +VW+ W WG + TVA +I
Sbjct: 165 ADQFDDTDEKEKESKSSFFNWFYFVINVGAMIASSVLVWIQMNVGWGWGLGVPTVAMAIA 224
Query: 182 FVTLAIGKPFYRIKTPGESPILRIIQVIVVAFKNRKLQLPESNEQLYEVYKDAT-----I 236
V G FYR++ PG SP+ R++QVIV + + K+++PE LYE +DA
Sbjct: 225 VVFFFAGSNFYRLQKPGGSPLTRMLQVIVASCRKSKVKIPEDESLLYE-NQDAESSIIGS 283
Query: 237 EKIVHTNQMRFLDKAAILQEN----SESQQPWKVCTVTQVEEVKILTRMLPILASTIVMN 292
K+ HT + F DKAA+ E+ + WK+CTVTQVEE+K L R+LPI A+ IV
Sbjct: 284 RKLEHTKILTFFDKAAVETESDNKGAAKSSSWKLCTVTQVEELKALIRLLPIWATGIVFA 343
Query: 293 TCLAQLQTFSVQQGNSMNLKLG-SFTVPASSIPVIPLIFLCTLIPIYELFFVPFIRKITH 351
+ +Q+ T V QGN+++ +G +F +P++S+ + + + P+Y+ VPF RK T
Sbjct: 344 SVYSQMGTVFVLQGNTLDQHMGPNFKIPSASLSLFDTLSVLFWAPVYDKLIVPFARKYTG 403
Query: 352 HPSGVTQLQRVGVGLVLSAISMTIAGFIEVKRRDQGR-----KDPSKPISLFWLSFQYAI 406
H G TQLQR+G+GLV+S SM AG +EV R + + + + P+++FW QY +
Sbjct: 404 HERGFTQLQRIGIGLVISIFSMVSAGILEVARLNYVQTHNLYNEETIPMTIFWQVPQYFL 463
Query: 407 FGIADMFTLVGLLEFFYREAPSTMKSLSTSFTFLSMSLGYFLSTVFVNVINTVTKRITPS 466
G A++FT +G LEFFY +AP M+SL ++ + +++ G +LST V ++ VT+ +
Sbjct: 464 VGCAEVFTFIGQLEFFYDQAPDAMRSLCSALSLTAIAFGNYLSTFLVTLVTKVTR--SGG 521
Query: 467 KQGWLQGFDLNQNNLNLFYWFLAILSCLNFFNFLYWASWYKYK 509
+ GW+ +LN +L+ F+W LA LS LNF +L+ A WY YK
Sbjct: 522 RPGWI-AKNLNNGHLDYFFWLLAGLSFLNFLVYLWIAKWYTYK 563
>At2g26690 putative nitrate transporter
Length = 586
Score = 348 bits (894), Expect = 4e-96
Identities = 196/520 (37%), Positives = 300/520 (57%), Gaps = 19/520 (3%)
Query: 8 VSLVLYFIGVMHFDLSSSANTLTNFMGSTFLLSLVGAFISDTYLNRFTTCLVFGSLEVMA 67
V+LV Y + MH S+SAN +T+FMG++FLL L+G F++D++L RF T +F +++ +
Sbjct: 59 VNLVTYLMETMHLPSSTSANIVTDFMGTSFLLCLLGGFLADSFLGRFKTIGIFSTIQALG 118
Query: 68 LVMITVQAALDNLHPKACGKS-SCVEGGIAVM--FYTSLCLYALGMGGVRGSLTAFGANQ 124
+ V L L P C +C+ M Y SL L ALG GG++ S++ FG++Q
Sbjct: 119 TGALAVATKLPELRPPTCHHGEACIPATAFQMTILYVSLYLIALGTGGLKSSISGFGSDQ 178
Query: 125 FDEKDPNEAKALATFFNWLLLSSTLGSVIGVTGIVWVSTQKAWHWGFFIITVASSIGFVT 184
FD+KDP E +A FFN ++G+++ VT +V++ + W + I TV+ +I V
Sbjct: 179 FDDKDPKEKAHMAFFFNRFFFFISMGTLLAVTVLVYMQDEVGRSWAYGICTVSMAIAIVI 238
Query: 185 LAIGKPFYRIKTPGESPILRIIQVIVVAFKNRKLQLPESNEQLYEVYKDATIEKIVHTNQ 244
G YR K SP+++I QVI AF+ RK++LP+S LYE + +I HT+Q
Sbjct: 239 FLCGTKRYRYKKSQGSPVVQIFQVIAAAFRKRKMELPQSIVYLYEDNPEGI--RIEHTDQ 296
Query: 245 MRFLDKAAILQENSESQ--------QPWKVCTVTQVEEVKILTRMLPILASTIVMNTCLA 296
LDKAAI+ E Q PWK+ +VT+VEEVK++ R+LPI A+TI+ T A
Sbjct: 297 FHLLDKAAIVAEGDFEQTLDGVAIPNPWKLSSVTKVEEVKMMVRLLPIWATTIIFWTTYA 356
Query: 297 QLQTFSVQQGNSMNLKLGSFTVPASSIPVIPLIFLCTLIPIYELFFVPFIRKITHHPSGV 356
Q+ TFSV+Q ++M +GSF +PA S+ V + + + +Y+ +PF +K P G
Sbjct: 357 QMITFSVEQASTMRRNIGSFKIPAGSLTVFFVAAILITLAVYDRAIMPFWKKWKGKP-GF 415
Query: 357 TQLQRVGVGLVLSAISMTIAGFIEVKRRDQGRKDPSK--PISLFWLSFQYAIFGIADMFT 414
+ LQR+ +GLVLS M A +E KR + K PIS+F L Q+ + G + F
Sbjct: 416 SSLQRIAIGLVLSTAGMAAAALVEQKRLSVAKSSSQKTLPISVFLLVPQFFLVGAGEAFI 475
Query: 415 LVGLLEFFYREAPSTMKSLSTSFTFLSMSLGYFLSTVFVNVINTVTKRITPSKQGWLQGF 474
G L+FF ++P MK++ST ++SLG+F+S+ V+++ VT T + GWL
Sbjct: 476 YTGQLDFFITQSPKGMKTMSTGLFLTTLSLGFFVSSFLVSIVKRVTS--TSTDVGWLAD- 532
Query: 475 DLNQNNLNLFYWFLAILSCLNFFNFLYWASWYKYKSEEDS 514
++N L+ FYW L ILS +NF ++ A W+K +DS
Sbjct: 533 NINHGRLDYFYWLLVILSGINFVVYIICALWFKPTKGKDS 572
>At1g62200 unknown protein (At1g62200)
Length = 590
Score = 348 bits (894), Expect = 4e-96
Identities = 191/500 (38%), Positives = 287/500 (57%), Gaps = 18/500 (3%)
Query: 9 SLVLYFIGVMHFDLSSSANTLTNFMGSTFLLSLVGAFISDTYLNRFTTCLVFGSLEVMAL 68
+L+ Y+ +H S+A+ + + G+ ++ L+GA I+D+Y R+ T F ++ + +
Sbjct: 79 NLITYYTSELHESNVSAASDVMIWQGTCYITPLIGAVIADSYWGRYWTIASFSAIYFIGM 138
Query: 69 VMITVQAALDNLHPKACGKSSCVEGGIAV-----MFYTSLCLYALGMGGVRGSLTAFGAN 123
++T+ A+L L P AC + A +F+T L L ALG GG++ +++FGA+
Sbjct: 139 ALLTLSASLPVLKPAACAGVAAALCSPATTVQYAVFFTGLYLIALGTGGIKPCVSSFGAD 198
Query: 124 QFDEKDPNEAKALATFFNWLLLSSTLGSVIGVTGIVWVSTQKAWHWGFFIITVASSIGFV 183
QFD+ DP E A+FFNW S +GS I T +VWV W GF I TV +
Sbjct: 199 QFDDTDPRERVRKASFFNWFYFSINIGSFISSTLLVWVQENVGWGLGFLIPTVFMGVSIA 258
Query: 184 TLAIGKPFYRIKTPGESPILRIIQVIVVAFKNRKLQLPESNEQLYEVYKD----ATIEKI 239
+ IG P YR + PG SPI R+ QV+V A++ KL LPE LYE + A KI
Sbjct: 259 SFFIGTPLYRFQKPGGSPITRVCQVLVAAYRKLKLNLPEDISFLYETREKNSMIAGSRKI 318
Query: 240 VHTNQMRFLDKAAILQE----NSESQQPWKVCTVTQVEEVKILTRMLPILASTIVMNTCL 295
HT+ +FLDKAA++ E + PWK+CTVTQVEEVK L RM PI AS IV +
Sbjct: 319 QHTDGYKFLDKAAVISEYESKSGAFSNPWKLCTVTQVEEVKTLIRMFPIWASGIVYSVLY 378
Query: 296 AQLQTFSVQQGNSMNLKLGSFTVPASSIPVIPLIFLCTLIPIYELFFVPFIRKITHHPSG 355
+Q+ T VQQG SMN + SF +P +S V + + IPIY+ F VPF+R+ T P G
Sbjct: 379 SQISTLFVQQGRSMNRIIRSFEIPPASFGVFDTLIVLISIPIYDRFLVPFVRRFTGIPKG 438
Query: 356 VTQLQRVGVGLVLSAISMTIAGFIEVKRRDQGRKDPSKPISLFWLSFQYAIFGIADMFTL 415
+T LQR+G+GL LS +S+ A +E R + + +S+FW QY + GIA++F
Sbjct: 439 LTDLQRMGIGLFLSVLSIAAAAIVETVRLQLAQDFVA--MSIFWQIPQYILMGIAEVFFF 496
Query: 416 VGLLEFFYREAPSTMKSLSTSFTFLSMSLGYFLSTVFVNVINTVTKRITPSKQGWLQGFD 475
+G +EFFY E+P M+S+ ++ L+ ++G +LS++ + ++ T K GW+ D
Sbjct: 497 IGRVEFFYDESPDAMRSVCSALALLNTAVGSYLSSLILTLVAYFT--ALGGKDGWVPD-D 553
Query: 476 LNQNNLNLFYWFLAILSCLN 495
LN+ +L+ F+W L L +N
Sbjct: 554 LNKGHLDYFFWLLVSLGLVN 573
>At1g68570 peptide transporter like
Length = 596
Score = 342 bits (878), Expect = 3e-94
Identities = 209/547 (38%), Positives = 302/547 (55%), Gaps = 35/547 (6%)
Query: 1 MGFVTNMVSLVLYFIGVMHFDLSSSANTLTNFMGSTFLLSLVGAFISDTYLNRFTTCLVF 60
+GF NM+S Y +H L+ +ANTLTNF G++ L L+GAFI+D++ RF T
Sbjct: 45 VGFHANMIS---YLTTQLHLPLTKAANTLTNFAGTSSLTPLLGAFIADSFAGRFWTITFA 101
Query: 61 GSLEVMALVMITVQAALDNLHPKAC-GKSSCVEGGIAVM--FYTSLCLYALGMGGVRGSL 117
+ + + ++T+ A + L P C G+ CV A + Y +L L ALG GG+R +
Sbjct: 102 SIIYQIGMTLLTISAIIPTLRPPPCKGEEVCVVADTAQLSILYVALLLGALGSGGIRPCV 161
Query: 118 TAFGANQFDEKDPNEAKALATFFNWLLLSSTLGSVIGVTGIVWVSTQKAWHWGFFIITVA 177
AFGA+QFDE DPN+ +FNW ++ VT +VW+ W G I TVA
Sbjct: 162 VAFGADQFDESDPNQTTKTWNYFNWYYFCMGAAVLLAVTVLVWIQDNVGWGLGLGIPTVA 221
Query: 178 SSIGFVTLAIGKPFYRIKTPGESPILRIIQVIVVAFKNRKLQLPESNEQLY---EVYKDA 234
+ + G YR P SP R+IQV V AF+ RKL++ LY E+ DA
Sbjct: 222 MFLSVIAFVGGFQLYRHLVPAGSPFTRLIQVGVAAFRKRKLRMVSDPSLLYFNDEI--DA 279
Query: 235 TIE---KIVHTNQMRFLDKAAILQENSE---SQQP--WKVCTVTQVEEVKILTRMLPILA 286
I K+ HT M FLDKAAI+ E Q P W++ TV +VEE+K + RM PI A
Sbjct: 280 PISLGGKLTHTKHMSFLDKAAIVTEEDNLKPGQIPNHWRLSTVHRVEELKSVIRMGPIGA 339
Query: 287 STIVMNTCLAQLQTFSVQQGNSMNLKL-GSFTVPASSIPVIPLIFLCTLIPIYELFFVPF 345
S I++ T AQ TFS+QQ +MN L SF +PA S+ V + + T I Y+ FV
Sbjct: 340 SGILLITAYAQQGTFSLQQAKTMNRHLTNSFQIPAGSMSVFTTVAMLTTIIFYDRVFVKV 399
Query: 346 IRKITHHPSGVTQLQRVGVGLVLSAISMTIAGFIEVKRR----DQGRKDPSK---PISLF 398
RK T G+T L R+G+G V+S I+ +AGF+EVKR+ + G D PIS
Sbjct: 400 ARKFTGLERGITFLHRMGIGFVISIIATLVAGFVEVKRKSVAIEHGLLDKPHTIVPISFL 459
Query: 399 WLSFQYAIFGIADMFTLVGLLEFFYREAPSTMKSLSTSFTFLSMSLGYFLSTVFVNVINT 458
WL QY + G+A+ F +G LEFFY +AP +M+S +T+ ++++S+G ++ST+ V +++
Sbjct: 460 WLIPQYGLHGVAEAFMSIGHLEFFYDQAPESMRSTATALFWMAISIGNYVSTLLVTLVHK 519
Query: 459 VTKRITPSKQGWLQGFDLNQNNLNLFYWFLAILSCLNFFNFLYWASWYKYK------SEE 512
+ + P WL +LN+ L FYW + +L +N +L+ A Y YK S+E
Sbjct: 520 FSAK--PDGSNWLPDNNLNRGRLEYFYWLITVLQAVNLVYYLWCAKIYTYKPVQVHHSKE 577
Query: 513 DSNNSKE 519
DS+ KE
Sbjct: 578 DSSPVKE 584
>At5g13400 peptide transporter - like protein
Length = 624
Score = 340 bits (872), Expect = 1e-93
Identities = 206/542 (38%), Positives = 302/542 (55%), Gaps = 32/542 (5%)
Query: 2 GFVTNMVSLVLYFIGVMHFDLSSSANTLTNFMGSTFLLSLVGAFISDTYLNRFTTCLVFG 61
G NMV+ + Y VMH SS+N + NF+G + S++G F++D YL R+ T +F
Sbjct: 87 GLSVNMVAFMFY---VMHRPFESSSNAVNNFLGISQASSVLGGFLADAYLGRYWTIAIFT 143
Query: 62 SLEVMALVMITVQAALDNLHPKA--CGKSS-----CVEGGIAVMFYTSLCLYALGMG--G 112
++ ++ L+ IT+ A+L P CG+ S C E M Y LY G G G
Sbjct: 144 TMYLVGLIGITLGASLKMFVPDQSNCGQLSLLLGNCEEAKSWQMLYLYTVLYITGFGAAG 203
Query: 113 VRGSLTAFGANQFDEKDPNEAKALATFFNWLLLSSTLGSVIGVTGIVWVSTQKAWHWGFF 172
+R +++FGA+QFDEK + L FFN+ LS TLG++I T +V+V + W F
Sbjct: 204 IRPCVSSFGADQFDEKSKDYKTHLDRFFNFFYLSVTLGAIIAFTLVVYVQMELGWGMAFG 263
Query: 173 IITVASSIGFVTLAIGKPFYRIKTPGESPILRIIQVIVVAFKNRKLQLPESNE-QLYEV- 230
+ VA I G P YR + PG SP+ R+ QV+V AF+ R S LYEV
Sbjct: 264 TLAVAMGISNALFFAGTPLYRHRLPGGSPLTRVAQVLVAAFRKRNAAFTSSEFIGLYEVP 323
Query: 231 -YKDAT--IEKIVHTNQMRFLDKAAI-LQENSESQQPWKVCTVTQVEEVKILTRMLPILA 286
K A KI H+N +LDKAA+ L+E+ PWK+CTVTQVEEVKIL R++PI
Sbjct: 324 GLKSAINGSRKIPHSNDFIWLDKAALELKEDGLEPSPWKLCTVTQVEEVKILIRLIPIPT 383
Query: 287 STIVMNTCLAQLQTFSVQQGNSMNLKLGSFTVPASSIPVIPLIFLCTLIPIYELFFVPFI 346
TI+++ L + T SVQQ ++N + +P + +PV P + + ++ +Y FVP
Sbjct: 384 CTIMLSLVLTEYLTLSVQQAYTLNTHIQHLKLPVTCMPVFPGLSIFLILSLYYSVFVPIT 443
Query: 347 RKITHHPSGVTQLQRVGVGLVLSAISMTIAGFIEVKRRDQGRKD--------PSKPISLF 398
R+IT +P G +QLQRVG+GL +S IS+ AG E RR ++ ++ +
Sbjct: 444 RRITGNPHGASQLQRVGIGLAVSIISVAWAGLFENYRRHYAIQNGFEFNFLTQMPDLTAY 503
Query: 399 WLSFQYAIFGIADMFTLVGLLEFFYREAPSTMKSLSTSFTFLSMSLGYFLSTVFVNVINT 458
WL QY + GIA++F +VGLLEF Y EAP MKS+ +++ L+ LG F +T+ N++
Sbjct: 504 WLLIQYCLIGIAEVFCIVGLLEFLYEEAPDAMKSIGSAYAALAGGLGCFAATILNNIVKA 563
Query: 459 VTKRITPSKQGWLQGFDLNQNNLNLFYWFLAILSCLNFFNFLYWASWYKYK---SEEDSN 515
T+ + WL ++N + YW L +LS LNF FL+ A YKY+ SEED +
Sbjct: 564 ATR--DSDGKSWLSQ-NINTGRFDCLYWLLTLLSFLNFCVFLWSAHRYKYRAIESEEDKS 620
Query: 516 NS 517
++
Sbjct: 621 SA 622
>At3g21670 nitrate transporter
Length = 590
Score = 339 bits (869), Expect = 3e-93
Identities = 191/518 (36%), Positives = 291/518 (55%), Gaps = 26/518 (5%)
Query: 8 VSLVLYFIGVMHFDLSSSANTLTNFMGSTFLLSLVGAFISDTYLNRFTTCLVFGSLEVMA 67
++LV Y +G +H + SA +TNFMG+ LL L+G F++D L R+ + S+ +
Sbjct: 55 MNLVTYLVGDLHISSAKSATIVTNFMGTLNLLGLLGGFLADAKLGRYKMVAISASVTALG 114
Query: 68 LVMITVQAALDNLHPKACGK-----SSCVE--GGIAVMFYTSLCLYALGMGGVRGSLTAF 120
++++TV + ++ P C C+E G + Y +L ALG GG++ +++ F
Sbjct: 115 VLLLTVATTISSMRPPICDDFRRLHHQCIEANGHQLALLYVALYTIALGGGGIKSNVSGF 174
Query: 121 GANQFDEKDPNEAKALATFFNWLLLSSTLGSVIGVTGIVWVSTQKAWHWGFFIITVASSI 180
G++QFD DP E K + FFN S ++GS+ V +V+V WG+ I +
Sbjct: 175 GSDQFDTSDPKEEKQMIFFFNRFYFSISVGSLFAVIALVYVQDNVGRGWGYGISAATMVV 234
Query: 181 GFVTLAIGKPFYRIKTPGESPILRIIQVIVVAFKNRKLQLPESNEQLYEVYKDATIEKIV 240
+ L G YR K P SP I +V +A+K RK P ++ L Y + T+
Sbjct: 235 AAIVLLCGTKRYRFKKPKGSPFTTIWRVGFLAWKKRKESYP-AHPSLLNGYDNTTVP--- 290
Query: 241 HTNQMRFLDKAAILQENS-------ESQQPWKVCTVTQVEEVKILTRMLPILASTIVMNT 293
HT ++ LDKAAI + S E + PW V TVTQVEEVK++ +++PI A+ I+ T
Sbjct: 291 HTEMLKCLDKAAISKNESSPSSKDFEEKDPWIVSTVTQVEEVKLVMKLVPIWATNILFWT 350
Query: 294 CLAQLQTFSVQQGNSMNLKLGSFTVPASSIPVIPLIFLCTLIPIYELFFVPFIRKITHHP 353
+Q+ TF+V+Q M+ KLGSFTVPA S ++ + + E FVP R++T P
Sbjct: 351 IYSQMTTFTVEQATFMDRKLGSFTVPAGSYSAFLILTILLFTSLNERVFVPLTRRLTKKP 410
Query: 354 SGVTQLQRVGVGLVLSAISMTIAGFIEVKRRDQGRKDPSKPISLFWLSFQYAIFGIADMF 413
G+T LQR+GVGLV S +M +A IE RR+ + K IS FWL QY + G + F
Sbjct: 411 QGITSLQRIGVGLVFSMAAMAVAAVIENARREAAVNN-DKKISAFWLVPQYFLVGAGEAF 469
Query: 414 TLVGLLEFFYREAPSTMKSLSTSFTFLSMSLGYFLSTVFVNVINTVTKRITPSKQGWLQG 473
VG LEFF REAP MKS+ST ++S+G+F+S++ V++++ VT + WL+
Sbjct: 470 AYVGQLEFFIREAPERMKSMSTGLFLSTISMGFFVSSLLVSLVDRVTDK------SWLRS 523
Query: 474 FDLNQNNLNLFYWFLAILSCLNFFNFLYWASWYKYKSE 511
+LN+ LN FYW L +L LNF F+ +A ++YK++
Sbjct: 524 -NLNKARLNYFYWLLVVLGALNFLIFIVFAMKHQYKAD 560
>At2g37900 putative peptide/amino acid transporter
Length = 575
Score = 339 bits (869), Expect = 3e-93
Identities = 189/521 (36%), Positives = 294/521 (56%), Gaps = 18/521 (3%)
Query: 2 GFVTNMVSLVLYFIGVMHFDLSSSANTLTNFMGSTFLLSLVGAFISDTYLNRFTTCLVFG 61
G TN LV+Y +++ DL + + + G T L+ L+G FI+D YL R+ T LV
Sbjct: 61 GLATN---LVVYLTTILNQDLKMAIRNVNYWSGVTTLMPLLGGFIADAYLGRYATVLVAT 117
Query: 62 SLEVMALVMITVQAALDNLHPKACGKSSCVEGGIA--VMFYTSLCLYALGMGGVRGSLTA 119
++ +M LV++T+ + L P C + CVE A V F+ ++ L ++G GG + SL +
Sbjct: 118 TIYLMGLVLLTMSWFIPGLKP--CHQEVCVEPRKAHEVAFFIAIYLISIGTGGHKPSLES 175
Query: 120 FGANQFDEKDPNEAKALATFFNWLLLSSTLGSVIGVTGIVWVSTQKAWHWGFFIITVASS 179
FGA+QFD+ E K +FFNW +S G + VT + ++ + W I+TV +
Sbjct: 176 FGADQFDDDHVEERKMKMSFFNWWNVSLCAGILTAVTAVAYIEDRVGWGVAGIILTVVMA 235
Query: 180 IGFVTLAIGKPFYRIKTPGESPILRIIQVIVVAFKNRKLQLPESNEQLYEVYKD--ATIE 237
I + IGKPFYR +TP SP+ I+QV V A R L P L+EV K +
Sbjct: 236 ISLIIFFIGKPFYRYRTPSGSPLTPILQVFVAAIAKRNLPYPSDPSLLHEVSKTEFTSGR 295
Query: 238 KIVHTNQMRFLDKAAILQENS----ESQQPWKVCTVTQVEEVKILTRMLPILASTIVMNT 293
+ HT ++FLDKAAI+++ + E Q PW++ T+T+VEE K++ ++PI ST+
Sbjct: 296 LLCHTEHLKFLDKAAIIEDKNPLALEKQSPWRLLTLTKVEETKLIINVIPIWFSTLAFGI 355
Query: 294 CLAQLQTFSVQQGNSMNLKLGSFTVPASSIPVIPLIFLCTLIPIYELFFVPFIRKITHHP 353
C Q TF ++Q +M+ +G FTVP +S+ + + L + +YE VP +R IT +
Sbjct: 356 CATQASTFFIKQAITMDRHIGGFTVPPASMFTLTALTLIISLTVYEKLLVPLLRSITRNQ 415
Query: 354 SGVTQLQRVGVGLVLSAISMTIAGFIEVKRRDQGRKDPSKPISLFWLSFQYAIFGIADMF 413
G+ LQR+G G++ S I+M IA +E +R D R + +KP+S+ WL+ Q+ + G AD F
Sbjct: 416 RGINILQRIGTGMIFSLITMIIAALVEKQRLD--RTNNNKPMSVIWLAPQFMVIGFADAF 473
Query: 414 TLVGLLEFFYREAPSTMKSLSTSFTFLSMSLGYFLSTVFVNVINTVTKRITPSKQGWLQG 473
TLVGL E+FY + P +M+SL +F + FL+ + + ++T+ + S + W G
Sbjct: 474 TLVGLQEYFYHQVPDSMRSLGIAFYLSVIGAASFLNNLLITAVDTLAENF--SGKSWF-G 530
Query: 474 FDLNQNNLNLFYWFLAILSCLNFFNFLYWASWYKYKSEEDS 514
DLN + L+ FYWFLA + N F+ A YKS + S
Sbjct: 531 KDLNSSRLDRFYWFLAGVIAANICVFVIVAKRCPYKSVQPS 571
>At3g25260 nitrate transporter, putative
Length = 515
Score = 338 bits (867), Expect = 5e-93
Identities = 186/507 (36%), Positives = 296/507 (57%), Gaps = 32/507 (6%)
Query: 1 MGFVTNMVSLVLYFIGVMHFDLSSSANTLTNFMGSTFLLSLVGAFISDTYLNRFTTCLVF 60
M F+ + ++YF M++ +A +TNF+G++FLL++ G F++D++L RF ++F
Sbjct: 40 MVFLACSTNFMMYFTKSMNYSTPKAATMVTNFVGTSFLLTIFGGFVADSFLTRFAAFVLF 99
Query: 61 GSLEVMALVMITVQAALDNLHPKACGKSSCVEGGIAVMFYTSLCLYALGMGGVRGSLTAF 120
GS+E++ L+M+T+QA + L P+ K S + + + +T L A+G+GGV+GSL A
Sbjct: 100 GSIELLGLIMLTLQAHITKLQPQGGKKPSTPQ---STVLFTGLYAIAIGVGGVKGSLPAH 156
Query: 121 GANQFDEKDPNEAKALATFFNWLLLSSTLGSVIGVTGIVWVSTQKAWHWGFFIITVASSI 180
G +Q ++ + ++ FFNW S LG + VT +VW+ W F I T +
Sbjct: 157 GGDQIGTRNQ---RLISGFFNWYFFSVCLGGFLAVTLMVWIEENIGWSSSFTISTAVLAS 213
Query: 181 GFVTLAIGKPFYRIKTPGESPILRIIQVIVVAFKNRKLQLPESNEQLYEVYKDATIEKIV 240
G P YR K P SP+ RI+ V V A +NR + ++ + +K +
Sbjct: 214 AIFVFVAGCPMYRFKRPAGSPLTRIVNVFVSAARNRNRFVTDAE---VVTQNHNSTDKSI 270
Query: 241 HTNQMRFLDKAAILQENSESQQPWKVCTVTQVEEVKILTRMLPILASTIVMNTCLAQLQT 300
H N+ +FL+KA + + S TQVEE + +LPI STI+MN C+AQ+ T
Sbjct: 271 HHNKFKFLNKAKLNNKIS----------ATQVEETRTFLALLPIFGSTIIMNCCVAQMGT 320
Query: 301 FSVQQGNSMNLKLG-SFTVPASSIPVIPLIFLCTLIPIYELFFVPFIRKITHHPSGVTQL 359
FSVQQG N KL SF +P +S+ IPL+ + + + +YELF + S L
Sbjct: 321 FSVQQGMVTNRKLSRSFEIPVASLNAIPLLCMLSSLALYELFGKRILSNSERSSS--FNL 378
Query: 360 QRVGVGLVLSAISMTIAGFIEVKRRDQGRKDPSKPISLFWLSFQYAIFGIADMFTLVGLL 419
+R+G GL L++ISM +A +EVKR+ + + K IS+FWL Q+ + ++DM T+ G+L
Sbjct: 379 KRIGYGLALTSISMAVAAIVEVKRKHEAVHNNIK-ISVFWLELQFVMLSLSDMLTVGGML 437
Query: 420 EFFYREAPSTMKSLSTSFTFLSMSLGYFLSTVFVNVINTVTKRITPSKQGWLQGFDLNQN 479
EFF+RE+P++M+S+ST+ + S ++G+FLS+V V V+N +T GWL+ DLN++
Sbjct: 438 EFFFRESPASMRSMSTALGWCSTAMGFFLSSVLVEVVNGIT--------GWLRD-DLNES 488
Query: 480 NLNLFYWFLAILSCLNFFNFLYWASWY 506
L LFY L +L+ LN FN+++WA Y
Sbjct: 489 RLELFYLVLCVLNTLNLFNYIFWAKRY 515
>At2g02040 histidine transport protein (PTR2-B)
Length = 585
Score = 338 bits (867), Expect = 5e-93
Identities = 185/517 (35%), Positives = 292/517 (55%), Gaps = 19/517 (3%)
Query: 9 SLVLYFIGVMHFDLSSSANTLTNFMGSTFLLSLVGAFISDTYLNRFTTCLVFGSLEVMAL 68
+L+ Y +H S+A +T + G+ +L L+GA ++D Y R+ T F + + +
Sbjct: 68 NLITYLTTKLHQGNVSAATNVTTWQGTCYLTPLIGAVLADAYWGRYWTIACFSGIYFIGM 127
Query: 69 VMITVQAALDNLHPKACGKSSCVEGGIA--VMFYTSLCLYALGMGGVRGSLTAFGANQFD 126
+T+ A++ L P C C A MF+ L L ALG GG++ +++FGA+QFD
Sbjct: 128 SALTLSASVPALKPAECIGDFCPSATPAQYAMFFGGLYLIALGTGGIKPCVSSFGADQFD 187
Query: 127 EKDPNEAKALATFFNWLLLSSTLGSVIGVTGIVWVSTQKAWHWGFFIITVASSIGFVTLA 186
+ D E A+FFNW S +G+++ + +VW+ + W GF I TV + +
Sbjct: 188 DTDSRERVRKASFFNWFYFSINIGALVSSSLLVWIQENRGWGLGFGIPTVFMGLAIASFF 247
Query: 187 IGKPFYRIKTPGESPILRIIQVIVVAFKNRKLQLPESNEQLYEVY-KDATI---EKIVHT 242
G P YR + PG SPI RI QV+V +F+ +++PE LYE K++ I KI HT
Sbjct: 248 FGTPLYRFQKPGGSPITRISQVVVASFRKSSVKVPEDATLLYETQDKNSAIAGSRKIEHT 307
Query: 243 NQMRFLDKAAILQE----NSESQQPWKVCTVTQVEEVKILTRMLPILASTIVMNTCLAQL 298
+ ++LDKAA++ E + + W++CTVTQVEE+KIL RM PI AS I+ + AQ+
Sbjct: 308 DDCQYLDKAAVISEEESKSGDYSNSWRLCTVTQVEELKILIRMFPIWASGIIFSAVYAQM 367
Query: 299 QTFSVQQGNSMNLKLGSFTVPASSIPVIPLIFLCTLIPIYELFFVPFIRKITHHPSGVTQ 358
T VQQG +MN K+GSF +P +++ + +P+Y+ F VP RK T G T+
Sbjct: 368 STMFVQQGRAMNCKIGSFQLPPAALGTFDTASVIIWVPLYDRFIVPLARKFTGVDKGFTE 427
Query: 359 LQRVGVGLVLSAISMTIAGFIEVKR----RDQGRKDPSK--PISLFWLSFQYAIFGIADM 412
+QR+G+GL +S + M A +E+ R D G + PIS+ W QY I G A++
Sbjct: 428 IQRMGIGLFVSVLCMAAAAIVEIIRLHMANDLGLVESGAPVPISVLWQIPQYFILGAAEV 487
Query: 413 FTLVGLLEFFYREAPSTMKSLSTSFTFLSMSLGYFLSTVFVNVINTVTKRITPSKQGWLQ 472
F +G LEFFY ++P M+SL ++ L+ +LG +LS++ + ++ T R ++GW+
Sbjct: 488 FYFIGQLEFFYDQSPDAMRSLCSALALLTNALGNYLSSLILTLVTYFTTR--NGQEGWIS 545
Query: 473 GFDLNQNNLNLFYWFLAILSCLNFFNFLYWASWYKYK 509
+LN +L+ F+W LA LS +N + + A+ YK K
Sbjct: 546 D-NLNSGHLDYFFWLLAGLSLVNMAVYFFSAARYKQK 581
>At2g40460 putative PTR2 family peptide transporter
Length = 583
Score = 328 bits (841), Expect = 5e-90
Identities = 190/530 (35%), Positives = 307/530 (57%), Gaps = 28/530 (5%)
Query: 1 MGFVTNMVSLVLYFIGVMHFDLSSSANTLTNFMGSTFLLSLVGAFISDTYLNRFTTCLVF 60
M F +LV Y +H D SS + N+ G+ ++ + GA+I+D+Y+ RF T
Sbjct: 43 MAFYGIASNLVNYLTKRLHEDTISSVRNVNNWSGAVWITPIAGAYIADSYIGRFWTFTAS 102
Query: 61 GSLEVMALVMITVQAALDNLHPKA----CGKSSCVEGGIAVMFYTSLCLYALGMGGVRGS 116
+ V+ ++++T+ + +L P C K+S ++ FY SL A+G GG + +
Sbjct: 103 SLIYVLGMILLTMAVTVKSLRPTCENGVCNKASSLQ---VTFFYISLYTIAIGAGGTKPN 159
Query: 117 LTAFGANQFDEKDPNEAKALATFFNWLLLSSTLGSVIGVTGIVWVSTQKAWHWGFFIITV 176
++ FGA+QFD E K +FFNW + SS LG++ G+V++ W G+ I TV
Sbjct: 160 ISTFGADQFDSYSIEEKKQKVSFFNWWMFSSFLGALFATLGLVYIQENLGWGLGYGIPTV 219
Query: 177 ASSIGFVTLAIGKPFYRIKTPGESPILR-IIQVIVVAFKNRKLQLPESNEQLYEV----Y 231
+ V IG PFYR K + + ++QV + AFKNRKLQ P+ + +LYE+ Y
Sbjct: 220 GLLVSLVVFYIGTPFYRHKVIKTDNLAKDLVQVPIAAFKNRKLQCPDDHLELYELDSHYY 279
Query: 232 KDATIEKIVHTNQMRFLDKAAILQENSESQQPWKVCTVTQVEEVKILTRMLPILASTIVM 291
K ++ HT RFLDKAAI + S+ P CTVT+VE K + ++ I T++
Sbjct: 280 KSNGKHQVHHTPVFRFLDKAAI---KTSSRVP---CTVTKVEVAKRVLGLIFIWLVTLIP 333
Query: 292 NTCLAQLQTFSVQQGNSMNLKLGS-FTVPASSIPVIPLIFLCTLIPIYELFFVPFIRKIT 350
+T AQ+ T V+QG +++ K+GS F +PA+S+ + + +P+Y+ FVPF+RK T
Sbjct: 334 STLWAQVNTLFVKQGTTLDRKIGSNFQIPAASLGSFVTLSMLLSVPMYDQSFVPFMRKKT 393
Query: 351 HHPSGVTQLQRVGVGLVLSAISMTIAGFIEVKR----RDQGRKDPSK--PISLFWLSFQY 404
+P G+T LQR+GVG + +++ IA +EVKR ++ P++ P+S+FWL QY
Sbjct: 394 GNPRGITLLQRLGVGFAIQIVAIAIASAVEVKRMRVIKEFHITSPTQVVPMSIFWLLPQY 453
Query: 405 AIFGIADMFTLVGLLEFFYREAPSTMKSLSTSFTFLSMSLGYFLSTVFVNVINTVTKRIT 464
++ GI D+F +GLLEFFY ++P M+SL T+F + LG FL++ V +I+ +T +
Sbjct: 454 SLLGIGDVFNAIGLLEFFYDQSPEEMQSLGTTFFTSGIGLGNFLNSFLVTMIDKITSK-- 511
Query: 465 PSKQGWLQGFDLNQNNLNLFYWFLAILSCLNFFNFLYWASWYKYKSEEDS 514
+ W+ G +LN + L+ +Y FL ++S +N F++ AS Y YKS++D+
Sbjct: 512 GGGKSWI-GNNLNDSRLDYYYGFLVVISIVNMGLFVWAASKYVYKSDDDT 560
>At1g12110 nitrate/chlorate transporter CHL1
Length = 590
Score = 328 bits (840), Expect = 6e-90
Identities = 191/542 (35%), Positives = 294/542 (54%), Gaps = 25/542 (4%)
Query: 8 VSLVLYFIGVMHFDLSSSANTLTNFMGSTFLLSLVGAFISDTYLNRFTTCLVFGSLEVMA 67
V+LV Y G MH +++ANT+TNF+G++F+L L+G FI+DT+L R+ T +F +++
Sbjct: 53 VNLVTYLTGTMHLGNATAANTVTNFLGTSFMLCLLGGFIADTFLGRYLTIAIFAAIQATG 112
Query: 68 LVMITVQAALDNLHPKACG---KSSCVEG-GIAV-MFYTSLCLYALGMGGVRGSLTAFGA 122
+ ++T+ + L P C S C + GI + + Y +L L ALG GGV+ S++ FG+
Sbjct: 113 VSILTLSTIIPGLRPPRCNPTTSSHCEQASGIQLTVLYLALYLTALGTGGVKASVSGFGS 172
Query: 123 NQFDEKDPNEAKALATFFNWLLLSSTLGSVIGVTGIVWVSTQKAWHWGFFIITVASSIGF 182
+QFDE +P E + FFN +GS++ VT +V+V WG+ I A +
Sbjct: 173 DQFDETEPKERSKMTYFFNRFFFCINVGSLLAVTVLVYVQDDVGRKWGYGICAFAIVLAL 232
Query: 183 VTLAIGKPFYRIKTPGESPILRIIQVIVVAFKNRKLQLPESNEQLYEVYKDATIE----- 237
G YR K SP+ ++ VIV A++NRKL+LP LY+V E
Sbjct: 233 SVFLAGTNRYRFKKLIGSPMTQVAAVIVAAWRNRKLELPADPSYLYDVDDIIAAEGSMKG 292
Query: 238 --KIVHTNQMRFLDKAAILQE----NSESQQPWKVCTVTQVEEVKILTRMLPILASTIVM 291
K+ HT Q R LDKAAI + S W + T+T VEEVK + RMLPI A+ I+
Sbjct: 293 KQKLPHTEQFRSLDKAAIRDQEAGVTSNVFNKWTLSTLTDVEEVKQIVRMLPIWATCILF 352
Query: 292 NTCLAQLQTFSVQQGNSMNLKLGSFTVPASSIPVIPLIFLCTLIPIYELFFVPFIRKITH 351
T AQL T SV Q +++ +GSF +P +S+ V + L +Y+ + +K+ +
Sbjct: 353 WTVHAQLTTLSVAQSETLDRSIGSFEIPPASMAVFYVGGLLLTTAVYDRVAIRLCKKLFN 412
Query: 352 HPSGVTQLQRVGVGLVLSAISMTIAGFIEVKR----RDQGRKDPSKPISLFWLSFQYAIF 407
+P G+ LQR+G+GL +++M +A +E+KR G + P+ + L QY I
Sbjct: 413 YPHGLRPLQRIGLGLFFGSMAMAVAALVELKRLRTAHAHGPTVKTLPLGFYLLIPQYLIV 472
Query: 408 GIADMFTLVGLLEFFYREAPSTMKSLSTSFTFLSMSLGYFLSTVFVNVINTVTKRITPSK 467
GI + G L+FF RE P MK +ST +++LG+F S+V V ++ T + P
Sbjct: 473 GIGEALIYTGQLDFFLRECPKGMKGMSTGLLLSTLALGFFFSSVLVTIVEKFTGKAHP-- 530
Query: 468 QGWLQGFDLNQNNLNLFYWFLAILSCLNFFNFLYWASWYKYKSEEDSNNSKELGETHLLM 527
W+ DLN+ L FYW +A+L LNF FL ++ WY YK + + EL + +
Sbjct: 531 --WIAD-DLNKGRLYNFYWLVAVLVALNFLIFLVFSKWYVYKEKRLAEVGIELDDEPSIP 587
Query: 528 VG 529
+G
Sbjct: 588 MG 589
>At3g53960 transporter like protein
Length = 620
Score = 315 bits (806), Expect = 6e-86
Identities = 177/520 (34%), Positives = 290/520 (55%), Gaps = 19/520 (3%)
Query: 9 SLVLYFIGVMHFDLSSSANTLTNFMGSTFLLSLVGAFISDTYLNRFTTCLVFGSLEVMAL 68
+LV+Y ++H DL + + G T L+ L+G F++D YL R+ T L+ ++ +M L
Sbjct: 64 NLVVYLTTILHQDLKMAVKNTNYWSGVTTLMPLLGGFVADAYLGRYGTVLLATTIYLMGL 123
Query: 69 VMITVQAALDNLHPKACGKSSCVEGGIA--VMFYTSLCLYALGMGGVRGSLTAFGANQFD 126
+++T+ + L KAC + CVE A + F+ ++ L ++G GG + SL +FGA+QF+
Sbjct: 124 ILLTLSWFIPGL--KACHEDMCVEPRKAHEIAFFIAIYLISIGTGGHKPSLESFGADQFE 181
Query: 127 EKDPNEAKALATFFNWLLLSSTLGSVIGVTGIVWVSTQKAWHWGFFIITVASSIGFVTLA 186
+ P E K ++FNW G + VT IV++ + W I+T+ + F
Sbjct: 182 DGHPEERKMKMSYFNWWNAGLCAGILTAVTVIVYIEDRIGWGVASIILTIVMATSFFIFR 241
Query: 187 IGKPFYRIKTPGESPILRIIQVIVVAFKNRKLQLPESNEQLYEVYKDATIE--KIVHTNQ 244
IGKPFYR + P SP+ ++QV V A R L P + L+E+ + + + +
Sbjct: 242 IGKPFYRYRAPSGSPLTPMLQVFVAAIAKRNLPCPSDSSLLHELTNEEYTKGRLLSSSKN 301
Query: 245 MRFLDKAAILQENSES-----QQPWKVCTVTQVEEVKILTRMLPILASTIVMNTCLAQLQ 299
++FLDKAA++++ +E+ Q PW++ TVT+VEEVK+L M+PI T+ C Q
Sbjct: 302 LKFLDKAAVIEDRNENTKAEKQSPWRLATVTKVEEVKLLINMIPIWFFTLAFGVCATQSS 361
Query: 300 TFSVQQGNSMNLKL--GSFTVPASSIPVIPLIFLCTLIPIYELFFVPFIRKITHHPSGVT 357
T ++Q M+ + SF VP +S+ + + + + IYE VP +R+ T + G++
Sbjct: 362 TLFIKQAIIMDRHITGTSFIVPPASLFSLIALSIIITVTIYEKLLVPLLRRATGNERGIS 421
Query: 358 QLQRVGVGLVLSAISMTIAGFIEVKRRDQGRK---DPSKPISLFWLSFQYAIFGIADMFT 414
LQR+GVG+V S +M IA IE KR D ++ + + +S WL+ Q+ + G+AD FT
Sbjct: 422 ILQRIGVGMVFSLFAMIIAALIEKKRLDYAKEHHMNKTMTLSAIWLAPQFLVLGVADAFT 481
Query: 415 LVGLLEFFYREAPSTMKSLSTSFTFLSMSLGYFLSTVFVNVINTVTKRITPSKQGWLQGF 474
LVGL E+FY + P +M+SL +F + F++ + + V + + + I S +GW G
Sbjct: 482 LVGLQEYFYDQVPDSMRSLGIAFYLSVLGAASFVNNLLITVSDHLAEEI--SGKGWF-GK 538
Query: 475 DLNQNNLNLFYWFLAILSCLNFFNFLYWASWYKYKSEEDS 514
DLN + L+ FYW LA L+ N F+ A Y YK+ + S
Sbjct: 539 DLNSSRLDRFYWMLAALTAANICCFVIVAMRYTYKTVQPS 578
>At4g21680 peptide transporter - like protein
Length = 589
Score = 311 bits (798), Expect = 5e-85
Identities = 185/530 (34%), Positives = 295/530 (54%), Gaps = 28/530 (5%)
Query: 8 VSLVLYFIGVMHFDLSSSANTLTNFMGSTFLLSLVGAFISDTYLNRFTTCLVFGSLEVMA 67
V+LVL+ VM D + +AN ++ + G+ ++ SL+GAF+SD+Y R+ TC +F + V
Sbjct: 54 VNLVLFLTRVMGQDNAEAANNVSKWTGTVYIFSLLGAFLSDSYWGRYKTCAIFQASFVAG 113
Query: 68 LVMITVQAALDNLHPKACG--KSSCVEGGI--AVMFYTSLCLYALGMGGVRGSLTAFGAN 123
L+M+++ L P CG S C V+FY S+ L ALG GG + ++ FGA+
Sbjct: 114 LMMLSLSTGALLLEPSGCGVEDSPCKPHSTFKTVLFYLSVYLIALGYGGYQPNIATFGAD 173
Query: 124 QFDEKDPNEAKALATFFNWLLLSSTLGSVIGVTGIVWVSTQKAWHWGFFIITVASSIGFV 183
QFD +D E + FF++ L+ LGS+ T + + Q W GF+ ++ G V
Sbjct: 174 QFDAEDSVEGHSKIAFFSYFYLALNLGSLFSNTVLGYFEDQGEWPLGFWASAGSAFAGLV 233
Query: 184 TLAIGKPFYRIKTPGESPILRIIQVIVVAFKNRKLQLPESNEQLYEVYKDAT-IEKIVHT 242
IG P YR TP ESP R QV+V A + K+ + LY+ T +KI+HT
Sbjct: 234 LFLIGTPKYRHFTPRESPWSRFCQVLVAATRKAKIDVHHEELNLYDSETQYTGDKKILHT 293
Query: 243 NQMRFLDKAAILQENSESQQ--------PWKVCTVTQVEEVKILTRMLPILASTIVMNTC 294
RFLD+AAI+ + E+++ PW++C+VTQVEEVK + R+LPI TI+ +
Sbjct: 294 KGFRFLDRAAIVTPDDEAEKVESGSKYDPWRLCSVTQVEEVKCVLRLLPIWLCTILYSVV 353
Query: 295 LAQLQTFSVQQGNSMNLKLGSFTVPASSIPVIPLIFLCTLIPIYELFFVPFIRKI--THH 352
Q+ + V QG +M + +F +PASS+ ++ + I Y F P ++ T
Sbjct: 354 FTQMASLFVVQGAAMKTNIKNFRIPASSMSSFDILSVAFFIFAYRRFLDPLFARLNKTER 413
Query: 353 PSGVTQLQRVGVGLVLSAISMTIAGFIEVKRRDQGRKDP--------SKPISLFWLSFQY 404
G+T+LQR+G+GLV++ ++M AG +E+ R K+P S +S+FW QY
Sbjct: 414 NKGLTELQRMGIGLVIAIMAMISAGIVEIHRLK--NKEPESATSISSSSTLSIFWQVPQY 471
Query: 405 AIFGIADMFTLVGLLEFFYREAPSTMKSLSTSFTFLSMSLGYFLSTVFVNVINTVTKRIT 464
+ G +++F VG LEFF +AP+ +KS +++ S+SLG ++S++ V+++ ++ T
Sbjct: 472 MLIGASEVFMYVGQLEFFNSQAPTGLKSFASALCMASISLGNYVSSLLVSIVMKIS--TT 529
Query: 465 PSKQGWLQGFDLNQNNLNLFYWFLAILSCLNFFNFLYWASWYKYKSEEDS 514
GW+ +LN+ +L FY+ LA L+ +F +L A WYKY E S
Sbjct: 530 DDVHGWIPE-NLNKGHLERFYFLLAGLTAADFVVYLICAKWYKYIKSEAS 578
>At5g46040 peptide transporter
Length = 586
Score = 306 bits (783), Expect = 3e-83
Identities = 166/535 (31%), Positives = 306/535 (57%), Gaps = 24/535 (4%)
Query: 9 SLVLYFIGVMHFDLSSSANTLTNFMGSTFLLSLVGAFISDTYLNRFTTCLVFGSLEVMAL 68
+LV+Y +H S+N +TN++G+++L ++GA+++D + R+ T ++ ++ ++ +
Sbjct: 54 NLVIYMTTKLHQGTVKSSNNVTNWVGTSWLTPILGAYVADAHFGRYITFVISSAIYLLGM 113
Query: 69 VMITVQAALDNLHPKACGKSS---CVEGGIA--VMFYTSLCLYALGMGGVRGSLTAFGAN 123
++T+ +L L P C ++ C + + +F+ +L A+G GG + +++ GA+
Sbjct: 114 ALLTLSVSLPGLKPPKCSTANVENCEKASVIQLAVFFGALYTLAIGTGGTKPNISTIGAD 173
Query: 124 QFDEKDPNEAKALATFFNWLLLSSTLGSVIGVTGIVWVSTQKAWHWGFFIITVASSIGFV 183
QFDE DP + +FFNW + S G+ T +V+V W G+ + T+ +
Sbjct: 174 QFDEFDPKDKIHKHSFFNWWMFSIFFGTFFATTVLVYVQDNVGWAIGYGLSTLGLAFSIF 233
Query: 184 TLAIGKPFYRIKTPGESPILRIIQVIVVAFKNRKLQLPESNEQLYEV----YKDATIEKI 239
+G YR K P SP ++ +VIV + + + + + + YE+ Y I
Sbjct: 234 IFLLGTRLYRHKLPMGSPFTKMARVIVASLRKAREPMSSDSTRFYELPPMEYASKRAFPI 293
Query: 240 VHTNQMRFLDKAAILQENSESQQPWKVCTVTQVEEVKILTRMLPILASTIVMNTCLAQLQ 299
T+ +RFL++A++ + S W++CT+T+VEE K + +MLP+L T V + LAQ+
Sbjct: 294 HSTSSLRFLNRASL---KTGSTHKWRLCTITEVEETKQMLKMLPVLFVTFVPSMMLAQIM 350
Query: 300 TFSVQQGNSMNLKL-GSFTVPASSIPVIPLIFLCTLIPIYELFFVPFIRKITHHPSGVTQ 358
T ++QG +++ +L +F++P +S+ + I IY+ FV F+RK+T +P G+T
Sbjct: 351 TLFIKQGTTLDRRLTNNFSIPPASLLGFTTFSMLVSIVIYDRVFVKFMRKLTGNPRGITL 410
Query: 359 LQRVGVGLVLSAISMTIAGFIEVKR----RDQGRKDPSK---PISLFWLSFQYAIFGIAD 411
LQR+G+G++L + M IA E R + G + P+S+F L QY + G+AD
Sbjct: 411 LQRMGIGMILHILIMIIASITERYRLKVAAEHGLTHQTAVPIPLSIFTLLPQYVLMGLAD 470
Query: 412 MFTLVGLLEFFYREAPSTMKSLSTSFTFLSMSLGYFLSTVFVNVINTVTKRITPSKQGWL 471
F + LEFFY +AP +MKSL TS+T SM++GYF+S++ ++ ++ +TK+ +GW+
Sbjct: 471 AFIEIAKLEFFYDQAPESMKSLGTSYTSTSMAVGYFMSSILLSSVSQITKK---QGRGWI 527
Query: 472 QGFDLNQNNLNLFYWFLAILSCLNFFNFLYWASWYKYKSEEDSNNSKELGETHLL 526
Q +LN++ L+ +Y F A+L+ LNF FL +Y+Y+++ + + E E +++
Sbjct: 528 QN-NLNESRLDNYYMFFAVLNLLNFILFLVVIRFYEYRADVTQSANVEQKEPNMV 581
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.137 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,117,096
Number of Sequences: 26719
Number of extensions: 502969
Number of successful extensions: 2073
Number of sequences better than 10.0: 75
Number of HSP's better than 10.0 without gapping: 58
Number of HSP's successfully gapped in prelim test: 17
Number of HSP's that attempted gapping in prelim test: 1684
Number of HSP's gapped (non-prelim): 107
length of query: 569
length of database: 11,318,596
effective HSP length: 105
effective length of query: 464
effective length of database: 8,513,101
effective search space: 3950078864
effective search space used: 3950078864
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 63 (28.9 bits)
Medicago: description of AC148406.6


