

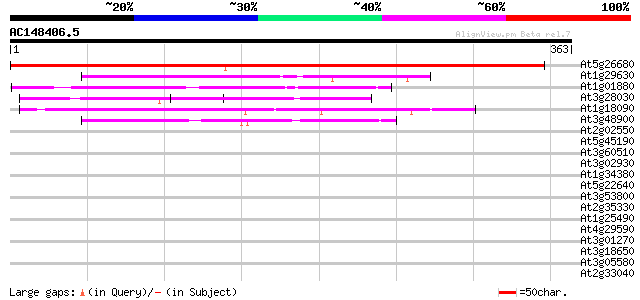
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148406.5 - phase: 0 /pseudo
(363 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g26680 flap endonuclease - like protein 591 e-169
At1g29630 exonuclease, putative 94 1e-19
At1g01880 hypothetical protein 78 7e-15
At3g28030 UV hypersensitive protein (UVH3) 74 1e-13
At1g18090 unknown protein 72 4e-13
At3g48900 putative protein 55 8e-08
At2g02550 hypothetical protein 34 0.11
At5g45190 unknown protein 31 1.2
At3g60510 enoyl-CoA-hydratase - like protein 31 1.2
At3g02930 unknown protein 30 1.6
At1g34380 DNA polymerase type I like protein 30 1.6
At5g22640 unknown protein 30 2.1
At3g53800 unknown protein 30 2.1
At2g35330 unknown protein 30 2.1
At1g25490 phosphoprotein phosphatase 2A regulatory subunit A (R... 30 2.7
At4g29590 unknown protein 29 3.5
At3g01270 pectate lyase like protein 29 3.5
At3g18650 hypothetical protein 29 4.6
At3g05580 putative serine/threonine protein phosphatase type one 29 4.6
At2g33040 mitochondrial F1-ATPase, gamma subunit (ATP3_ARATH) 29 4.6
>At5g26680 flap endonuclease - like protein
Length = 465
Score = 591 bits (1524), Expect = e-169
Identities = 297/358 (82%), Positives = 321/358 (88%), Gaps = 12/358 (3%)
Query: 1 MKENKFESYFGRKIAVDASMSIYQFLIVVGRSGTEMLTNEAGEVTSHLQGMFARTIRLLE 60
MKE KFESYFGRKIAVDASMSIYQFLIVVGR+GTEMLTNEAGEVTSHLQGMF RTIRLLE
Sbjct: 18 MKEQKFESYFGRKIAVDASMSIYQFLIVVGRTGTEMLTNEAGEVTSHLQGMFNRTIRLLE 77
Query: 61 AGMKPVYVFDGKPPEMKNQELKKRLSKRAEATAGLTEALEADNKEDIEKFSKRTVKVTKQ 120
AG+KPVYVFDGKPPE+K QEL KR SKRA+ATA LT A+EA NKEDIEK+SKRTVKVTKQ
Sbjct: 78 AGIKPVYVFDGKPPELKRQELAKRYSKRADATADLTGAIEAGNKEDIEKYSKRTVKVTKQ 137
Query: 121 HNDDCKRLLRLMGVPVVE------------APSEAEAQCAALCKAGKVYAVASEDMDSLT 168
HNDDCKRLLRLMGVPVVE A SEAEAQCAALCK+GKVY VASEDMDSLT
Sbjct: 138 HNDDCKRLLRLMGVPVVEFEKKTDVASYIQATSEAEAQCAALCKSGKVYGVASEDMDSLT 197
Query: 169 FGAPKFLRHLMDPSSKKIPVMEFDVAKILEELDLTMDQFIDLCILSGCDYCDNIRGIGGM 228
FGAPKFLRHLMDPSS+KIPVMEF+VAKILEEL LTMDQFIDLCILSGCDYCD+IRGIGG
Sbjct: 198 FGAPKFLRHLMDPSSRKIPVMEFEVAKILEELQLTMDQFIDLCILSGCDYCDSIRGIGGQ 257
Query: 229 TALKLIRQHGSIEKILENISKERYQVPDDWPYQEARRLFKEPEVSTDDEVLNLKWSPPDE 288
TALKLIRQHGSIE ILEN++KERYQ+P++WPY EAR+LFKEP+V TD+E L++KW+ PDE
Sbjct: 258 TALKLIRQHGSIETILENLNKERYQIPEEWPYNEARKLFKEPDVITDEEQLDIKWTSPDE 317
Query: 289 EGLITFLVNENGFNSDRVTKAIEKIKAAKNKSSQGRLESFFKPTANPSVPIKRKQLII 346
EG++ FLVNENGFN DRVTKAIEKIK AKNKSSQGRLESFFKP AN SVP KRK +I
Sbjct: 318 EGIVQFLVNENGFNIDRVTKAIEKIKTAKNKSSQGRLESFFKPVANSSVPAKRKGNLI 375
>At1g29630 exonuclease, putative
Length = 317
Score = 94.0 bits (232), Expect = 1e-19
Identities = 69/233 (29%), Positives = 111/233 (47%), Gaps = 11/233 (4%)
Query: 47 HLQGMFARTIRLLEAGMKPVYVFDGKPPEMKNQELKKRLSKRAEATAGLTEALEADNKED 106
H+Q R L G+KP+ VFDG P MK ++ KR R E A E N
Sbjct: 55 HIQYCMHRVNLLRHHGVKPIMVFDGGPLPMKLEQENKRARSRKENLARALEHEANGNSSA 114
Query: 107 IEKFSKRTVKVTKQHNDDCKRLLRLMGVPVVEAPSEAEAQCAALCKAGKVYAVASEDMDS 166
+ + V ++ + ++LR V V AP EA+AQ A L +V A+ +ED D
Sbjct: 115 AYECYSKAVDISPSIAHELIQVLRQENVDYVVAPYEADAQMAFLAITKQVDAIITEDSDL 174
Query: 167 LTFGAPKFLRHLMDPSSKKIPVMEFDVAKILEELDLTMDQF-----IDLCILSGCDYCDN 221
+ FG + + MD + EF +K+ + DL++ F +++CILSGCDY +
Sbjct: 175 IPFGCLRII-FKMDKFGHGV---EFQASKLPKNKDLSLSGFSSQMLLEMCILSGCDYLQS 230
Query: 222 IRGIGGMTALKLIRQHGSIEKILENISKERYQVPD--DWPYQEARRLFKEPEV 272
+ G+G A LI + S +++++++ VP + ++ A FK V
Sbjct: 231 LPGMGLKRAHALITKFKSYDRVIKHLKYSTVSVPPLYEESFKRALLTFKHQRV 283
>At1g01880 hypothetical protein
Length = 570
Score = 78.2 bits (191), Expect = 7e-15
Identities = 73/249 (29%), Positives = 118/249 (47%), Gaps = 24/249 (9%)
Query: 2 KENKFESYFGRKIAVDASMSIYQFLIVVGRSGTEMLTNEAGEVTSHLQGMFARTIRLLEA 61
++ F+ +++AVD S I Q V + + HL+ F RTI L
Sbjct: 16 QQQGFDFLRNKRVAVDLSFWIVQHETAV----------KGFVLKPHLRLTFFRTINLFSK 65
Query: 62 -GMKPVYVFDGKPPEMKNQELKKRLSKRAEA-TAGLTEALEADNKEDIEKFSKRTVKVTK 119
G PV+V DG P +K+Q R + + T L + + E + FS+
Sbjct: 66 FGAYPVFVVDGTPSPLKSQARISRFFRSSGIDTCNLPVIKDGVSVERNKLFSEWV----- 120
Query: 120 QHNDDCKRLLRLMGVPVVEAPSEAEAQCAALCKAGKVYAVASEDMDSLTFGAPKFLRHLM 179
+C LL L+G+PV++A EAEA CA L G V A + D D+ FGA ++ +
Sbjct: 121 ---RECVELLELLGIPVLKANGEAEALCAQLNSQGFVDACITPDSDAFLFGAMCVIKDI- 176
Query: 180 DPSSKKIPVMEFDVAKILEELDLTMDQFIDLCILSGCDY-CDNIRGIGGMTALKLIRQHG 238
P+S++ P + ++ I L L I + +L G DY + GIG AL+++R+
Sbjct: 177 KPNSRE-PFECYHMSHIESGLGLKRKHLIAISLLVGNDYDSGGVLGIGVDKALRIVREF- 234
Query: 239 SIEKILENI 247
S +++LE +
Sbjct: 235 SEDQVLERL 243
>At3g28030 UV hypersensitive protein (UVH3)
Length = 1479
Score = 73.9 bits (180), Expect = 1e-13
Identities = 39/130 (30%), Positives = 72/130 (55%), Gaps = 3/130 (2%)
Query: 105 EDIEKFSKRTVKVTKQHNDDCKRLLRLMGVPVVEAPSEAEAQCAALCKAGKVYAVASEDM 164
++ K + V+ + +C+ LL++ G+P + AP EAEAQCA + ++ V + ++D
Sbjct: 909 DEQRKLERNAESVSSEMFAECQELLQIFGIPYIIAPMEAEAQCAFMEQSNLVDGIVTDDS 968
Query: 165 DSLTFGAPKFLRHLMDPSSKKIPVMEFDVAKILEELDLTMDQFIDLCILSGCDYCDNIRG 224
D FGA +++ D V + + I +EL L+ D+ I + +L G DY + I G
Sbjct: 969 DVFLFGARSVYKNIFDDRKY---VETYFMKDIEKELGLSRDKIIRMAMLLGSDYTEGISG 1025
Query: 225 IGGMTALKLI 234
IG + A++++
Sbjct: 1026 IGIVNAIEVV 1035
Score = 52.4 bits (124), Expect = 4e-07
Identities = 38/136 (27%), Positives = 66/136 (47%), Gaps = 10/136 (7%)
Query: 7 ESYFGRKIAVDASMSIYQFLIVVGRSGTEMLTNEAGEVTSHLQGMFARTIRLLEAGMKPV 66
E+ +++A+DAS+ + QF+ + +M+ N +HL G F R +LL KP+
Sbjct: 20 ETLANKRLAIDASIWMVQFIKAMRDEKGDMVQN------AHLIGFFRRICKLLFLRTKPI 73
Query: 67 YVFDGKPPEMKNQELKKRLSKRAEATAGL---TEALEADNKEDIE-KFSKRTVKVTKQHN 122
+VFDG P +K + + R +R A + E L + +DI K + +K +
Sbjct: 74 FVFDGATPALKRRTVIARRRQRENAQTKIRKTAEKLLLNRLKDIRLKEQAKDIKNQRLKQ 133
Query: 123 DDCKRLLRLMGVPVVE 138
DD R+ + + VE
Sbjct: 134 DDSDRVKKRVSSDSVE 149
>At1g18090 unknown protein
Length = 577
Score = 72.4 bits (176), Expect = 4e-13
Identities = 79/308 (25%), Positives = 135/308 (43%), Gaps = 19/308 (6%)
Query: 7 ESYFGRKIAVDASMSIYQFLIVVGRS-GTEMLTNEAGEVTSHLQGMFARTIRLLEA-GMK 64
+ Y G+++ +DA Y +L S E+ + G+ F + LL+ +
Sbjct: 20 QKYAGKRVGIDA----YSWLHKGAYSCSMELCLDTDGKKKLRYIDYFMHRVNLLQHYEII 75
Query: 65 PVYVFDGKPPEMKNQELKKRLSKRAEATAGLTEALEADNKEDIEKFSKRTVKVTKQHNDD 124
PV V DG K +R KR L+ N + +R V VT
Sbjct: 76 PVVVLDGGNMPCKAATGDERHRKRKANFDAAMVKLKEGNVAAATELFQRAVSVTSSMAHQ 135
Query: 125 CKRLLRLMGVPVVEAPSEAEAQCAALC----KAGKVYAVASEDMDSLTFGAPKFLRHLMD 180
++L+ V + AP EA+AQ A L + G + AV +ED D L +G K + MD
Sbjct: 136 LIQVLKSENVEFIVAPYEADAQLAYLSSLELEQGGIAAVITEDSDLLAYGC-KAVIFKMD 194
Query: 181 PSSKKIPVMEFDVAKILEEL----DLTMDQFIDLCILSGCDYCDNIRGIGGMTALKLIRQ 236
K ++ +V + +++ + + F +C+L+GCD+ ++ G+G A I +
Sbjct: 195 RYGKGEELVLDNVFQAVDQKPSFQNFDQELFTAMCVLAGCDFLPSVPGVGISRAHAFISK 254
Query: 237 HGSIEKILENI-SKERYQVPDDW--PYQEARRLFKEPEVSTDDEVLNLKWSPPDEEGLIT 293
+ S+E +L + +K+ VPDD+ + EA +F+ V D + LK P L+
Sbjct: 255 YQSVELVLSFLKTKKGKLVPDDYSSSFTEAVSVFQHARV-YDFDAKKLKHLKPLSHNLLN 313
Query: 294 FLVNENGF 301
V + F
Sbjct: 314 LPVEQLEF 321
>At3g48900 putative protein
Length = 337
Score = 54.7 bits (130), Expect = 8e-08
Identities = 56/219 (25%), Positives = 94/219 (42%), Gaps = 27/219 (12%)
Query: 47 HLQGMFARTIRLLEAGMKPVYVFDGKPPEMKNQELKKRLSKRAE-ATAGLTEALEADNKE 105
+L+G F R L+ + V DG P +K K+RL R E A G+ + E K
Sbjct: 53 YLRGFFHRLRALIALNCSIILVSDGAIPGIKVPTYKRRLKARFEIADDGVEPSKETSLKR 112
Query: 106 DIEKFSKRTVKVTKQHNDDCKRLLRLMGVPVVEAPSEAEAQCA-----ALCK-------- 152
++ +K + K + +G+ ++ EAEAQCA +LC+
Sbjct: 113 NMGSEFSCIIK-------EAKVIASTLGILCLDGIEEAEAQCALLNSESLCRNMPWSVER 165
Query: 153 -AGKVYAVASEDMDSLTFGAPKFLRHLMDPSSKKIPVMEFDVAKILEELDLTMDQFIDLC 211
+ + + DM + KF L + V+ +++ I ++L L + I L
Sbjct: 166 DSLVCWFLLPSDMVNSIISPNKFANELGEGGY----VVCYEMDDIKKKLGLGRNSLIALA 221
Query: 212 ILSGCDYCDNIRGIGGMTALKLIRQHGSIEKILENISKE 250
+L G DY +RG+ A +L+R G ILE ++ E
Sbjct: 222 LLLGSDYSQGVRGLRQEKACELVRSIGD-NVILEKVASE 259
>At2g02550 hypothetical protein
Length = 216
Score = 34.3 bits (77), Expect = 0.11
Identities = 25/121 (20%), Positives = 53/121 (43%), Gaps = 4/121 (3%)
Query: 1 MKENKFESYFGRKIAVDASMSIYQFLIVVGRSGTEMLTNEAGEVTSHLQGMFARTIRLLE 60
+K F + + +A+DASM +Y+ + R E T E H+ + + + ++
Sbjct: 16 LKSISFSALNSKTLAIDASMWLYRGALNNARKFLEEPTTPNSE---HIMFVIKKVKQFID 72
Query: 61 AGMKPVYVFDGKPPEMKNQELKKRLSKRAEATAGLTEALEADNKEDIEKFSKRTVKVTKQ 120
+ P+ VFDG +K+ + + A+ + + D+ + +K K K+ +
Sbjct: 73 RNVNPILVFDGHRLPIKSPRNPIKTLEYAKDLDASAQLMAPDDPKRKDK-EKNATKIFQS 131
Query: 121 H 121
H
Sbjct: 132 H 132
>At5g45190 unknown protein
Length = 579
Score = 30.8 bits (68), Expect = 1.2
Identities = 30/106 (28%), Positives = 49/106 (45%), Gaps = 5/106 (4%)
Query: 85 LSKRAEATAGLTEALEADNKEDIEKFSKRTVKVTKQH--NDDCKRLLRLMGVPVVEAPSE 142
L++ E G + DNKE+IE+ +K + + H + D R P+VE P +
Sbjct: 332 LNQNNENGGGEAANVSVDNKEEIERETKESSLHLESHPAHKDNVREAPHNSRPLVEGPGK 391
Query: 143 --AEAQCAALCKAGKVYAVASEDM-DSLTFGAPKFLRHLMDPSSKK 185
+E + L G V+ + D+ D+L +PK L+ L D K
Sbjct: 392 DNSEREGGELQDDGAVHKSRNVDVGDALISQSPKDLKLLRDKVKAK 437
>At3g60510 enoyl-CoA-hydratase - like protein
Length = 401
Score = 30.8 bits (68), Expect = 1.2
Identities = 20/91 (21%), Positives = 45/91 (48%), Gaps = 2/91 (2%)
Query: 183 SKKIPVMEFDVAKILEELDLTMDQFIDLCILSGCDYCDNIRGIGGMTALKLIRQHGSIEK 242
S+++PVME + K+L + ++ ++ C + + + I + L+ H ++E+
Sbjct: 220 SEEVPVMEEQLKKLLTDDPSVVESCLEKC--AEVAHPEKTGVIRRIDLLEKCFSHDTVEE 277
Query: 243 ILENISKERYQVPDDWPYQEARRLFKEPEVS 273
I++++ E + D W RRL + +S
Sbjct: 278 IIDSLEIEASRRKDTWCITTLRRLKESSPLS 308
>At3g02930 unknown protein
Length = 806
Score = 30.4 bits (67), Expect = 1.6
Identities = 41/155 (26%), Positives = 69/155 (44%), Gaps = 30/155 (19%)
Query: 80 ELKKRLSKRAEATAGLTEALEADNKE----DIEKF-----------------SKRTVKVT 118
+LK+ + EA+ L EALEA K +IEKF K V
Sbjct: 116 QLKEARKEAEEASEKLDEALEAQKKSLENFEIEKFEVVEAGIEAVQRKEEELKKELENVK 175
Query: 119 KQHNDDCKRLLRL---MGVPVVEAPSEAEAQCAALCK---AGKVYAVASEDMDSLTFGAP 172
QH + LL + + E + +A+ ALC+ A K+ A+ +E ++ L+
Sbjct: 176 NQHASESATLLLVTQELENVNQELANAKDAKSKALCRADDASKMAAIHAEKVEILSSELI 235
Query: 173 KFLRHLMDPSSKKIPVMEFDVAKIL--EELDLTMD 205
+ L+ L+D + +K + + ++A L E +DL D
Sbjct: 236 R-LKALLDSTREKEIISKNEIALKLGAEIVDLKRD 269
Score = 28.1 bits (61), Expect = 7.8
Identities = 26/131 (19%), Positives = 56/131 (41%), Gaps = 7/131 (5%)
Query: 75 EMKNQELKKRLSKRAEATAGLTEALEADNKEDIEKFSKRTVKVTKQHNDDCKRLLRLMGV 134
E N+E + L K +AT+ + LE K E ++ +K+ + K+ + +
Sbjct: 407 ETVNEEKTQALKKEQDATSSVQRLLEEKKKILSE------LESSKEEEEKSKKAMESLAS 460
Query: 135 PVVEAPSEA-EAQCAALCKAGKVYAVASEDMDSLTFGAPKFLRHLMDPSSKKIPVMEFDV 193
+ E SE+ E + L + + Y ED+ + +++D + +I V+ V
Sbjct: 461 ALHEVSSESRELKEKLLSRGDQNYETQIEDLKLVIKATNNKYENMLDEARHEIDVLVNAV 520
Query: 194 AKILEELDLTM 204
+ ++ + M
Sbjct: 521 EQTKKQFESAM 531
>At1g34380 DNA polymerase type I like protein
Length = 343
Score = 30.4 bits (67), Expect = 1.6
Identities = 17/41 (41%), Positives = 24/41 (58%), Gaps = 9/41 (21%)
Query: 211 CILSGCDYCDNIRGI-------GGMTALKLIRQHGSIEKIL 244
CI+ D D + GI G TA+KL+R+HGS+E +L
Sbjct: 238 CIMG--DEVDGVPGIQHMVPAFGRKTAMKLVRKHGSLESLL 276
>At5g22640 unknown protein
Length = 871
Score = 30.0 bits (66), Expect = 2.1
Identities = 24/121 (19%), Positives = 50/121 (40%), Gaps = 6/121 (4%)
Query: 6 FESYFGRKIAVDASMSIYQFLIVVGRS------GTEMLTNEAGEVTSHLQGMFARTIRLL 59
F+ ++G+++ V I F++ + +S G E T E + + M + + L+
Sbjct: 557 FDEFYGKEVVVKKEHPIKSFVLGIEKSVKPMLDGLEKWTEEKKKAYEERKEMIQQELELV 616
Query: 60 EAGMKPVYVFDGKPPEMKNQELKKRLSKRAEATAGLTEALEADNKEDIEKFSKRTVKVTK 119
EA + + E+K +E ++ T + L KE+ +K ++ K
Sbjct: 617 EAEICLEEAIEDMDEELKKKEQEEEKKTEMGLTEEDEDVLVPVYKEEKVVTAKEKIQENK 676
Query: 120 Q 120
Q
Sbjct: 677 Q 677
>At3g53800 unknown protein
Length = 363
Score = 30.0 bits (66), Expect = 2.1
Identities = 32/126 (25%), Positives = 56/126 (44%), Gaps = 6/126 (4%)
Query: 89 AEATAGLTEALEADNKEDIEKFSKRTVKVTKQHNDDCKRLLRLMGVP--VVEAPSEAEAQ 146
A AGL +AL +D K + ++ N DCK ++R +G P ++ S + +
Sbjct: 183 ANGYAGLRDALVSDTVRFQRKALNLLHYLLQESNSDCK-IVRDLGFPRIMIHLASNQDFE 241
Query: 147 CAALCKAGKVYAVASEDMDSLTFGAPKFLRHLMDPSSKKIPVM-EFDVAKILEELDLTMD 205
G + E + +L G LR L++ +++I VM + D+ EE L +D
Sbjct: 242 VREFALRGLLELAREESVRNLDRGDVN-LRQLLEERTRRIIVMSDEDLCAAREERQL-VD 299
Query: 206 QFIDLC 211
+C
Sbjct: 300 SLWTVC 305
>At2g35330 unknown protein
Length = 738
Score = 30.0 bits (66), Expect = 2.1
Identities = 15/52 (28%), Positives = 27/52 (51%)
Query: 77 KNQELKKRLSKRAEATAGLTEALEADNKEDIEKFSKRTVKVTKQHNDDCKRL 128
+ Q+ K+++ + E EA+EA NK +E + ++H DD +RL
Sbjct: 572 QEQKAKEQVLAQVEEEQRSKEAIEASNKRKVESLRLKIEIDFQRHKDDLQRL 623
>At1g25490 phosphoprotein phosphatase 2A regulatory subunit A
(RCN1)
Length = 588
Score = 29.6 bits (65), Expect = 2.7
Identities = 13/36 (36%), Positives = 25/36 (69%)
Query: 173 KFLRHLMDPSSKKIPVMEFDVAKILEELDLTMDQFI 208
KFL +++ S ++P ++F+VAK+L+ L +DQ +
Sbjct: 514 KFLPVVVEASKDRVPNIKFNVAKLLQSLIPIVDQSV 549
>At4g29590 unknown protein
Length = 317
Score = 29.3 bits (64), Expect = 3.5
Identities = 15/50 (30%), Positives = 26/50 (52%), Gaps = 3/50 (6%)
Query: 162 EDMDSLTFGAPKFLRHLMDPSSKKIPVMEFDVAKILEELDLTMDQFIDLC 211
E DS + AP+F+ H+ DP+ I + +K+L + D +D+C
Sbjct: 118 ESSDSTFYEAPRFVTHIDDPA---IAALTKYYSKVLPQSDTPGVSILDMC 164
>At3g01270 pectate lyase like protein
Length = 475
Score = 29.3 bits (64), Expect = 3.5
Identities = 18/59 (30%), Positives = 30/59 (50%), Gaps = 1/59 (1%)
Query: 152 KAGKVYAVASEDMDSLTFGAPKFLRHLMDPSSKKIPVMEFDVA-KILEELDLTMDQFID 209
K G++Y V S D + P LRH + V + D++ ++ +EL +T D+ ID
Sbjct: 159 KRGRIYVVTSPRDDDMVNPRPGTLRHAVIQKEPLWIVFKHDMSIRLSQELMITSDKTID 217
>At3g18650 hypothetical protein
Length = 386
Score = 28.9 bits (63), Expect = 4.6
Identities = 14/34 (41%), Positives = 21/34 (61%)
Query: 86 SKRAEATAGLTEALEADNKEDIEKFSKRTVKVTK 119
+KR + + L E LE NK+D EK K+ +KV +
Sbjct: 92 TKRRKGSVDLHEFLEKMNKDDPEKEEKKKIKVRR 125
>At3g05580 putative serine/threonine protein phosphatase type one
Length = 318
Score = 28.9 bits (63), Expect = 4.6
Identities = 20/87 (22%), Positives = 36/87 (40%), Gaps = 20/87 (22%)
Query: 219 CDNIRGIGGMTALKLIRQHGSIEKILENISKERYQVPDDWPYQEARRLFKEPEVSTDDEV 278
C N + + K++ HG + LEN+ + R + + E+ + +
Sbjct: 157 CFNCLPVAALIDEKILCMHGGLSPELENLG-------------QIREIQRPTEIPDNGLL 203
Query: 279 LNLKWSPPDEEGLITFLVNENGFNSDR 305
+L WS PD++ NE +SDR
Sbjct: 204 CDLLWSDPDQK-------NEGWTDSDR 223
>At2g33040 mitochondrial F1-ATPase, gamma subunit (ATP3_ARATH)
Length = 325
Score = 28.9 bits (63), Expect = 4.6
Identities = 15/52 (28%), Positives = 31/52 (58%), Gaps = 8/52 (15%)
Query: 302 NSDRVTKAIEKIKAAKNKSSQGRLES---FFKPTA-----NPSVPIKRKQLI 345
N ++TKA++ + A+K ++ QGR E+ ++P NPS+ +K+ ++
Sbjct: 57 NIQKITKAMKMVAASKLRAVQGRAENSRGLWQPFTALLGDNPSIDVKKSVVV 108
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.135 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,083,987
Number of Sequences: 26719
Number of extensions: 343701
Number of successful extensions: 1106
Number of sequences better than 10.0: 29
Number of HSP's better than 10.0 without gapping: 13
Number of HSP's successfully gapped in prelim test: 16
Number of HSP's that attempted gapping in prelim test: 1078
Number of HSP's gapped (non-prelim): 36
length of query: 363
length of database: 11,318,596
effective HSP length: 101
effective length of query: 262
effective length of database: 8,619,977
effective search space: 2258433974
effective search space used: 2258433974
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC148406.5


