

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148406.1 + phase: 0
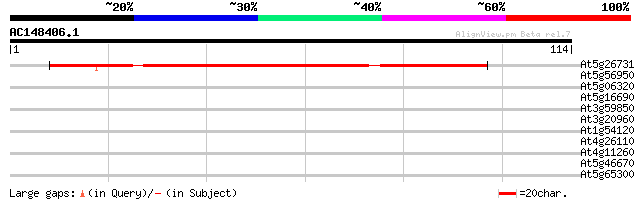
(114 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g26731 unknown protein 78 6e-16
At5g56950 nucleosome assembly protein 28 0.74
At5g06320 NDR1/HIN1-like protein 3 (NHL3) 28 0.74
At5g16690 unknown protein 28 0.97
At3g59850 polygalacturonase-like protein 28 0.97
At3g20960 cytochrome P450 like protein 28 0.97
At1g54120 unknown protein 27 1.7
At4g26110 nucleosome assembly protein I-like protein 26 3.7
At4g11260 Sgt1 like protein 26 3.7
At5g46670 putative protein 25 4.8
At5g65300 unknown protein 25 6.3
>At5g26731 unknown protein
Length = 99
Score = 78.2 bits (191), Expect = 6e-16
Identities = 46/90 (51%), Positives = 60/90 (66%), Gaps = 5/90 (5%)
Query: 9 LFRTESFG-YYNFPDQNYTMIEKRQLFLRSYQFCRKKSLTERIKGSLVRAKKVVWLKLRY 67
+FR+ S G YY P Y +EKRQLFLRSYQF RK+S E++ S+ R K+VV +LR
Sbjct: 6 VFRSTSGGDYYGAPYGEY--MEKRQLFLRSYQFSRKQSFAEKVSRSVTRVKRVVLTRLRS 63
Query: 68 ARGLRRKLVFFPRFKCGFYYRRRRFSQLLN 97
A L+R V + R + FYYRRRRF +LL+
Sbjct: 64 APKLKR--VVWSRLRSAFYYRRRRFFRLLH 91
>At5g56950 nucleosome assembly protein
Length = 374
Score = 28.1 bits (61), Expect = 0.74
Identities = 16/43 (37%), Positives = 24/43 (55%), Gaps = 6/43 (13%)
Query: 44 KSLTERIKGSLVRAKKVVWLKLRYARGLRRKLVFF----PRFK 82
+ +TER +G+L+ K + W K+ +G KL FF P FK
Sbjct: 142 EEITERDEGALIYLKDIKWCKIEEPKGF--KLEFFFDQNPYFK 182
>At5g06320 NDR1/HIN1-like protein 3 (NHL3)
Length = 231
Score = 28.1 bits (61), Expect = 0.74
Identities = 13/41 (31%), Positives = 21/41 (50%)
Query: 43 KKSLTERIKGSLVRAKKVVWLKLRYARGLRRKLVFFPRFKC 83
+K L E + + R + LK+R+ GL + F P+ KC
Sbjct: 166 RKDLNEDVNSQIYRIDAKLRLKIRFKFGLIKSWRFKPKIKC 206
>At5g16690 unknown protein
Length = 734
Score = 27.7 bits (60), Expect = 0.97
Identities = 16/44 (36%), Positives = 23/44 (51%), Gaps = 5/44 (11%)
Query: 21 PDQNYTMIEKRQLFLRSYQFCRKKSLTERIKGSLVRAKKVVWLK 64
P +T+ K LF+RSY C+ +LT S VR K+ L+
Sbjct: 318 PCSGFTVSHKVALFMRSYFLCQDGTLT-----SFVRTLKIACLQ 356
>At3g59850 polygalacturonase-like protein
Length = 388
Score = 27.7 bits (60), Expect = 0.97
Identities = 17/56 (30%), Positives = 30/56 (53%), Gaps = 11/56 (19%)
Query: 27 MIEKRQLFLRSYQF----CRKKSLTERIKGSLVR-------AKKVVWLKLRYARGL 71
++ K + LRS F C++KS+T RI+G+LV K+ W+ ++ G+
Sbjct: 59 LVPKGRFLLRSIIFDGSKCKRKSVTFRIQGTLVAPSDYRVIGKENYWILFQHLDGI 114
>At3g20960 cytochrome P450 like protein
Length = 418
Score = 27.7 bits (60), Expect = 0.97
Identities = 22/71 (30%), Positives = 33/71 (45%), Gaps = 3/71 (4%)
Query: 40 FCRKKSLTERIKGSLVRAKKVVWLKLRYARGLRR--KLVFFPRFKCGFYYRRRRFSQLLN 97
F + ER++G LV + K+ +A LRR K + P FK RF++LL
Sbjct: 99 FSEENGEAERVRG-LVAESYALAKKIFFASVLRRLLKKLRIPLFKKDIMDVSNRFNELLE 157
Query: 98 NSGSSHHRKVD 108
H+ K+D
Sbjct: 158 KILVEHNEKLD 168
>At1g54120 unknown protein
Length = 124
Score = 26.9 bits (58), Expect = 1.7
Identities = 11/25 (44%), Positives = 17/25 (68%)
Query: 54 LVRAKKVVWLKLRYARGLRRKLVFF 78
+VR K+ WLKL Y R +++ VF+
Sbjct: 49 MVRRMKLRWLKLHYVRVVKKIKVFY 73
>At4g26110 nucleosome assembly protein I-like protein
Length = 372
Score = 25.8 bits (55), Expect = 3.7
Identities = 16/43 (37%), Positives = 23/43 (53%), Gaps = 6/43 (13%)
Query: 44 KSLTERIKGSLVRAKKVVWLKLRYARGLRRKLVFF----PRFK 82
+ +TER +G+L K + W K+ +G KL FF P FK
Sbjct: 143 EEVTERDEGALKYLKDIKWCKIEEPKGF--KLEFFFDTNPYFK 183
>At4g11260 Sgt1 like protein
Length = 358
Score = 25.8 bits (55), Expect = 3.7
Identities = 9/30 (30%), Positives = 18/30 (60%)
Query: 41 CRKKSLTERIKGSLVRAKKVVWLKLRYARG 70
CR + L+ +++ L +A+ + W L Y +G
Sbjct: 221 CRFEVLSTKVEIRLAKAEIITWASLEYGKG 250
>At5g46670 putative protein
Length = 332
Score = 25.4 bits (54), Expect = 4.8
Identities = 10/20 (50%), Positives = 14/20 (70%)
Query: 14 SFGYYNFPDQNYTMIEKRQL 33
SFG Y+F D++Y I+ R L
Sbjct: 270 SFGVYSFEDRDYNWIDVRAL 289
>At5g65300 unknown protein
Length = 150
Score = 25.0 bits (53), Expect = 6.3
Identities = 10/27 (37%), Positives = 16/27 (59%)
Query: 44 KSLTERIKGSLVRAKKVVWLKLRYARG 70
K TE++K + V+ K W KL + +G
Sbjct: 106 KKKTEKLKSTTVKKKTGFWTKLLHLKG 132
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.329 0.140 0.442
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,572,387
Number of Sequences: 26719
Number of extensions: 91002
Number of successful extensions: 222
Number of sequences better than 10.0: 11
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 2
Number of HSP's that attempted gapping in prelim test: 211
Number of HSP's gapped (non-prelim): 11
length of query: 114
length of database: 11,318,596
effective HSP length: 90
effective length of query: 24
effective length of database: 8,913,886
effective search space: 213933264
effective search space used: 213933264
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 52 (24.6 bits)
Medicago: description of AC148406.1


