

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148404.8 - phase: 0
(416 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
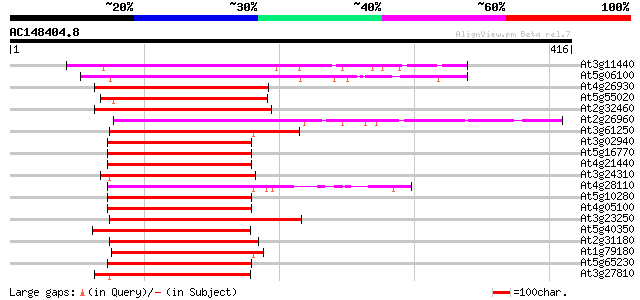
Score E
Sequences producing significant alignments: (bits) Value
At3g11440 transcription factor like protein (MYB65) 204 7e-53
At5g06100 transcription factor MYB33 - like protein 203 2e-52
At4g26930 putative myb-related protein 196 2e-50
At5g55020 putative transcription factor MYB120 (MYB120) 192 2e-49
At2g32460 putative MYB family transcription factor 191 6e-49
At2g26960 putative transcription factor (MYB81) 187 1e-47
At3g61250 putative transcription factor (MYB17) 160 1e-39
At3g02940 MYB family transcription factor like protein 154 9e-38
At5g16770 putative transcription factor (MYB9) 154 1e-37
At4g21440 myb-related protein M4 154 1e-37
At3g24310 putative transcription factor (MYB71) 151 7e-37
At4g28110 putative transcription factor (MYB41) 150 1e-36
At5g10280 putative transcription factor MYB92 149 4e-36
At4g05100 MYB - like protein 149 4e-36
At3g23250 myb-related transcription factor like protein 149 4e-36
At5g40350 putative transcription factor MYB24 148 5e-36
At2g31180 putative transcription factor (MYB14) 147 8e-36
At1g79180 ranscription factor like protein (MYB63) 147 8e-36
At5g65230 transcription factor-like protein 147 1e-35
At3g27810 ATMYB3 (Atmyb3) 147 1e-35
>At3g11440 transcription factor like protein (MYB65)
Length = 553
Score = 204 bits (519), Expect = 7e-53
Identities = 130/339 (38%), Positives = 178/339 (52%), Gaps = 50/339 (14%)
Query: 43 SSGDDDRKRQSILSADQETDEGDTTG--GEGGEGGLKKGPWTAAEDEILVAHVQKYGEGN 100
++ D D S + + + + G G LKKGPWT+ ED IL+ +V+K+GEGN
Sbjct: 6 ATADSDDGMHSSIHNESPAPDSISNGCRSRGKRSVLKKGPWTSTEDGILIDYVKKHGEGN 65
Query: 101 WNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAITAEEERRIIELHHKMGNKWAQMAALL 160
WN+V+K T LARCGKSCRLRWANHLRP+L+KGA + EEE+ I+E+H KMGNKWAQMA L
Sbjct: 66 WNAVQKHTSLARCGKSCRLRWANHLRPNLKKGAFSQEEEQLIVEMHAKMGNKWAQMAEHL 125
Query: 161 PGRTDNEIKNFWNTRCKKRGRANLPIYPEDLSSECL-----LNQENADMLTNEASQHD-- 213
PGRTDNEIKN+WNTR K+R RA LP+YP ++ + L + N + D
Sbjct: 126 PGRTDNEIKNYWNTRIKRRQRAGLPLYPPEIYVDDLHWSEEYTKSNIIRVDRRRRHQDFL 185
Query: 214 EAENKVPEVIFKDYKLRPDILPPCFDKLLAGL---------------------LLRPSKR 252
+ N V+F D +LP D L+ L +L K+
Sbjct: 186 QLGNSKDNVLFDDLNFAASLLPAASD--LSDLVACNMLGTGASSSRYESYMPPILPSPKQ 243
Query: 253 PWKSDMNLYSYSNNA----AAPAAFDQ--LRKYPMPSPSSP------PWEDINEPHSYNQ 300
W+S S+N +P F ++K P SP P+E+ HS +
Sbjct: 244 IWESGSRFPMCSSNIKHEFQSPEHFQNTAVQKNPRSCSISPCDVDHHPYEN---QHSSHM 300
Query: 301 FNEYDNPMVGIHMAPKTSSFEPIYGFMKTEPPSLQYSQT 339
D+ V M P + +P++G +K E PS QYS+T
Sbjct: 301 MMVPDSHTVTYGMHPTS---KPLFGAVKLELPSFQYSET 336
>At5g06100 transcription factor MYB33 - like protein
Length = 520
Score = 203 bits (516), Expect = 2e-52
Identities = 128/329 (38%), Positives = 180/329 (53%), Gaps = 49/329 (14%)
Query: 53 SILSADQETDEGDTTGGEGGE-------GGLKKGPWTAAEDEILVAHVQKYGEGNWNSVR 105
S S D + +E G + LKKGPW++AED+IL+ +V K+GEGNWN+V+
Sbjct: 2 SYTSTDSDHNESPAADDNGSDCRSRWDGHALKKGPWSSAEDDILIDYVNKHGEGNWNAVQ 61
Query: 106 KCTGLARCGKSCRLRWANHLRPDLRKGAITAEEERRIIELHHKMGNKWAQMAALLPGRTD 165
K T L RCGKSCRLRWANHLRP+L+KGA + EEE+ I+ELH KMGN+WA+MAA LPGRTD
Sbjct: 62 KHTSLFRCGKSCRLRWANHLRPNLKKGAFSQEEEQLIVELHAKMGNRWARMAAHLPGRTD 121
Query: 166 NEIKNFWNTRCKKRGRANLPIYPEDLSSECL-LNQENA-DMLTNEASQHDE--AENKVPE 221
NEIKN+WNTR K+R RA LP+YP ++ E L +QE A + E +H +
Sbjct: 122 NEIKNYWNTRIKRRQRAGLPLYPPEMHVEALEWSQEYAKSRVMGEDRRHQDFLQLGSCES 181
Query: 222 VIFKDYKLRPDILPPCFD-------KLLAGLLLRP------------SKRPWKSDMNLYS 262
+F D D++P FD K + P SKR W+S++ LY
Sbjct: 182 NVFFDTLNFTDMVPGTFDLADMTAYKNMGNCASSPRYENFMTPTIPSSKRLWESEL-LYP 240
Query: 263 YSNNAAAPAAFDQLRKYPMPSPSSPPWEDINEPHSYNQFNEYDNPMVGIHMAPK------ 316
++ F ++ SP + I++ S++ + ++P+ G +P
Sbjct: 241 -GCSSTIKQEFSSPEQFRNTSP-----QTISKTCSFSVPCDVEHPLYGNRHSPVMIPDSH 294
Query: 317 ------TSSFEPIYGFMKTEPPSLQYSQT 339
+P+YG +K E PS QYS+T
Sbjct: 295 TPTDGIVPYSKPLYGAVKLELPSFQYSET 323
>At4g26930 putative myb-related protein
Length = 389
Score = 196 bits (499), Expect = 2e-50
Identities = 91/129 (70%), Positives = 106/129 (81%)
Query: 64 GDTTGGEGGEGGLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWAN 123
G + GEGG LKKGPWT AEDE L A+V++YGEGNWNSV+K T LARCGKSCRLRWAN
Sbjct: 7 GASEDGEGGGVVLKKGPWTVAEDETLAAYVREYGEGNWNSVQKKTWLARCGKSCRLRWAN 66
Query: 124 HLRPDLRKGAITAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKRGRAN 183
HLRP+LRKG+ T EEER II+LH ++GNKWA+MAA LPGRTDNEIKN+WNTR K+ R
Sbjct: 67 HLRPNLRKGSFTPEEERLIIQLHSQLGNKWARMAAQLPGRTDNEIKNYWNTRLKRFQRQG 126
Query: 184 LPIYPEDLS 192
LP+YP + S
Sbjct: 127 LPLYPPEYS 135
>At5g55020 putative transcription factor MYB120 (MYB120)
Length = 523
Score = 192 bits (489), Expect = 2e-49
Identities = 90/135 (66%), Positives = 108/135 (79%), Gaps = 11/135 (8%)
Query: 68 GGEGGEGG-----------LKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKS 116
GG G +GG LKKGPWTAAEDEIL A+V++ GEGNWN+V+K TGLARCGKS
Sbjct: 7 GGAGKDGGSTNHLSDGGVILKKGPWTAAEDEILAAYVRENGEGNWNAVQKNTGLARCGKS 66
Query: 117 CRLRWANHLRPDLRKGAITAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRC 176
CRLRWANHLRP+L+KG+ T +EER II+LH ++GNKWA+MAA LPGRTDNEIKN+WNTR
Sbjct: 67 CRLRWANHLRPNLKKGSFTGDEERLIIQLHAQLGNKWARMAAQLPGRTDNEIKNYWNTRL 126
Query: 177 KKRGRANLPIYPEDL 191
K+ R LP+YP D+
Sbjct: 127 KRLLRQGLPLYPPDI 141
>At2g32460 putative MYB family transcription factor
Length = 490
Score = 191 bits (485), Expect = 6e-49
Identities = 86/132 (65%), Positives = 108/132 (81%), Gaps = 1/132 (0%)
Query: 64 GDTTGGEGGEG-GLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWA 122
G+TT EG GLKKGPWT ED IL +V+K+GEGNWN+V+K +GL RCGKSCRLRWA
Sbjct: 5 GETTATATMEGRGLKKGPWTTTEDAILTEYVRKHGEGNWNAVQKNSGLLRCGKSCRLRWA 64
Query: 123 NHLRPDLRKGAITAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKRGRA 182
NHLRP+L+KG+ T +EE+ II+LH K+GNKWA+MA+ LPGRTDNEIKN+WNTR K+R RA
Sbjct: 65 NHLRPNLKKGSFTPDEEKIIIDLHAKLGNKWARMASQLPGRTDNEIKNYWNTRMKRRQRA 124
Query: 183 NLPIYPEDLSSE 194
LP+YP ++ +
Sbjct: 125 GLPLYPHEIQHQ 136
>At2g26960 putative transcription factor (MYB81)
Length = 427
Score = 187 bits (474), Expect = 1e-47
Identities = 121/353 (34%), Positives = 173/353 (48%), Gaps = 31/353 (8%)
Query: 78 KGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAITAE 137
KGPWT AED +L+A+V K+G+GNWN+V+ +GL+RCGKSCRLRW NHLRPDL+KGA T +
Sbjct: 22 KGPWTQAEDNLLIAYVDKHGDGNWNAVQNNSGLSRCGKSCRLRWVNHLRPDLKKGAFTEK 81
Query: 138 EERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKRGRANLPIYPEDLSSECLL 197
EE+R+IELH +GNKWA+MA LPGRTDNEIKNFWNTR K+ R LP+YP+++ +
Sbjct: 82 EEKRVIELHALLGNKWARMAEELPGRTDNEIKNFWNTRLKRLQRLGLPVYPDEVREHAMN 141
Query: 198 NQENADMLTNEASQHDEAEN------KVPEVIFKDYKLRPDILPPCFDKLLAGL----LL 247
++ + T+ H E ++PEV F+ L + +L + +
Sbjct: 142 AATHSGLNTDSLDGHHSQEYMEADTVEIPEVDFEHLPLNRS--SSYYQSMLRHVPPTNVF 199
Query: 248 RPSKRPWKSDMNLYS------YSNNAAAP----AAFDQLRKYPMPSPSSPPWEDINEPHS 297
K + N+Y+ Y + P AF Y M S W N P
Sbjct: 200 VRQKPCFFQPPNVYNLIPPSPYMSTGKRPREPETAFPCPGGYTMNEQSPRLW---NYPFV 256
Query: 298 YNQFNEYDNPMVGIHMAPKTSSFEPIYGFMKTEPPSLQYSQTQHRSYMIPYPQVEPVQSV 357
N + + + + A + ++G E PS QY + + + P P
Sbjct: 257 ENVSEQLPDSHLLGNAAYSSPPGPLVHGVENFEFPSFQYHE-EPGGWGADQPNPMPEHES 315
Query: 358 PTSLERSHFLPSNEANLSTQWDESDDQYDGLFPSFDYNNNNHMNTCSTDGHHS 410
+L +S ++ S YDGL S Y ++ TD S
Sbjct: 316 DNTLVQSPLTAQTPSDC-----PSSSLYDGLLESVVYGSSGEKPATDTDSESS 363
>At3g61250 putative transcription factor (MYB17)
Length = 299
Score = 160 bits (405), Expect = 1e-39
Identities = 77/143 (53%), Positives = 97/143 (66%), Gaps = 2/143 (1%)
Query: 75 GLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAI 134
GLKKGPWT EDE+LVAH++K G G+W ++ K GL RCGKSCRLRW N+LRPD+++G
Sbjct: 11 GLKKGPWTPEEDEVLVAHIKKNGHGSWRTLPKLAGLLRCGKSCRLRWTNYLRPDIKRGPF 70
Query: 135 TAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR--GRANLPIYPEDLS 192
TA+EE+ +I+LH +GN+WA +AA LPGRTDNEIKN WNT KKR P E L
Sbjct: 71 TADEEKLVIQLHAILGNRWAAIAAQLPGRTDNEIKNLWNTHLKKRLLSMGLDPRTHEPLP 130
Query: 193 SECLLNQENADMLTNEASQHDEA 215
S L Q + T +Q + A
Sbjct: 131 SYGLAKQAPSSPTTRHMAQWESA 153
>At3g02940 MYB family transcription factor like protein
Length = 321
Score = 154 bits (389), Expect = 9e-38
Identities = 65/107 (60%), Positives = 84/107 (77%)
Query: 73 EGGLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKG 132
E GLKKGPWT ED+ L+ H++K+G G+W ++ K GL RCGKSCRLRW N+LRPD+++G
Sbjct: 9 ESGLKKGPWTPEEDQKLINHIRKHGHGSWRALPKQAGLNRCGKSCRLRWTNYLRPDIKRG 68
Query: 133 AITAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
TAEEE+ II LH +GNKW+ +A LPGRTDNEIKN+WNT +K+
Sbjct: 69 NFTAEEEQTIINLHSLLGNKWSSIAGHLPGRTDNEIKNYWNTHIRKK 115
>At5g16770 putative transcription factor (MYB9)
Length = 336
Score = 154 bits (388), Expect = 1e-37
Identities = 65/107 (60%), Positives = 83/107 (76%)
Query: 73 EGGLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKG 132
E GLKKGPWT ED+ L+ H+QK+G G+W ++ K GL RCGKSCRLRW N+LRPD+++G
Sbjct: 9 ENGLKKGPWTQEEDDKLIDHIQKHGHGSWRALPKQAGLNRCGKSCRLRWTNYLRPDIKRG 68
Query: 133 AITAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
T EEE+ II LH +GNKW+ +A LPGRTDNEIKN+WNT +K+
Sbjct: 69 NFTEEEEQTIINLHSLLGNKWSSIAGNLPGRTDNEIKNYWNTHLRKK 115
>At4g21440 myb-related protein M4
Length = 350
Score = 154 bits (388), Expect = 1e-37
Identities = 66/107 (61%), Positives = 85/107 (78%)
Query: 73 EGGLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKG 132
+ GLKKGPWT+ ED+ LV ++QK+G GNW ++ K GL RCGKSCRLRW N+LRPD+++G
Sbjct: 9 KNGLKKGPWTSEEDQKLVDYIQKHGYGNWRTLPKNAGLQRCGKSCRLRWTNYLRPDIKRG 68
Query: 133 AITAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
+ EEE II+LH +GNKW+ +AA LPGRTDNEIKNFWNT +K+
Sbjct: 69 RFSFEEEETIIQLHSFLGNKWSAIAARLPGRTDNEIKNFWNTHIRKK 115
>At3g24310 putative transcription factor (MYB71)
Length = 269
Score = 151 bits (381), Expect = 7e-37
Identities = 70/120 (58%), Positives = 87/120 (72%), Gaps = 5/120 (4%)
Query: 68 GGEGG-----EGGLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWA 122
GG GG E G +KGPWTA ED +L+ +VQ +GEG WNSV + GL R GKSCRLRW
Sbjct: 5 GGMGGGWGMVEEGWRKGPWTAEEDRLLIDYVQLHGEGRWNSVARLAGLKRNGKSCRLRWV 64
Query: 123 NHLRPDLRKGAITAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKRGRA 182
N+LRPDL++G IT EE I+ELH K GN+W+ +A LPGRTDNEIKN+W T KK+ ++
Sbjct: 65 NYLRPDLKRGQITPHEETIILELHAKWGNRWSTIARSLPGRTDNEIKNYWRTHFKKKTKS 124
>At4g28110 putative transcription factor (MYB41)
Length = 282
Score = 150 bits (379), Expect = 1e-36
Identities = 98/238 (41%), Positives = 124/238 (51%), Gaps = 47/238 (19%)
Query: 73 EGGLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKG 132
+ G+KKGPWTA ED+ L+ +++ +G GNW ++ K GL RCGKSCRLRW N+LRPD+++G
Sbjct: 9 KNGVKKGPWTAEEDQKLIDYIRFHGPGNWRTLPKNAGLHRCGKSCRLRWTNYLRPDIKRG 68
Query: 133 AITAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR----GRANLPIYP 188
+ EEE II+LH MGNKW+ +AA LPGRTDNEIKN WNT +KR G + P
Sbjct: 69 RFSFEEEETIIQLHSVMGNKWSAIAARLPGRTDNEIKNHWNTHIRKRLVRSGIDPVTHSP 128
Query: 189 E----DLSS--ECLLNQENADMLTNEASQHDEAENKVPEVIFKDYKLRPDILPPCFDKLL 242
DLSS L NQ N + AS L PD+L L
Sbjct: 129 RLDLLDLSSLLSALFNQPNFSAVATHASS----------------LLNPDVL------RL 166
Query: 243 AGLLLRPSKRPWKSDMNLYSYSNNAAAPAAFDQLRKYPMPS--PSSPPWEDINEPHSY 298
A LLL P + P N P+ DQ + P S S P E P +Y
Sbjct: 167 ASLLL-PLQNP------------NPVYPSNLDQNLQTPNTSSESSQPQAETSTVPTNY 211
>At5g10280 putative transcription factor MYB92
Length = 334
Score = 149 bits (375), Expect = 4e-36
Identities = 64/107 (59%), Positives = 82/107 (75%)
Query: 73 EGGLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKG 132
+ GLKKGPWT EDE LV +VQK+G +W ++ K GL RCGKSCRLRW N+LRPD+++G
Sbjct: 9 DSGLKKGPWTPDEDEKLVNYVQKHGHSSWRALPKLAGLNRCGKSCRLRWTNYLRPDIKRG 68
Query: 133 AITAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
+ +EE+ I+ LH +GNKW+ +A LPGRTDNEIKNFWNT KK+
Sbjct: 69 RFSPDEEQTILNLHSVLGNKWSTIANQLPGRTDNEIKNFWNTHLKKK 115
>At4g05100 MYB - like protein
Length = 324
Score = 149 bits (375), Expect = 4e-36
Identities = 64/107 (59%), Positives = 82/107 (75%)
Query: 73 EGGLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKG 132
+ GLKKGPWT ED+ L+ ++ +G GNW ++ K GL RCGKSCRLRW N+LRPD+++G
Sbjct: 10 KNGLKKGPWTPEEDQKLIDYINIHGYGNWRTLPKNAGLQRCGKSCRLRWTNYLRPDIKRG 69
Query: 133 AITAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
+ EEE II+LH MGNKW+ +AA LPGRTDNEIKN+WNT +KR
Sbjct: 70 RFSFEEEETIIQLHSIMGNKWSAIAARLPGRTDNEIKNYWNTHIRKR 116
>At3g23250 myb-related transcription factor like protein
Length = 285
Score = 149 bits (375), Expect = 4e-36
Identities = 68/142 (47%), Positives = 94/142 (65%)
Query: 75 GLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAI 134
GLK+GPWT ED+ILV+ + +G NW ++ K GL RCGKSCRLRW N+L+PD+++G
Sbjct: 11 GLKRGPWTPEEDQILVSFILNHGHSNWRALPKQAGLLRCGKSCRLRWMNYLKPDIKRGNF 70
Query: 135 TAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKRGRANLPIYPEDLSSE 194
T EEE II LH +GN+W+ +AA LPGRTDNEIKN W+T KKR P P+ + +
Sbjct: 71 TKEEEDAIISLHQILGNRWSAIAAKLPGRTDNEIKNVWHTHLKKRLEDYQPAKPKTSNKK 130
Query: 195 CLLNQENADMLTNEASQHDEAE 216
++ ++T+ S E+E
Sbjct: 131 KGTKPKSESVITSSNSTRSESE 152
>At5g40350 putative transcription factor MYB24
Length = 214
Score = 148 bits (374), Expect = 5e-36
Identities = 66/118 (55%), Positives = 90/118 (75%), Gaps = 1/118 (0%)
Query: 62 DEGDTTGGEG-GEGGLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLR 120
++ +++GG G G+ ++KGPWT ED IL+ ++ +GEG WNS+ K GL R GKSCRLR
Sbjct: 2 EKRESSGGSGSGDAEVRKGPWTMEEDLILINYIANHGEGVWNSLAKSAGLKRTGKSCRLR 61
Query: 121 WANHLRPDLRKGAITAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKK 178
W N+LRPD+R+G IT EE+ I+ELH K GN+W+++A LPGRTDNEIKNFW T+ +K
Sbjct: 62 WLNYLRPDVRRGNITPEEQLTIMELHAKWGNRWSKIAKHLPGRTDNEIKNFWRTKIQK 119
>At2g31180 putative transcription factor (MYB14)
Length = 249
Score = 147 bits (372), Expect = 8e-36
Identities = 63/110 (57%), Positives = 82/110 (74%)
Query: 75 GLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAI 134
G+K+GPWT ED+IL+ ++ YG NW ++ K GL RCGKSCRLRW N+LRPD+++G
Sbjct: 11 GVKRGPWTPEEDQILINYIHLYGHSNWRALPKHAGLLRCGKSCRLRWINYLRPDIKRGNF 70
Query: 135 TAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKRGRANL 184
T +EE+ II LH +GN+W+ +AA LPGRTDNEIKN W+T KKR NL
Sbjct: 71 TPQEEQTIINLHESLGNRWSAIAAKLPGRTDNEIKNVWHTHLKKRLSKNL 120
>At1g79180 ranscription factor like protein (MYB63)
Length = 294
Score = 147 bits (372), Expect = 8e-36
Identities = 67/118 (56%), Positives = 87/118 (72%), Gaps = 5/118 (4%)
Query: 76 LKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAIT 135
+K+GPW+ ED L++ +QK+G NW S+ K +GL RCGKSCRLRW N+LRPDL++G T
Sbjct: 14 VKRGPWSPEEDIKLISFIQKFGHENWRSLPKQSGLLRCGKSCRLRWINYLRPDLKRGNFT 73
Query: 136 AEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR-----GRANLPIYP 188
+EEE II+LHH GNKW+++A+ LPGRTDNEIKN W+T KKR G A+ P P
Sbjct: 74 SEEEETIIKLHHNYGNKWSKIASQLPGRTDNEIKNVWHTHLKKRLAQSSGTADEPASP 131
>At5g65230 transcription factor-like protein
Length = 310
Score = 147 bits (371), Expect = 1e-35
Identities = 62/107 (57%), Positives = 82/107 (75%)
Query: 73 EGGLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKG 132
E GLKKGPW ED+ L+ ++ K+G +W+++ K GL RCGKSCRLRW N+LRPD+++G
Sbjct: 9 ETGLKKGPWLPEEDDKLINYIHKHGHSSWSALPKLAGLNRCGKSCRLRWTNYLRPDIKRG 68
Query: 133 AITAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
+AEEE I+ LH +GNKW+ +A+ LPGRTDNEIKNFWNT KK+
Sbjct: 69 KFSAEEEETILNLHAVLGNKWSMIASHLPGRTDNEIKNFWNTHLKKK 115
>At3g27810 ATMYB3 (Atmyb3)
Length = 226
Score = 147 bits (370), Expect = 1e-35
Identities = 67/117 (57%), Positives = 87/117 (74%), Gaps = 2/117 (1%)
Query: 64 GDTTGGEGG--EGGLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRW 121
G ++GG G E ++KGPWT ED IL+ ++ +G+G WNS+ K GL R GKSCRLRW
Sbjct: 6 GGSSGGSGSSAEAEVRKGPWTMEEDLILINYIANHGDGVWNSLAKSAGLKRTGKSCRLRW 65
Query: 122 ANHLRPDLRKGAITAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKK 178
N+LRPD+R+G IT EE+ I+ELH K GN+W+++A LPGRTDNEIKNFW TR +K
Sbjct: 66 LNYLRPDVRRGNITPEEQLIIMELHAKWGNRWSKIAKHLPGRTDNEIKNFWRTRIQK 122
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.312 0.131 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,954,539
Number of Sequences: 26719
Number of extensions: 532373
Number of successful extensions: 1751
Number of sequences better than 10.0: 206
Number of HSP's better than 10.0 without gapping: 175
Number of HSP's successfully gapped in prelim test: 31
Number of HSP's that attempted gapping in prelim test: 1425
Number of HSP's gapped (non-prelim): 257
length of query: 416
length of database: 11,318,596
effective HSP length: 102
effective length of query: 314
effective length of database: 8,593,258
effective search space: 2698283012
effective search space used: 2698283012
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC148404.8


