

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148404.4 - phase: 0
(385 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
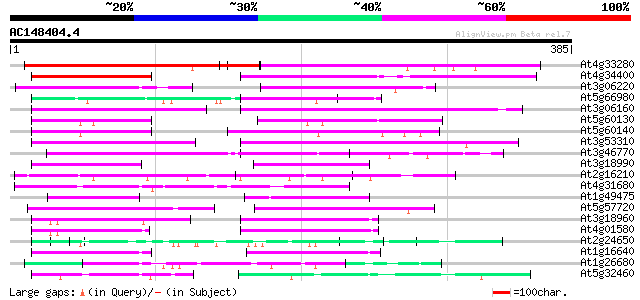
Score E
Sequences producing significant alignments: (bits) Value
At4g33280 hypothetical protein 126 2e-29
At4g34400 putative protein 91 1e-18
At3g06220 hypothetical protein 77 2e-14
At5g66980 putative protein 73 2e-13
At3g06160 hypothetical protein 72 4e-13
At5g60130 putative protein 69 3e-12
At5g60140 putative protein 67 1e-11
At3g53310 unknown protein 67 2e-11
At3g46770 putative protein 65 6e-11
At3g18990 unknown protein 64 1e-10
At2g16210 unknown protein 62 4e-10
At4g31680 putative protein 62 7e-10
At1g49475 putative protein 61 1e-09
At5g57720 putative protein 59 4e-09
At3g18960 unknown protein 57 2e-08
At4g01580 hypothetical protein 57 2e-08
At2g24650 hypothetical protein 56 3e-08
At1g16640 Unknown protein 54 2e-07
At1g26680 unknown protein 53 2e-07
At5g32460 meristem protein - like 51 9e-07
>At4g33280 hypothetical protein
Length = 461
Score = 126 bits (317), Expect = 2e-29
Identities = 70/147 (47%), Positives = 94/147 (63%), Gaps = 8/147 (5%)
Query: 11 NLLQALPKTFSDNLKKKLPGNVTLKGPSGVVWNVGLTARDD-TIYFTNGWQRFVKDHSLK 69
NLLQ +P+ FS + K+KLP VTLK PSGV +NVG+ D+ T+ F GW +FVKDHSL+
Sbjct: 103 NLLQVIPRKFSTHCKRKLPQIVTLKSPSGVTYNVGVEEDDEKTMAFRFGWDKFVKDHSLE 162
Query: 70 ENDFLVFKYNGESLFEVLIFHGDSFCEKAASYFVGKCGQAH-TEQVCNKGKSSNNS---- 124
END LVFK++G S FEVL+F G + CEK SYFV KCG A T+ + + +S+ S
Sbjct: 163 ENDLLVFKFHGVSEFEVLVFDGQTLCEKPTSYFVRKCGHAEKTKGIIDFNATSSRSPKRH 222
Query: 125 --VEEVSTPSNGSVECSLPEKSWEEDI 149
++V T N + S P + ED+
Sbjct: 223 FNPDDVETTPNQQLVISPPVDNELEDL 249
Score = 113 bits (283), Expect = 2e-25
Identities = 69/212 (32%), Positives = 112/212 (52%), Gaps = 20/212 (9%)
Query: 173 LPKTFSDNLNKKLPENVTLKGPGGVAWKIGLITRDD-TVYFLNGWQRFVNDHSLKDNDFL 231
+P+ FS + +KLP+ VTLK P GV + +G+ D+ T+ F GW +FV DHSL++ND L
Sbjct: 108 IPRKFSTHCKRKLPQIVTLKSPSGVTYNVGVEEDDEKTMAFRFGWDKFVKDHSLEENDLL 167
Query: 232 VFNYNGESHFDVLIFDGESFCEKEASYFVGKCSHTQTELG-------GSKANETNNSIEE 284
VF ++G S F+VL+FDG++ CEK SYFV KC H + G S++ + + + ++
Sbjct: 168 VFKFHGVSEFEVLVFDGQTLCEKPTSYFVRKCGHAEKTKGIIDFNATSSRSPKRHFNPDD 227
Query: 285 VNTASS-----DGGVECGLYEKFQ-----DLNNIGTPLAVPVET--TNEKTFNAGVESDS 332
V T + V+ L + D++ I PL V T E+ N+ +++ S
Sbjct: 228 VETTPNQQLVISPPVDNELEDLIDIDLDFDIDKILNPLLVASHTGYEQEEHINSDIDTAS 287
Query: 333 PELLIADTVTKTTAIQFPYQPTGKRSKRRRSS 364
+L + + + Y +G + RR S
Sbjct: 288 AQLPVISPTSTVRVSEGKYPLSGFKKMRRELS 319
Score = 44.7 bits (104), Expect = 9e-05
Identities = 17/27 (62%), Positives = 21/27 (76%)
Query: 145 WEEDIYWTHFQFIHFTQLLPEDFKQQL 171
WEE++YWTHFQ +HFTQLL F +L
Sbjct: 11 WEEELYWTHFQTLHFTQLLLPGFHNRL 37
Score = 33.5 bits (75), Expect = 0.20
Identities = 17/39 (43%), Positives = 25/39 (63%), Gaps = 4/39 (10%)
Query: 215 GWQRFVNDHSLKDNDFLVFN-YNGES---HFDVLIFDGE 249
GW++FV D++L++ D VF N E+ H +V IF GE
Sbjct: 407 GWKKFVQDNNLREGDVCVFEPANSETKPLHLNVYIFRGE 445
Score = 31.2 bits (69), Expect = 1.0
Identities = 16/41 (39%), Positives = 26/41 (63%), Gaps = 4/41 (9%)
Query: 56 TNGWQRFVKDHSLKENDFLVFK-YNGESL---FEVLIFHGD 92
+ GW++FV+D++L+E D VF+ N E+ V IF G+
Sbjct: 405 STGWKKFVQDNNLREGDVCVFEPANSETKPLHLNVYIFRGE 445
>At4g34400 putative protein
Length = 389
Score = 90.5 bits (223), Expect = 1e-18
Identities = 58/203 (28%), Positives = 97/203 (47%), Gaps = 13/203 (6%)
Query: 159 FTQLLPEDFKQQLALPKTFSDNLNKKLPENVTLKGPGGVAWKIGLITRDDTVYFLNGWQR 218
FT + + + +P ++ D++ + P+ V L+GPGG +WK+ +DD V F GW +
Sbjct: 17 FTVFVSHFSSEFMVIPVSYYDHIPHRFPKTVILRGPGGCSWKVATEIKDDEVLFSQGWPK 76
Query: 219 FVNDHSLKDNDFLVFNYNGESHFDVLIFDGESFCEKEASYFVGKCSHTQTELGGSKANET 278
FV D++L D DFL F YNG F+V IF G C KE S TEL E
Sbjct: 77 FVRDNTLNDGDFLTFAYNGAHIFEVSIFRGYDAC-KEIS--------EVTEL----EEEE 123
Query: 279 NNSIEEVNTASSDGGVECGLYEKFQDLNNIGTPLAVPVETTNEKTFNAGVESDSPELLIA 338
+S+ +++ +D G + + + + G VE +++ V S+S E
Sbjct: 124 EDSVISLSSEDTDTGAKSEMKNTVPEGRDKGKSKVEVVEDSDDDEEEDSVYSESSEETET 183
Query: 339 DTVTKTTAIQFPYQPTGKRSKRR 361
DT ++ + + K+ K++
Sbjct: 184 DTDSEFKVAKPTIPKSQKKGKKK 206
Score = 82.0 bits (201), Expect = 5e-16
Identities = 33/82 (40%), Positives = 51/82 (61%)
Query: 16 LPKTFSDNLKKKLPGNVTLKGPSGVVWNVGLTARDDTIYFTNGWQRFVKDHSLKENDFLV 75
+P ++ D++ + P V L+GP G W V +DD + F+ GW +FV+D++L + DFL
Sbjct: 31 IPVSYYDHIPHRFPKTVILRGPGGCSWKVATEIKDDEVLFSQGWPKFVRDNTLNDGDFLT 90
Query: 76 FKYNGESLFEVLIFHGDSFCEK 97
F YNG +FEV IF G C++
Sbjct: 91 FAYNGAHIFEVSIFRGYDACKE 112
>At3g06220 hypothetical protein
Length = 175
Score = 77.0 bits (188), Expect = 2e-14
Identities = 36/121 (29%), Positives = 69/121 (56%), Gaps = 8/121 (6%)
Query: 5 LNWKSINLLQALPKTFSDNLKKKLPGNVTLKGPSGVVWNVGLTARDDTIYFTNGWQRFVK 64
L+ S N L+ +P+++ ++L ++LP L G G VW V + ++ + +Y GW+ FV
Sbjct: 15 LSCVSSNSLKLIPRSYYEHLPRRLPKTAILTGTGGRVWKVAMMSKREQVYLARGWENFVA 74
Query: 65 DHSLKENDFLVFKYNGESLFEVLIFHGDSFCEKAASYFVGKCGQAHTEQVCNKGKSSNNS 124
D+ LK+ +FL F ++G +EV I+ G C++ + H E++ ++ +S N+S
Sbjct: 75 DNELKDGEFLTFVFDGYKSYEVSIY-GRGSCKETRAV-------VHVEEISDESESDNDS 126
Query: 125 V 125
+
Sbjct: 127 L 127
Score = 71.6 bits (174), Expect = 7e-13
Identities = 38/123 (30%), Positives = 68/123 (54%), Gaps = 4/123 (3%)
Query: 173 LPKTFSDNLNKKLPENVTLKGPGGVAWKIGLITRDDTVYFLNGWQRFVNDHSLKDNDFLV 232
+P+++ ++L ++LP+ L G GG WK+ ++++ + VY GW+ FV D+ LKD +FL
Sbjct: 26 IPRSYYEHLPRRLPKTAILTGTGGRVWKVAMMSKREQVYLARGWENFVADNELKDGEFLT 85
Query: 233 FNYNGESHFDVLIFDGESFCEKEASYFVGKC---SHTQTELGGSKANETNNSIEEVNTAS 289
F ++G ++V I+ S E A V + S + + GS + T EE N+
Sbjct: 86 FVFDGYKSYEVSIYGRGSCKETRAVVHVEEISDESESDNDSLGSLVDVTPMPAEE-NSDD 144
Query: 290 SDG 292
++G
Sbjct: 145 TEG 147
>At5g66980 putative protein
Length = 334
Score = 73.2 bits (178), Expect = 2e-13
Identities = 41/101 (40%), Positives = 53/101 (51%), Gaps = 5/101 (4%)
Query: 159 FTQLLPEDFKQQLALPKTFSDNLNKKLPENVTLKGPGGVAWKIGLITRDDT----VYFLN 214
F LPE +L +P F D L K LP+ L G W + T D V+F
Sbjct: 11 FKVFLPEFGSHELVIPPAFIDMLEKPLPKEAFLVDEIGRLWCVETKTEDTEERFCVFFTK 70
Query: 215 GWQRFVNDHSLKDNDFLVFNYNGESHFDVLIFDGESFCEKE 255
GWQ F ND SL+ DFLVF+Y+G+S F V IF + C+K+
Sbjct: 71 GWQSFANDQSLEFGDFLVFSYDGDSRFSVTIFANDG-CKKD 110
Score = 70.5 bits (171), Expect = 1e-12
Identities = 70/268 (26%), Positives = 100/268 (37%), Gaps = 62/268 (23%)
Query: 16 LPKTFSDNLKKKLPGNVTLKGPSGVVWNVGLTARDDT-----IYFTNGWQRFVKDHSLKE 70
+P F D L+K LP L G +W V T +DT ++FT GWQ F D SL+
Sbjct: 25 IPPAFIDMLEKPLPKEAFLVDEIGRLWCVE-TKTEDTEERFCVFFTKGWQSFANDQSLEF 83
Query: 71 NDFLVFKYNGESLFEVLIFHGDSFCEKAASYFVG-------------------------- 104
DFLVF Y+G+S F V IF D C+K
Sbjct: 84 GDFLVFSYDGDSRFSVTIFANDG-CKKDVGVVSTTDRSRVSLDEEEPDDIFTKPDRMRDC 142
Query: 105 KCGQA--------------------HTEQVCNKGKSSNNSVEEVSTPSNGSVECSL---P 141
CGQ+ E V +S + T + C + P
Sbjct: 143 DCGQSINRKRKRDSVNEDPHVLVDDKPEYVSTYKTKPEHSEKTQRTVNRAGDTCDISWFP 202
Query: 142 EK---SWEEDIYWTHFQFIHFTQLLPEDFKQQLALPKTFSDNLNKKLPENVTLKGPGGVA 198
EK +EE +Y + HF + + Q+L LP TF + +L E++ L G
Sbjct: 203 EKKHNGFEESVYKP--KHPHFVRNITRGSLQKLELPLTFLRSNGIELEEDIELCDESGKK 260
Query: 199 WKIGLITRDDTVYFLN-GWQRFVNDHSL 225
W + ++ D F + W F H +
Sbjct: 261 WPLKILNHDRGFKFSHESWLCFCKSHEM 288
>At3g06160 hypothetical protein
Length = 330
Score = 72.4 bits (176), Expect = 4e-13
Identities = 49/194 (25%), Positives = 88/194 (45%), Gaps = 5/194 (2%)
Query: 159 FTQLLPEDFKQQLALPKTFSDNLNKKLPENVTLKGPGGVAWKIGLITRDDTVYFLNGWQR 218
FT L + + +P+++ + L + LP+ L G GG WK+ + ++ + VYF GW
Sbjct: 26 FTVFLSHCSSESMVIPRSYYNLLPRPLPKTAILIGTGGRFWKVAMTSKQEQVYFEQGWGN 85
Query: 219 FVNDHSLKDNDFLVFNYNGESHFDVLIFDGESFCEKEASYFVGKCSHTQTELGGSKANET 278
FV D+ LK+ +FL F ++G ++V I+ E A V + S + S + +
Sbjct: 86 FVADNQLKEGEFLTFVFDGHKSYEVSIYGRGDCKETRAVIQVEEISDDTEDDNVSLHSPS 145
Query: 279 NNSIEEVNTASSDGGVECGLYEKFQDLNNIGTPLAVPVETTNEKTFNAGVESDSPELLIA 338
N S++ ++ S L D + + E + E +A + +S E+
Sbjct: 146 NVSLDSLSNDSHHSTSNVSLRSLSNDSLHGDAEIESDSEYSPENLPSASISVESVEV--- 202
Query: 339 DTVTKTTAIQFPYQ 352
V TT+ Q Y+
Sbjct: 203 --VNPTTSRQRSYK 214
Score = 68.2 bits (165), Expect = 7e-12
Identities = 36/120 (30%), Positives = 60/120 (50%)
Query: 16 LPKTFSDNLKKKLPGNVTLKGPSGVVWNVGLTARDDTIYFTNGWQRFVKDHSLKENDFLV 75
+P+++ + L + LP L G G W V +T++ + +YF GW FV D+ LKE +FL
Sbjct: 40 IPRSYYNLLPRPLPKTAILIGTGGRFWKVAMTSKQEQVYFEQGWGNFVADNQLKEGEFLT 99
Query: 76 FKYNGESLFEVLIFHGDSFCEKAASYFVGKCGQAHTEQVCNKGKSSNNSVEEVSTPSNGS 135
F ++G +EV I+ E A V + + + SN S++ +S S+ S
Sbjct: 100 FVFDGHKSYEVSIYGRGDCKETRAVIQVEEISDDTEDDNVSLHSPSNVSLDSLSNDSHHS 159
>At5g60130 putative protein
Length = 300
Score = 69.3 bits (168), Expect = 3e-12
Identities = 41/132 (31%), Positives = 64/132 (48%), Gaps = 6/132 (4%)
Query: 171 LALPKTFSDNLNKKLPENVTLKGPGGVAWKIGL---ITRDDTVYFL--NGWQRFVNDHSL 225
+ LP +F+ L K LPE VT++ G WK+ L DDT F+ NGW+R V D L
Sbjct: 1 MVLPISFTRFLPKSLPETVTVRSISGNIWKLELKKCCGDDDTEKFVMVNGWKRIVKDEDL 60
Query: 226 KDNDFLVFNYNGESHFDVLIFDGESFCEKEASYFVGKCSHTQTELGGSKANETNNSIEEV 285
K DFL F ++G S F I++ + C K+ + + E + N+ ++ + +
Sbjct: 61 KGGDFLEFEFDGSSCFHFCIYEHRTMC-KKIRRSSDQSEEIKVESDSDEQNQASDDVLSL 119
Query: 286 NTASSDGGVECG 297
+ D CG
Sbjct: 120 DEDDDDSDYNCG 131
Score = 65.1 bits (157), Expect = 6e-11
Identities = 36/87 (41%), Positives = 49/87 (55%), Gaps = 5/87 (5%)
Query: 16 LPKTFSDNLKKKLPGNVTLKGPSGVVWNVGLT---ARDDTIYFT--NGWQRFVKDHSLKE 70
LP +F+ L K LP VT++ SG +W + L DDT F NGW+R VKD LK
Sbjct: 3 LPISFTRFLPKSLPETVTVRSISGNIWKLELKKCCGDDDTEKFVMVNGWKRIVKDEDLKG 62
Query: 71 NDFLVFKYNGESLFEVLIFHGDSFCEK 97
DFL F+++G S F I+ + C+K
Sbjct: 63 GDFLEFEFDGSSCFHFCIYEHRTMCKK 89
>At5g60140 putative protein
Length = 328
Score = 67.4 bits (163), Expect = 1e-11
Identities = 48/166 (28%), Positives = 75/166 (44%), Gaps = 20/166 (12%)
Query: 150 YWTHFQFIHFTQLLPEDFKQQLALPKTFSDNLNKKLPENVTLKGPGGVAWKIGLITRDDT 209
Y F F LP + L LP +F+ L K LP+ VT++ G WK+G
Sbjct: 7 YSNGFTSKFFKPYLPSESGDDLVLPISFNSCLPKSLPKTVTVRSISGNIWKLGFKKCGGE 66
Query: 210 V---YFLNGWQRFVNDHSLKDNDFLVFNYNGESHFDVLIFDGESFCEK-----EASYFVG 261
V ++GW++ V D +L D L F ++G F+ IFD E+ C++ E S +
Sbjct: 67 VERFVMVSGWKKIVRDENLNGGDLLSFEFDGSHFFNFSIFDHETTCKRLKRSSEQSKDII 126
Query: 262 KCSHTQTE----------LGGSKANETNN--SIEEVNTASSDGGVE 295
K E L +++++N S+E+ N A D G+E
Sbjct: 127 KVGSDCEEESQASDDVIVLNSDDSDDSDNDYSVEDDNVAEDDDGLE 172
Score = 55.5 bits (132), Expect = 5e-08
Identities = 26/85 (30%), Positives = 45/85 (52%), Gaps = 3/85 (3%)
Query: 16 LPKTFSDNLKKKLPGNVTLKGPSGVVWNVGLT---ARDDTIYFTNGWQRFVKDHSLKEND 72
LP +F+ L K LP VT++ SG +W +G + +GW++ V+D +L D
Sbjct: 30 LPISFNSCLPKSLPKTVTVRSISGNIWKLGFKKCGGEVERFVMVSGWKKIVRDENLNGGD 89
Query: 73 FLVFKYNGESLFEVLIFHGDSFCEK 97
L F+++G F IF ++ C++
Sbjct: 90 LLSFEFDGSHFFNFSIFDHETTCKR 114
>At3g53310 unknown protein
Length = 286
Score = 66.6 bits (161), Expect = 2e-11
Identities = 48/205 (23%), Positives = 85/205 (41%), Gaps = 14/205 (6%)
Query: 159 FTQLLPEDFKQQLALPKTFSDNLNKKLPENVTLKGPGGVAWKIGLITRDDTVYFLNGWQR 218
F L E + L +P F L LP+ V L+G GG W + L Y GW +
Sbjct: 12 FKVFLVESASESLMIPLPFMAFLADPLPKTVKLQGLGGKLWTVSLKKISGAAYLTRGWPK 71
Query: 219 FVNDHSLKDNDFLVFNYNGESHFDVLIFDGESFCEKEASYFVGKCSHTQTELGGSKANET 278
F +H LK+ +F+ F Y+G F+V +FD E A S + ++ ++
Sbjct: 72 FAEEHELKNGEFMTFVYDGHRTFEVSVFDRWGSKEVRAEIQAIPLSDSDSDSVVEDEKDS 131
Query: 279 NNSIEEVNTASSDGGV-ECGLYEKFQDLNNIGTPL-------------AVPVETTNEKTF 324
+ +E+ + D + G +++ ++++ P+ + VE F
Sbjct: 132 TDVVEDDDDEDEDEDEDDDGSFDEDEEISQSLYPIDEETATDAAVFEGNLDVEALTNPHF 191
Query: 325 NAGVESDSPELLIADTVTKTTAIQF 349
+++ ELLI V K ++F
Sbjct: 192 PTTLKNRIYELLIPANVVKDNNLEF 216
Score = 63.9 bits (154), Expect = 1e-10
Identities = 35/112 (31%), Positives = 50/112 (44%)
Query: 16 LPKTFSDNLKKKLPGNVTLKGPSGVVWNVGLTARDDTIYFTNGWQRFVKDHSLKENDFLV 75
+P F L LP V L+G G +W V L Y T GW +F ++H LK +F+
Sbjct: 26 IPLPFMAFLADPLPKTVKLQGLGGKLWTVSLKKISGAAYLTRGWPKFAEEHELKNGEFMT 85
Query: 76 FKYNGESLFEVLIFHGDSFCEKAASYFVGKCGQAHTEQVCNKGKSSNNSVEE 127
F Y+G FEV +F E A + ++ V K S + VE+
Sbjct: 86 FVYDGHRTFEVSVFDRWGSKEVRAEIQAIPLSDSDSDSVVEDEKDSTDVVED 137
>At3g46770 putative protein
Length = 276
Score = 65.1 bits (157), Expect = 6e-11
Identities = 51/210 (24%), Positives = 94/210 (44%), Gaps = 5/210 (2%)
Query: 26 KKLPGNVTLKG-PSGVVWNVGLTARDDTIYFTNGWQRFVKDHSLKENDFLVFKYNGESLF 84
K LP VT+K SG +W + + A +T++ +GW++ VKD ++ E FL F+++G +F
Sbjct: 48 KPLPRKVTVKSVSSGNIWRMEMKANGNTVFLRDGWKKIVKDENVTEPIFLEFEFDGYGVF 107
Query: 85 EVLIFHGDSFCEKAASYFVGKCGQAHTEQVCNKGKSSNNSVEEVSTPSNGSVECSLPEKS 144
++ S C++ S + + +E+ G + E S G+ K+
Sbjct: 108 HFCVYEYGSMCKRMRSPMEKEVIKVDSEEDVLVGNEESTKGLEESPRRGGTSRRRAKLKT 167
Query: 145 WEEDIYWTHFQFIHFTQLLPEDFKQ-QLALPKTFSDNLNKKLPENVTLKGPGGVAWKIGL 203
IY H P D Q + +P + N P V ++ G+ + +
Sbjct: 168 KSHKIY-EHLD-NKLNPSFPVDMTQNRTRIPSLLIKDYNLTFPNMVIMRDKIGILKRRIV 225
Query: 204 ITRDDTVYFLNGWQRFVNDHSLKDNDFLVF 233
I ++ +VY LNG + + +K + +VF
Sbjct: 226 IWKNRSVY-LNGIGSIIRRNHVKPGNEVVF 254
Score = 53.5 bits (127), Expect = 2e-07
Identities = 46/189 (24%), Positives = 83/189 (43%), Gaps = 18/189 (9%)
Query: 159 FTQLLPEDFKQQLALPKTFSDNLNKKLPENVTLKG-PGGVAWKIGLITRDDTVYFLNGWQ 217
F +P + LP K LP VT+K G W++ + +TV+ +GW+
Sbjct: 24 FRVYIPNQTADDMNLPLVSDKISGKPLPRKVTVKSVSSGNIWRMEMKANGNTVFLRDGWK 83
Query: 218 RFVNDHSLKDNDFLVFNYNGESHFDVLIFDGESFCEKEASYF---VGKCSHTQTELGGSK 274
+ V D ++ + FL F ++G F +++ S C++ S V K + L G++
Sbjct: 84 KIVKDENVTEPIFLEFEFDGYGVFHFCVYEYGSMCKRMRSPMEKEVIKVDSEEDVLVGNE 143
Query: 275 ANETNNSIEEV----NTASSDGGVECGLYEKFQDLNNIGTPLAVPVETTNEKTFNAGVES 330
E+ +EE T+ ++ ++ ++ L+N P + PV+ T +T
Sbjct: 144 --ESTKGLEESPRRGGTSRRRAKLKTKSHKIYEHLDNKLNP-SFPVDMTQNRT------- 193
Query: 331 DSPELLIAD 339
P LLI D
Sbjct: 194 RIPSLLIKD 202
>At3g18990 unknown protein
Length = 341
Score = 64.3 bits (155), Expect = 1e-10
Identities = 31/75 (41%), Positives = 42/75 (55%)
Query: 16 LPKTFSDNLKKKLPGNVTLKGPSGVVWNVGLTARDDTIYFTNGWQRFVKDHSLKENDFLV 75
+P F K +L V L P G VW VGL D+ I+F +GWQ FV +S++ L+
Sbjct: 22 VPDKFVSKFKDELSVAVALTVPDGHVWRVGLRKADNKIWFQDGWQEFVDRYSIRIGYLLI 81
Query: 76 FKYNGESLFEVLIFH 90
F+Y G S F V IF+
Sbjct: 82 FRYEGNSAFSVYIFN 96
Score = 61.6 bits (148), Expect = 7e-10
Identities = 28/80 (35%), Positives = 45/80 (56%)
Query: 168 KQQLALPKTFSDNLNKKLPENVTLKGPGGVAWKIGLITRDDTVYFLNGWQRFVNDHSLKD 227
+++L +P F +L V L P G W++GL D+ ++F +GWQ FV+ +S++
Sbjct: 17 EKRLRVPDKFVSKFKDELSVAVALTVPDGHVWRVGLRKADNKIWFQDGWQEFVDRYSIRI 76
Query: 228 NDFLVFNYNGESHFDVLIFD 247
L+F Y G S F V IF+
Sbjct: 77 GYLLIFRYEGNSAFSVYIFN 96
>At2g16210 unknown protein
Length = 291
Score = 62.4 bits (150), Expect = 4e-10
Identities = 44/157 (28%), Positives = 79/157 (50%), Gaps = 14/157 (8%)
Query: 156 FIHFTQLLPEDFKQQLALPKTFSDNL-NKKLPENVTLKGPGGVAWKIGLITRDDTVYFL- 213
F Q + + + ALP FS + +K+L + ++ G +W++G I+++ YF+
Sbjct: 19 FFKVVQSINVSSENKRALPHDFSRSFTDKELSRKMKIRAQWGNSWEVG-ISKNPRFYFME 77
Query: 214 -NGWQRFVNDHSLKDNDFLVFNYNGESHFDVLIF--DGESFCE-KEASYFVGKCSHTQTE 269
+GW++FV D++L +++ L F + G+ HF V IF DG+ + ++ F S +TE
Sbjct: 78 KSGWEKFVRDNALGNSELLTFTHKGKMHFTVNIFKLDGKEMMQPPQSRSFFASSSRIKTE 137
Query: 270 LGGSKANETNNSIEEVNTASSDGGVECGLYEKFQDLN 306
+ N I+E SS+ G K + LN
Sbjct: 138 -------QEENDIKEEVVVSSNRGQTTAAESKGRKLN 167
Score = 50.4 bits (119), Expect = 2e-06
Identities = 63/262 (24%), Positives = 113/262 (43%), Gaps = 38/262 (14%)
Query: 4 SLNWKSINLLQALPKTFSDNLK-KKLPGNVTLKGPSGVVWNVGLTARDDTIYFT--NGWQ 60
S+N S N +ALP FS + K+L + ++ G W VG++ ++ YF +GW+
Sbjct: 25 SINVSSENK-RALPHDFSRSFTDKELSRKMKIRAQWGNSWEVGIS-KNPRFYFMEKSGWE 82
Query: 61 RFVKDHSLKENDFLVFKYNGESLFEVLIFHGDS---FCEKAASYFVGKCGQAHTEQVCNK 117
+FV+D++L ++ L F + G+ F V IF D + F + TEQ N
Sbjct: 83 KFVRDNALGNSELLTFTHKGKMHFTVNIFKLDGKEMMQPPQSRSFFASSSRIKTEQEEND 142
Query: 118 GKS----SNNSVEEVSTPSNGSVECSLPEKSWEEDIYWTHFQFI---------------- 157
K S+N + + S G + +L +++ +E + +
Sbjct: 143 IKEEVVVSSNRGQTTAAESKGR-KLNLGKRAAKESQSSKRTEKVVRARSDYAGASSSTAA 201
Query: 158 HFTQLLPEDFKQQLALPKTFSDNLNKKLPENVT---LKGPGG-VAWKIGLITRDDTVYFL 213
FT L + + L +P + S + ++P+ T + P G +W + + R F
Sbjct: 202 AFTILFKQGYLVFLRIPNSVSKD---QVPDEKTVFKIHHPNGKKSWNVVYLERFGA--FS 256
Query: 214 NGWQRFVNDHSLKDNDFLVFNY 235
GW+R V ++ L D F +
Sbjct: 257 GGWRRVVKEYPLAVGDTCKFTF 278
>At4g31680 putative protein
Length = 478
Score = 61.6 bits (148), Expect = 7e-10
Identities = 61/238 (25%), Positives = 105/238 (43%), Gaps = 27/238 (11%)
Query: 4 SLNWKSINLLQALP-KTFSDNLKKKLPGN-VTLKGPSGV-VWNVGLTARDDTIYFTNGWQ 60
+L+ S++LLQ +P K FS++++ K G V L+ + W V + D T GW+
Sbjct: 25 NLSHGSVSLLQNIPVKFFSEHIEGKHEGETVKLRADASKRTWEVKM----DGNRLTEGWK 80
Query: 61 RFVKDHSLKENDFLVFKYNGESLFEVLIFHGDSFCE----KAASYFVGKCGQAHTEQVCN 116
FV+ H L+ DF+VF++ G+ +F V G S CE ++ S+ G+ + +
Sbjct: 81 EFVEAHDLRIRDFVVFRHEGDMVFHVTAL-GPSCCEIQYPQSISHEEGE-ESGEVGMIES 138
Query: 117 KGKSSNNSVEEVSTPSNGSVECSLPEKSWEEDIYWTHFQFIHFTQLLPEDFKQQLALPKT 176
K S VEE L + + + TH + LP DF ++ L K
Sbjct: 139 SFKISEREVEENLQKEYDQSSADL--NCFSQSV--THSNISNDLVTLPRDFAKRNGLDKG 194
Query: 177 FSDNLNKKLPENVTLKGPGGVAWKIGLITR-DDTVYFLNGWQRFVNDHSLKDNDFLVF 233
+ + L G +W+ + +R V+ + GW+ +++ LK D F
Sbjct: 195 MHE---------IVLMNEEGKSWESEVRSRMSGQVFIVGGWKSLCSENKLKGGDSCTF 243
Score = 37.7 bits (86), Expect = 0.011
Identities = 30/100 (30%), Positives = 45/100 (45%), Gaps = 10/100 (10%)
Query: 175 KTFSDNLNKKLP-ENVTLKGPGGV-AWKIGLITRDDTVYFLNGWQRFVNDHSLKDNDFLV 232
K FS+++ K E V L+ W++ + D GW+ FV H L+ DF+V
Sbjct: 40 KFFSEHIEGKHEGETVKLRADASKRTWEVKM----DGNRLTEGWKEFVEAHDLRIRDFVV 95
Query: 233 FNYNGESHFDVLIFDGESFCEKEASYFVGKCSHTQTELGG 272
F + G+ F V G S CE + + SH + E G
Sbjct: 96 FRHEGDMVFHVTAL-GPSCCEIQ---YPQSISHEEGEESG 131
>At1g49475 putative protein
Length = 250
Score = 60.8 bits (146), Expect = 1e-09
Identities = 34/86 (39%), Positives = 45/86 (51%), Gaps = 1/86 (1%)
Query: 162 LLPEDFKQQLALPKTFSDNLNKKLPENVTLKGPGGVAWKIGLITRDDTVYFLNGWQRFVN 221
LL ++ + +P F KL +NVTL+ P G I + D V+F GW F
Sbjct: 39 LLSRIIEKMMKVPARFV-RFGPKLTDNVTLQTPVGFKRSIRIKRIGDEVWFEKGWSEFAE 97
Query: 222 DHSLKDNDFLVFNYNGESHFDVLIFD 247
HSL D FL F+Y G+S F V+IFD
Sbjct: 98 AHSLSDGHFLFFHYEGDSCFRVVIFD 123
Score = 56.6 bits (135), Expect = 2e-08
Identities = 26/63 (41%), Positives = 36/63 (56%)
Query: 27 KLPGNVTLKGPSGVVWNVGLTARDDTIYFTNGWQRFVKDHSLKENDFLVFKYNGESLFEV 86
KL NVTL+ P G ++ + D ++F GW F + HSL + FL F Y G+S F V
Sbjct: 60 KLTDNVTLQTPVGFKRSIRIKRIGDEVWFEKGWSEFAEAHSLSDGHFLFFHYEGDSCFRV 119
Query: 87 LIF 89
+IF
Sbjct: 120 VIF 122
>At5g57720 putative protein
Length = 300
Score = 58.9 bits (141), Expect = 4e-09
Identities = 40/129 (31%), Positives = 54/129 (41%), Gaps = 4/129 (3%)
Query: 13 LQALPKTFSDNLKKKLPGNVTLKGPSGVVWNVGLTARDDTIY-FTNGWQRFVKDHSLKEN 71
L +P++++ LP LK P G WNV T + I GW +FVKD+ L +
Sbjct: 25 LLVIPRSYNRYYPNPLPQTAVLKNPEGRFWNVQWTKSQEVIISLQEGWVKFVKDNGLIDR 84
Query: 72 DFLVFKYNGESLFEVLIFHGDSFCEKAASYFVGKCGQAHTEQVCNKGKSSNNSVEEVSTP 131
DFL+F Y+G F V I E A + + E G + EE T
Sbjct: 85 DFLLFTYDGSRSFWVRIHRNGLPLEPTAPIKIQEISDDEDE---TNGDGDPHMEEEGDTD 141
Query: 132 SNGSVECSL 140
N V SL
Sbjct: 142 ENMIVSLSL 150
Score = 57.0 bits (136), Expect = 2e-08
Identities = 37/131 (28%), Positives = 57/131 (43%), Gaps = 8/131 (6%)
Query: 169 QQLALPKTFSDNLNKKLPENVTLKGPGGVAWKIGLITRDDTVYFLN-GWQRFVNDHSLKD 227
Q L +P++++ LP+ LK P G W + + + L GW +FV D+ L D
Sbjct: 24 QLLVIPRSYNRYYPNPLPQTAVLKNPEGRFWNVQWTKSQEVIISLQEGWVKFVKDNGLID 83
Query: 228 NDFLVFNYNGESHFDVLIFDGESFCEKEASYFVGKCSHTQTELGG-------SKANETNN 280
DFL+F Y+G F V I E A + + S + E G + + N
Sbjct: 84 RDFLLFTYDGSRSFWVRIHRNGLPLEPTAPIKIQEISDDEDETNGDGDPHMEEEGDTDEN 143
Query: 281 SIEEVNTASSD 291
I ++ SSD
Sbjct: 144 MIVSLSLGSSD 154
>At3g18960 unknown protein
Length = 209
Score = 57.0 bits (136), Expect = 2e-08
Identities = 34/96 (35%), Positives = 48/96 (49%), Gaps = 2/96 (2%)
Query: 159 FTQLLPEDFKQQLA-LPKTFSDNLNKKLPENVTLKGPGGVAWKIGLITRDDTVYFLNGWQ 217
F +LP K ++ +P F KL E VTL+ P G I L + ++F GW
Sbjct: 31 FKLVLPSTMKDKMMKIPPRFVKLQGSKLSEVVTLETPAGFKRSIKLKRIGEEIWFHEGWS 90
Query: 218 RFVNDHSLKDNDFLVFNYNGESHFDVLIFDGESFCE 253
F HS+++ FL+F Y S F V+IF+ S CE
Sbjct: 91 EFAEAHSIEEGHFLLFEYKENSSFRVIIFN-VSACE 125
Score = 56.2 bits (134), Expect = 3e-08
Identities = 40/138 (28%), Positives = 58/138 (41%), Gaps = 29/138 (21%)
Query: 16 LPKTFSDNLKK------KLPGN-----VTLKGPSGVVWNVGLTARDDTIYFTNGWQRFVK 64
LP T D + K KL G+ VTL+ P+G ++ L + I+F GW F +
Sbjct: 35 LPSTMKDKMMKIPPRFVKLQGSKLSEVVTLETPAGFKRSIKLKRIGEEIWFHEGWSEFAE 94
Query: 65 DHSLKENDFLVFKYNGESLFEVLIFH------------------GDSFCEKAASYFVGKC 106
HS++E FL+F+Y S F V+IF+ D + FVG
Sbjct: 95 AHSIEEGHFLLFEYKENSSFRVIIFNVSACETKYPLDAVHIIDSDDDIIDITGKEFVGTQ 154
Query: 107 GQAHTEQVCNKGKSSNNS 124
G Q N G + + S
Sbjct: 155 GTRKNNQSVNGGDTEHKS 172
>At4g01580 hypothetical protein
Length = 190
Score = 56.6 bits (135), Expect = 2e-08
Identities = 34/96 (35%), Positives = 47/96 (48%), Gaps = 2/96 (2%)
Query: 159 FTQLLPEDFKQQLA-LPKTFSDNLNKKLPENVTLKGPGGVAWKIGLITRDDTVYFLNGWQ 217
F +LP K ++ +P F KL E VTL P G I L + ++F GW
Sbjct: 31 FKLVLPSTMKDKMMRIPPRFVKLQGSKLSEVVTLVTPAGYKRSIKLKRIGEEIWFHEGWS 90
Query: 218 RFVNDHSLKDNDFLVFNYNGESHFDVLIFDGESFCE 253
F HS+++ FL+F Y S F V+IF+ S CE
Sbjct: 91 EFAEAHSIEEGHFLLFEYKKNSSFRVIIFNA-SACE 125
Score = 53.9 bits (128), Expect = 1e-07
Identities = 33/92 (35%), Positives = 48/92 (51%), Gaps = 12/92 (13%)
Query: 16 LPKTFSDNLKK------KLPGN-----VTLKGPSGVVWNVGLTARDDTIYFTNGWQRFVK 64
LP T D + + KL G+ VTL P+G ++ L + I+F GW F +
Sbjct: 35 LPSTMKDKMMRIPPRFVKLQGSKLSEVVTLVTPAGYKRSIKLKRIGEEIWFHEGWSEFAE 94
Query: 65 DHSLKENDFLVFKYNGESLFEVLIFHGDSFCE 96
HS++E FL+F+Y S F V+IF+ S CE
Sbjct: 95 AHSIEEGHFLLFEYKKNSSFRVIIFNA-SACE 125
>At2g24650 hypothetical protein
Length = 1440
Score = 56.2 bits (134), Expect = 3e-08
Identities = 83/336 (24%), Positives = 121/336 (35%), Gaps = 42/336 (12%)
Query: 16 LPKTFSDNLKKKLPGNVTLKGPSGVVWNVGLT--ARDDTIYFTNGWQRFVKDHSLKENDF 73
LPK F+ PG +TL G G+ L + + +GW+ FVKD+ LK D
Sbjct: 466 LPKVFTRENGINKPGRITLLGKDGIKQQTNLLFDKANGAMSLGHGWKDFVKDNGLKTGDS 525
Query: 74 LVFKYNGESLFEVL-IFHGDSFCEKAASYFVGKCGQAHT----EQVCNKGKSSNNSVEEV 128
K E VL + D ++ A + Q + C K + N ++
Sbjct: 526 FTLKLIWEDQTPVLSLCPADCSIDREAGGGRSETNQKKSLPIEPSTCKKIRKDVNIKDDN 585
Query: 129 STPSNGSVECSL--PEKSWEEDIYWTHFQFIHFTQL--LPEDFKQ-QLALPKTFSDNLNK 183
S N E E+ + Y T HF L P K+ +L L F+ N
Sbjct: 586 SKEKNDKEESKSVDGERKYLRGTYLTPSSQKHFVTLTITPSSIKKDRLILSPQFARKNNI 645
Query: 184 KLPENVTLKGPGGVAWKIGL-ITRDDTVYFLNGWQRFVNDHSLKDNDFLVFNYNGESHFD 242
P + L G W I L + T+ GW+ F + NDF + GES
Sbjct: 646 DKPGMIYLLDTDGTKWLISLQRDKKGTMSLGKGWKEFA-----EANDFKL----GESFTM 696
Query: 243 VLIFDGESFCEKEASYFVGKCSHTQTELGGSKANETNNSIEEVNTASSDGGVECGLYEKF 302
L+++ + S +TE SKANE + E T S ++
Sbjct: 697 ELVWEDTT----------PMLSLLRTEFRSSKANEKESISSEHKTRESSPTIK------- 739
Query: 303 QDLNNIGTPLAVPVETTNEKTFNAGVESDSPELLIA 338
N I TP P + K + S E+L A
Sbjct: 740 ---NRIVTPALTPEDVKACKLVTSSNYSCFSEILSA 772
Score = 46.2 bits (108), Expect = 3e-05
Identities = 47/183 (25%), Positives = 73/183 (39%), Gaps = 27/183 (14%)
Query: 52 TIYFTNGWQRFVKDHSLKENDFLVFKYNGESLFEVLIFHGDSFCEKAASYFVGKCGQAHT 111
T Y + GW+ F ++ K F +FK G+ E L+ SFC + + +
Sbjct: 188 TFYISRGWRNFCDENGQKAGGFFLFKLVGKG--ETLVL---SFCPTESINGEENITREDS 242
Query: 112 EQVCNKGKSSNNSVEE---VSTPSNGSVECSLPEKSWEEDIYWTHFQFIHFTQLLPEDF- 167
+ C+ S N VE+ + P P +H QF+ FT LP D+
Sbjct: 243 KDECSSLDSLMNIVEKKKYIPKPRGSPYSSYSP----------SHKQFVTFT--LPPDYA 290
Query: 168 ---KQQLALPKTFSDNLNKKLPENVTLKGPGGVAWKIG-LITRDDTVYFLNGWQRFVNDH 223
K L+ P + +NK P + L G W L+ T+ GW+ FV +
Sbjct: 291 RIGKLSLSAPFVRENGINK--PGEICLLDKHGRKWLTSLLLDSKGTMSLGKGWKEFVKAN 348
Query: 224 SLK 226
SL+
Sbjct: 349 SLE 351
Score = 42.7 bits (99), Expect = 3e-04
Identities = 65/272 (23%), Positives = 92/272 (32%), Gaps = 42/272 (15%)
Query: 29 PGNVTLKGPSGVVWNVGLTARDD-TIYFTNGWQRFVKDHSLKENDFLVFKYNGESLFEVL 87
PG + L G W L T+ GW+ FVK +SL+ K E VL
Sbjct: 310 PGEICLLDKHGRKWLTSLLLDSKGTMSLGKGWKEFVKANSLETG--FTLKLIWEETTPVL 367
Query: 88 IFHGDSFCEKAASYFVGKCGQAHTEQV--CNKGKSSNNSVEEVSTPSNGSVECSLPEKSW 145
+ K + H+ + N+ K SNN EE N S E
Sbjct: 368 SLCSPESNSDREQEEISKAIEKHSLFIDPSNRDKISNNDKEE-----NMSWERKKDHLKS 422
Query: 146 EEDIYWTHFQFIHFTQLLPEDFKQQLA----------------LPKTFSDNLNKKLPENV 189
+ + QF+ T D L+ LPK F+ P +
Sbjct: 423 RDSTLSSQKQFVTITITPSSDRLVSLSNDSCLVVVSLLYFDMRLPKVFTRENGINKPGRI 482
Query: 190 TLKGPGGVAWKIGLI--TRDDTVYFLNGWQRFVNDHSLKDNDFLVFNYNGESHFDVLIFD 247
TL G G+ + L+ + + +GW+ FV D+ LK D E VL
Sbjct: 483 TLLGKDGIKQQTNLLFDKANGAMSLGHGWKDFVKDNGLKTGDSFTLKLIWEDQTPVL--- 539
Query: 248 GESFCEKEASYFVGKCSHTQTELGGSKANETN 279
S C + S E GG + +ETN
Sbjct: 540 --SLCPADCS--------IDREAGGGR-SETN 560
Score = 42.0 bits (97), Expect = 6e-04
Identities = 53/220 (24%), Positives = 88/220 (39%), Gaps = 44/220 (20%)
Query: 42 WNVGLTARDDTIYFTNGWQRFVKDHSLKENDFLVFKYNGESLFEVLIFHGDSFCEKAASY 101
W V L+ R T+GW+ FV + + D + F+Y G L+FH + +Y
Sbjct: 50 WEVKLSDRR----ITDGWEEFVVANDFRIGDVVAFRYVGN-----LVFHVSNL---GPNY 97
Query: 102 FVGKCGQAHTEQVCNKGKSSNNSVE--EVSTPSNGSVECSLPEKSWEEDIYWTHFQFIHF 159
+ + H +++ + K+ NS E VS+ S+ C + T
Sbjct: 98 YEIE----HNDELPKEKKAKTNSEEADAVSSSSSADKSCFM--------AIITALDLTTD 145
Query: 160 TQLLPEDFKQQLALPKTFSDNLNKKLPENVTLKGPGGVAWKIGLITRDD---TVYFLNGW 216
T LP F T ++ L +K E + GG ++ + D+ T Y GW
Sbjct: 146 TLYLPLHF--------TSANGLTRKNREIILT--DGGERSRVLDLRFDESSGTFYISRGW 195
Query: 217 QRFVNDHSLKDNDFLVFNYNGESHFDVLIFDGESFCEKEA 256
+ F +++ K F +F G+ VL SFC E+
Sbjct: 196 RNFCDENGQKAGGFFLFKLVGKGETLVL-----SFCPTES 230
Score = 40.8 bits (94), Expect = 0.001
Identities = 50/228 (21%), Positives = 77/228 (32%), Gaps = 39/228 (17%)
Query: 38 SGVVWNVGLTARDDTIYFTNGWQRFVKDHSLKENDFLVFKYNGESLFEVLIFHGDSFCEK 97
S + W V + D T+GW+ F H L+ D +VF+ + F V + G S CE
Sbjct: 995 SKITWEVKI----DGQRLTDGWKEFALSHDLRIGDIVVFRQERDMSFHVTML-GPSCCEI 1049
Query: 98 AASYFVGKCGQAHTEQVCNKGKSSNNSVEEVSTPSNGSVECSLPEKSWEEDIYWTHFQFI 157
G C ++E+ P+ + SL + ++
Sbjct: 1050 Q----YGSCSD------------EERNLEKKKNPNGEAKSSSLDPSCFSANV-------- 1085
Query: 158 HFTQLLPEDFKQQLAL-PKTFSDNLNKKLPENVTLKGPGGVAWKIGLITRDD--TVYFLN 214
P + L L P F + L G +W L + TVY
Sbjct: 1086 -----APSSLRYDLMLFPMGFVRENGVVGSGKIVLMNEKGRSWNFNLRQKPSCGTVYVRG 1140
Query: 215 GWQRFVNDHSLKDNDFLVFNY--NGESHFDVLIFDGESFCEKEASYFV 260
GW F + + L+ D F G + L+ G C + S FV
Sbjct: 1141 GWVSFCDANGLQAGDIYTFKLIKRGGTLVLRLLPKGAESCSLDPSCFV 1188
Score = 38.1 bits (87), Expect = 0.008
Identities = 24/68 (35%), Positives = 37/68 (54%), Gaps = 4/68 (5%)
Query: 162 LLPEDFKQ-QLALPKTF--SDNLNKKLPENVTLKGPGGVAWKIGLITRDDTVYFLNGWQR 218
L PED +L LP F ++ +N KL + +TL G GV W +++ D T+ NGW+
Sbjct: 883 LTPEDVTACKLILPSQFMKANGINNKLGK-ITLLGENGVEWPGYMLSLDGTLALGNGWEG 941
Query: 219 FVNDHSLK 226
F + +K
Sbjct: 942 FCEANGVK 949
Score = 35.4 bits (80), Expect = 0.053
Identities = 20/68 (29%), Positives = 34/68 (49%), Gaps = 3/68 (4%)
Query: 16 LPKTF--SDNLKKKLPGNVTLKGPSGVVWNVGLTARDDTIYFTNGWQRFVKDHSLKENDF 73
LP F ++ + KL G +TL G +GV W + + D T+ NGW+ F + + +K
Sbjct: 895 LPSQFMKANGINNKL-GKITLLGENGVEWPGYMLSLDGTLALGNGWEGFCEANGVKLGQT 953
Query: 74 LVFKYNGE 81
++ E
Sbjct: 954 FTLEFVNE 961
Score = 34.3 bits (77), Expect = 0.12
Identities = 26/101 (25%), Positives = 42/101 (40%), Gaps = 16/101 (15%)
Query: 197 VAWKIGLITRDDTVYFLNGWQRFVNDHSLKDNDFLVFNYNGESHFDVLIFDGESFCEKEA 256
+ W++ + D +GW+ F H L+ D +VF + F V + G S CE +
Sbjct: 997 ITWEVKI----DGQRLTDGWKEFALSHDLRIGDIVVFRQERDMSFHVTML-GPSCCEIQ- 1050
Query: 257 SYFVGKCSHTQTEL-------GGSKANETNNSIEEVNTASS 290
G CS + L G +K++ + S N A S
Sbjct: 1051 ---YGSCSDEERNLEKKKNPNGEAKSSSLDPSCFSANVAPS 1088
Score = 33.5 bits (75), Expect = 0.20
Identities = 23/78 (29%), Positives = 30/78 (37%), Gaps = 4/78 (5%)
Query: 30 GNVTLKGPSGVVWNVGLTARDD--TIYFTNGWQRFVKDHSLKENDFLVFKY--NGESLFE 85
G + L G WN L + T+Y GW F + L+ D FK G +L
Sbjct: 1111 GKIVLMNEKGRSWNFNLRQKPSCGTVYVRGGWVSFCDANGLQAGDIYTFKLIKRGGTLVL 1170
Query: 86 VLIFHGDSFCEKAASYFV 103
L+ G C S FV
Sbjct: 1171 RLLPKGAESCSLDPSCFV 1188
>At1g16640 Unknown protein
Length = 134
Score = 53.5 bits (127), Expect = 2e-07
Identities = 26/82 (31%), Positives = 45/82 (54%), Gaps = 1/82 (1%)
Query: 16 LPKTFSDNLKKKLPGNVTLKGPSGVVWNVGLTARDDTIYFTNGWQRFVKDHSLKENDFLV 75
+P F++ P V L SG W V + R + ++ T GW+ FVKD++L++ +L
Sbjct: 24 IPLGFNEYFPAPFPITVDLLDYSGRSWTVRMKKRGEKVFLTVGWENFVKDNNLEDGKYLQ 83
Query: 76 FKYNGESLFEVLIFHGDSFCEK 97
F Y+ + F V+I+ G + C +
Sbjct: 84 FIYDRDRTFYVIIY-GHNMCSE 104
Score = 51.6 bits (122), Expect = 7e-07
Identities = 26/92 (28%), Positives = 47/92 (50%), Gaps = 1/92 (1%)
Query: 163 LPEDFKQQLALPKTFSDNLNKKLPENVTLKGPGGVAWKIGLITRDDTVYFLNGWQRFVND 222
+ E + L +P F++ P V L G +W + + R + V+ GW+ FV D
Sbjct: 14 ISEKSSKSLEIPLGFNEYFPAPFPITVDLLDYSGRSWTVRMKKRGEKVFLTVGWENFVKD 73
Query: 223 HSLKDNDFLVFNYNGESHFDVLIFDGESFCEK 254
++L+D +L F Y+ + F V+I+ G + C +
Sbjct: 74 NNLEDGKYLQFIYDRDRTFYVIIY-GHNMCSE 104
>At1g26680 unknown protein
Length = 920
Score = 53.1 bits (126), Expect = 2e-07
Identities = 48/186 (25%), Positives = 78/186 (41%), Gaps = 24/186 (12%)
Query: 51 DTIYFTNGWQRFVKDHSLKENDFLVFKYNGESLFEVLIFHGDSFCEKAASYFVGKCGQAH 110
D + T+GW+ F H L+ D +VF+ GE +F V G S CE Q H
Sbjct: 64 DGLKLTDGWEDFAFAHDLRTGDIVVFRLEGEMVFHVTAL-GPSCCEI----------QYH 112
Query: 111 TEQVCNKGKSSNNSVEEVSTPSNGSVECSLPEKSWEEDIYWTHFQFIHFTQLLPEDFKQQ 170
T N+ + S N S P K E + + F L D +
Sbjct: 113 TSSHNINDDDRNDQINIAS--RNSSRVKKNPRKKVESSLDHSRFVAKVSAWCLSND---R 167
Query: 171 LALPKTFS--DNLNKKLPENVTLKGPGGVAWKIGLITRDD----TVYFLNGWQRFVNDHS 224
L +P +F+ + LNK + + L+ G +WK L+ R D + +GW+RF +++
Sbjct: 168 LYIPLSFARLNGLNKINSKKIYLQNEEGRSWK--LVLRHDKSGMQTFVQSGWRRFCSENG 225
Query: 225 LKDNDF 230
++ +
Sbjct: 226 IRQGQY 231
Score = 49.7 bits (117), Expect = 3e-06
Identities = 71/305 (23%), Positives = 114/305 (37%), Gaps = 37/305 (12%)
Query: 11 NLLQALPKTF-SDNLKKKLPGNVTLKGPSGVVWNVGLTARDDTIYFTNGWQRFVKDHSLK 69
N L LP++F + N K + LK GV W + L Y GW F + + +K
Sbjct: 279 NDLLYLPRSFVNSNRLDKRCSEIVLKNEQGVKWPLVLKRFKSVTYLPKGWTSFCQVNRIK 338
Query: 70 ENDFLVFKYNGESLFEVLIFHGDSFCEKAASYFVG--KCGQAH----TEQVC-------- 115
D FK G VL S C ++ +C + + +E+ C
Sbjct: 339 AGDSFKFKLVGTWKKPVL-----SLCPTQSNNHKTPLECSEGNKSEESEEDCLEVKKKKY 393
Query: 116 -NKGKSSNNSVEEVSTPSNGSVECSLPEKSWEEDIYWTHFQFIHFTQLLPED-FKQQLAL 173
++ ++S ++++ T S K+ E + + + P +K QL L
Sbjct: 394 WSRCRASVENMDDDQTNIGNSSRKKRVSKNPREKVESSSDHSSFVGSVNPSSLYKDQLYL 453
Query: 174 PKTF-SDNLNKKLPENVTLKGPGGVAWKIGL-ITRDDTVYFLNGWQRFVNDHSLKDNDFL 231
P+ F S N K + LK G + L + D + GW F + +K D
Sbjct: 454 PRNFVSSNFLDKRCSEIVLKNERGEKRTLVLKHFKKDLTFLKKGWTSFCQVNRIKAGDSF 513
Query: 232 VFNYNGESHFDVLIFDGESFCEKEASYFVGKCSHTQTELGGSKANETNNSIEEVNTASSD 291
F G + VL S C E +Y +T L S+ N++ S EE T +
Sbjct: 514 KFKLVGTWNKPVL-----SLCPTETNYH-------KTPLACSEGNKSEES-EEEGTEDKN 560
Query: 292 GGVEC 296
+C
Sbjct: 561 TSQDC 565
Score = 37.7 bits (86), Expect = 0.011
Identities = 28/97 (28%), Positives = 44/97 (44%), Gaps = 3/97 (3%)
Query: 158 HFTQLLPEDFKQQLALPKTF-SDNLNKKLPENVTLKGPGGVAWKIGLITRDDTVYFLNGW 216
HF Q + + + L +P F S ++ + +N T+ + K L+ D + +GW
Sbjct: 14 HFFQPILTESRTHLNIPVAFFSKHVEGRNNQNKTVTLRSDASDKTWLVKMDG-LKLTDGW 72
Query: 217 QRFVNDHSLKDNDFLVFNYNGESHFDVLIFDGESFCE 253
+ F H L+ D +VF GE F V G S CE
Sbjct: 73 EDFAFAHDLRTGDIVVFRLEGEMVFHVTAL-GPSCCE 108
>At5g32460 meristem protein - like
Length = 517
Score = 51.2 bits (121), Expect = 9e-07
Identities = 47/213 (22%), Positives = 82/213 (38%), Gaps = 38/213 (17%)
Query: 158 HFTQLLPEDFKQQLALPKTFSDNLNKKLPENVTLK---GPGGVAWKIGLITRDDTVYFLN 214
HF + + F+ + +PK F N + E + + WKI + R T
Sbjct: 13 HFIKPMLPGFETYIVIPKAFYSNYLEGRQEGNAAELRSDATEITWKIKIDGRRMT----K 68
Query: 215 GWQRFVNDHSLKDNDFLVFNYNGESHFDVLIFDGESFCEKEASYFVGKCSHTQTELGGSK 274
GW+ F H+L+ +D LVF + G F V F G SFCE ++
Sbjct: 69 GWEEFAVAHNLQVDDILVFRHEGNLLFHVTPF-GLSFCE-----------ILYSQRDEKD 116
Query: 275 ANETNNSIEEVNTASSDGGVECGLYEKFQDLNNIGTPLAVPVETTNEK----------TF 324
+T + T +G EC +++ T VPV +N++ T
Sbjct: 117 VKDTTGKVTRSRTVKKNGKNEC---------SSVDTDFVVPVTASNQRVDSFYLPRGFTT 167
Query: 325 NAGVESDSPELLIADTVTKTTAIQFPYQPTGKR 357
++G E+++ D + + ++ Y + R
Sbjct: 168 SSGSSKLCNEIILIDEKDRPSTLKLRYNKSSNR 200
Score = 47.4 bits (111), Expect = 1e-05
Identities = 38/122 (31%), Positives = 56/122 (45%), Gaps = 18/122 (14%)
Query: 16 LPKTFSDN-LKKKLPGNVT--LKGPSGVVWNVGLTARDDTIYFTNGWQRFVKDHSLKEND 72
+PK F N L+ + GN + + W + + R T GW+ F H+L+ +D
Sbjct: 28 IPKAFYSNYLEGRQEGNAAELRSDATEITWKIKIDGRR----MTKGWEEFAVAHNLQVDD 83
Query: 73 FLVFKYNGESLFEVLIFHGDSFC--------EKAASYFVGKCGQAHTEQVCNKGKSSNNS 124
LVF++ G LF V F G SFC EK GK ++ T V GK+ +S
Sbjct: 84 ILVFRHEGNLLFHVTPF-GLSFCEILYSQRDEKDVKDTTGKVTRSRT--VKKNGKNECSS 140
Query: 125 VE 126
V+
Sbjct: 141 VD 142
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.133 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,606,164
Number of Sequences: 26719
Number of extensions: 444899
Number of successful extensions: 1268
Number of sequences better than 10.0: 73
Number of HSP's better than 10.0 without gapping: 50
Number of HSP's successfully gapped in prelim test: 23
Number of HSP's that attempted gapping in prelim test: 1102
Number of HSP's gapped (non-prelim): 163
length of query: 385
length of database: 11,318,596
effective HSP length: 101
effective length of query: 284
effective length of database: 8,619,977
effective search space: 2448073468
effective search space used: 2448073468
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC148404.4


