

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148403.7 - phase: 0
(315 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
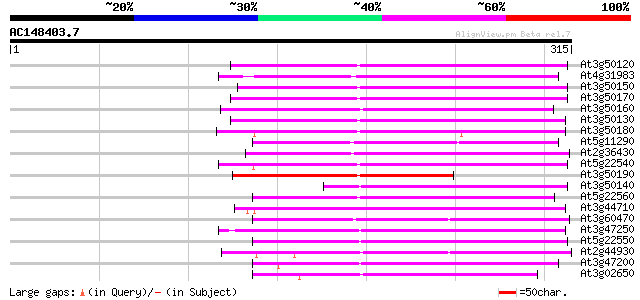
Score E
Sequences producing significant alignments: (bits) Value
At3g50120 putative protein 147 1e-35
At4g31983 putative protein 139 2e-33
At3g50150 putative protein 139 2e-33
At3g50170 putative protein 136 2e-32
At3g50160 putative protein 132 2e-31
At3g50130 putative protein 129 2e-30
At3g50180 putative protein 116 2e-26
At5g11290 putative protein 106 1e-23
At2g36430 unknown protein (At2g36430) 105 4e-23
At5g22540 unknown protein 100 8e-22
At3g50190 putative protein 97 9e-21
At3g50140 putative protein 97 1e-20
At5g22560 putative protein 93 2e-19
At3g44710 unknown protein 91 6e-19
At3g60470 putative protein 89 2e-18
At3g47250 unknown protein 89 4e-18
At5g22550 unknown protein 88 5e-18
At2g44930 unknown protein 85 6e-17
At3g47200 unknown protein 84 1e-16
At3g02650 hypothetical protein 80 1e-15
>At3g50120 putative protein
Length = 531
Score = 147 bits (370), Expect = 1e-35
Identities = 78/190 (41%), Positives = 112/190 (58%), Gaps = 2/190 (1%)
Query: 125 KVSKEDEDHLNRSAMELKEAGISFKKSKTRSLKDVSFNRGVLRLPKLVVDDHTECMFLNL 184
+V+ + L ELKEAGI F++ KT D+ F G L +P+L++ D T+ +FLNL
Sbjct: 340 RVADKRRQQLIHCVTELKEAGIKFRRRKTDRFWDMQFKNGYLEIPRLLIHDGTKSLFLNL 399
Query: 185 IAFELLHVDGTRKEVISFVCFIDTIIDSAVDVAILIRSGIIINYLESDKAVAKLFNSLAK 244
IAFE H+D + ++ S++ F+D +IDS DV+ L GII ++L SD VA LFN L +
Sbjct: 400 IAFEQCHIDSS-NDITSYIIFMDNLIDSHEDVSYLHYCGIIEHWLGSDSEVADLFNRLCQ 458
Query: 245 EIPMDREGE-LEEVTKSMISYCKKPWKSWRASLIQTYFRNPWAMVSLVAAFFLFALTIIQ 303
E+ D E L ++ + Y W +WRA+L YF NPWA+VS AA L LT Q
Sbjct: 459 EVVFDTEDSYLSRLSIEVNRYYDHKWNAWRATLKHKYFNNPWAIVSFCAAVILLVLTFSQ 518
Query: 304 TIYTVGQFYQ 313
+ Y V +Y+
Sbjct: 519 SFYAVYAYYK 528
>At4g31983 putative protein
Length = 414
Score = 139 bits (351), Expect = 2e-33
Identities = 75/192 (39%), Positives = 113/192 (58%), Gaps = 9/192 (4%)
Query: 118 KTSETSLKVSKEDEDHLNRSAMELKEAGISFKKSKTRS-LKDVSFNRGVLRLPKLVVDDH 176
K T++KV E A EL AG+ FK ++T S L D+SF GVL++P +VVDD
Sbjct: 228 KLEYTTVKVDNAPE------ATELHTAGVRFKPAETSSCLLDISFADGVLKIPTIVVDDL 281
Query: 177 TECMFLNLIAFELLHVDGTRKEVISFVCFIDTIIDSAVDVAILIRSGIIINYLESDKAVA 236
TE ++ N+I FE + K + ++ + I S D +LI SGII+NYL + V+
Sbjct: 282 TESLYKNIIGFEQCRC--SNKNFLDYIMLLGCFIKSPTDADLLIHSGIIVNYLGNSVDVS 339
Query: 237 KLFNSLAKEIPMDREGELEEVTKSMISYCKKPWKSWRASLIQTYFRNPWAMVSLVAAFFL 296
LFNS++KE+ DR +++++ +YC PW W+A L + YF NPWA+ S+ AA L
Sbjct: 340 NLFNSISKEVIYDRRFYFSMLSENLQAYCNTPWNRWKAILRRDYFHNPWAVASVFAALLL 399
Query: 297 FALTIIQTIYTV 308
LT IQ++ ++
Sbjct: 400 LLLTFIQSVCSI 411
>At3g50150 putative protein
Length = 509
Score = 139 bits (350), Expect = 2e-33
Identities = 72/186 (38%), Positives = 111/186 (58%), Gaps = 2/186 (1%)
Query: 129 EDEDHLNRSAMELKEAGISFKKSKTRSLKDVSFNRGVLRLPKLVVDDHTECMFLNLIAFE 188
E + L EL+ AG++F + +T L D+ F G L++PKL++ D T+ +F NLIAFE
Sbjct: 322 EKQQQLIHCVTELRGAGVNFMRKETGQLWDIEFKNGYLKIPKLLIHDGTKSLFSNLIAFE 381
Query: 189 LLHVDGTRKEVISFVCFIDTIIDSAVDVAILIRSGIIINYLESDKAVAKLFNSLAKEIPM 248
H + + S++ F+D +I+S+ DV+ L GII ++L SD VA LFN L KE+
Sbjct: 382 QCHTQSS-NNITSYIIFMDNLINSSQDVSYLHHDGIIEHWLGSDSEVADLFNRLCKEVIF 440
Query: 249 D-REGELEEVTKSMISYCKKPWKSWRASLIQTYFRNPWAMVSLVAAFFLFALTIIQTIYT 307
D ++G L ++++ + Y + W S +A+L Q YF NPWA S AA L LT Q+ +
Sbjct: 441 DPKDGYLSQLSREVNRYYSRKWNSLKATLRQKYFNNPWAYFSFSAAVILLFLTFFQSFFA 500
Query: 308 VGQFYQ 313
V +Y+
Sbjct: 501 VYAYYK 506
>At3g50170 putative protein
Length = 541
Score = 136 bits (342), Expect = 2e-32
Identities = 71/190 (37%), Positives = 110/190 (57%), Gaps = 2/190 (1%)
Query: 125 KVSKEDEDHLNRSAMELKEAGISFKKSKTRSLKDVSFNRGVLRLPKLVVDDHTECMFLNL 184
+V + + L EL+EAG+ F+K KT D+ F G L +PKL++ D T+ +F NL
Sbjct: 348 RVVDKRQQQLVHCVTELREAGVKFRKRKTDRFWDIEFKNGYLEIPKLLIHDGTKSLFSNL 407
Query: 185 IAFELLHVDGTRKEVISFVCFIDTIIDSAVDVAILIRSGIIINYLESDKAVAKLFNSLAK 244
IAFE H++ + + S++ F+D +I+S+ DV+ L GII ++L SD VA LFN L +
Sbjct: 408 IAFEQCHIESS-NHITSYIIFMDNLINSSEDVSYLHYCGIIEHWLGSDSEVADLFNRLCQ 466
Query: 245 EIPMD-REGELEEVTKSMISYCKKPWKSWRASLIQTYFRNPWAMVSLVAAFFLFALTIIQ 303
E+ D ++ L ++ + Y + W +A+L YF NPWA S AA L LT+ Q
Sbjct: 467 EVVFDPKDSHLSRLSGDVNRYYNRKWNVLKATLTHKYFNNPWAYFSFSAAVILLLLTLCQ 526
Query: 304 TIYTVGQFYQ 313
+ Y V +Y+
Sbjct: 527 SFYAVYAYYK 536
>At3g50160 putative protein
Length = 511
Score = 132 bits (333), Expect = 2e-31
Identities = 69/188 (36%), Positives = 110/188 (57%), Gaps = 2/188 (1%)
Query: 119 TSETSLKVSKEDEDHLNRSAMELKEAGISFKKSKTRSLKDVSFNRGVLRLPKLVVDDHTE 178
TS+ + + + L EL+ AG+ F + +T D+ F G L++PKL++ D T+
Sbjct: 304 TSDEDMSMVNKQPQQLIHCVTELRNAGVEFMRKETGHFWDIEFKNGYLKIPKLLIHDGTK 363
Query: 179 CMFLNLIAFELLHVDGTRKEVISFVCFIDTIIDSAVDVAILIRSGIIINYLESDKAVAKL 238
+FLNLIAFE H+ ++K + S++ F+D +I+S+ DV+ L GII N+L SD V+ L
Sbjct: 364 SLFLNLIAFEQCHIKSSKK-ITSYIIFMDNLINSSEDVSYLHHYGIIENWLGSDSEVSDL 422
Query: 239 FNSLAKEIPMD-REGELEEVTKSMISYCKKPWKSWRASLIQTYFRNPWAMVSLVAAFFLF 297
FN L KE+ D +G L +T + Y ++ W +A+L YF NPWA S +AA +
Sbjct: 423 FNGLGKEVIFDPNDGYLSALTGEVNIYYRRKWNYLKATLRHKYFNNPWAYFSFIAACSIR 482
Query: 298 ALTIIQTI 305
A I +++
Sbjct: 483 AFEIARSV 490
>At3g50130 putative protein
Length = 564
Score = 129 bits (325), Expect = 2e-30
Identities = 69/189 (36%), Positives = 107/189 (56%), Gaps = 2/189 (1%)
Query: 125 KVSKEDEDHLNRSAMELKEAGISFKKSKTRSLKDVSFNRGVLRLPKLVVDDHTECMFLNL 184
+V+ + + L EL+EAGI F+ KT D+ F G L +PKL++ D T+ +F NL
Sbjct: 373 RVADKRQQQLIHCVTELREAGIKFRTRKTDRFWDIRFKNGYLEIPKLLIHDGTKSLFSNL 432
Query: 185 IAFELLHVDGTRKEVISFVCFIDTIIDSAVDVAILIRSGIIINYLESDKAVAKLFNSLAK 244
IAFE H+D + ++ S++ F+D +IDS+ DV L GII ++L +D VA LFN L +
Sbjct: 433 IAFEQCHIDSS-NDITSYIIFMDNLIDSSEDVRYLHYCGIIEHWLGNDYEVADLFNRLCQ 491
Query: 245 EIPMDREGE-LEEVTKSMISYCKKPWKSWRASLIQTYFRNPWAMVSLVAAFFLFALTIIQ 303
E+ D + L +++ + + W +A L YF NPWA S AA L LT+ Q
Sbjct: 492 EVAFDPQNSYLSQLSNKVDRNYSRKWNVLKAILKHKYFNNPWAYFSFFAALVLLVLTLFQ 551
Query: 304 TIYTVGQFY 312
+ +T ++
Sbjct: 552 SFFTAYPYF 560
>At3g50180 putative protein
Length = 588
Score = 116 bits (290), Expect = 2e-26
Identities = 69/204 (33%), Positives = 108/204 (52%), Gaps = 9/204 (4%)
Query: 117 FKTSETSLKVSKEDEDHLNR---SAMELKEAGISFKKSKTRSLKDVSFNRGVLRLPKLVV 173
F S S+ + HL R + EL++AG FK +KT D+ F+ G L +P L++
Sbjct: 382 FPRSSGKANYSRVADKHLQRVIPTVTELRDAGFKFKLNKTDRFWDIKFSNGYLEIPGLLI 441
Query: 174 DDHTECMFLNLIAFELLHVDGTRKEVISFVCFIDTIIDSAVDVAILIRSGIIINYLESDK 233
D T+ +FLNLIAFE H++ + ++ S++ F+D +IDS D++ L GII + L S+
Sbjct: 442 HDGTKSLFLNLIAFEQCHIESS-NDITSYIIFMDNLIDSPEDISYLHHCGIIEHSLGSNS 500
Query: 234 AVAKLFNSLAKEIPMDREG-----ELEEVTKSMISYCKKPWKSWRASLIQTYFRNPWAMV 288
VA +FN L +E+ D + L EV + + S + +LI Y NPWA +
Sbjct: 501 EVADMFNQLCQEVVFDTKDIYLSQLLIEVHRCYKQNYSRKLNSLKTTLILKYLDNPWAYL 560
Query: 289 SLVAAFFLFALTIIQTIYTVGQFY 312
S AA L LT Q+ + ++
Sbjct: 561 SFFAAVILLILTFSQSYFAAYAYF 584
>At5g11290 putative protein
Length = 422
Score = 106 bits (265), Expect = 1e-23
Identities = 59/173 (34%), Positives = 97/173 (55%), Gaps = 4/173 (2%)
Query: 137 SAMELKEAGISFKKSKTRSLK-DVSFNRGVLRLPKLVVDDHTECMFLNLIAFELLHVDGT 195
SA E++ AG+ + + + D+SF GVL +PK+ ++D TE ++ N+I FE H
Sbjct: 250 SAKEIQNAGVKLQPADNNTCALDISFANGVLTIPKIKINDITESLYRNIILFEQCH--RL 307
Query: 196 RKEVISFVCFIDTIIDSAVDVAILIRSGIIINYLESDKAVAKLFNSLAKEIPMDREGELE 255
I ++ F+ I S +D + I GII+N + + V++LFNS+ KE +
Sbjct: 308 DAYFIHYMRFLSCFIRSPMDAELFIDHGIIVNRFGNAEDVSRLFNSILKETSYSGF-YYK 366
Query: 256 EVTKSMISYCKKPWKSWRASLIQTYFRNPWAMVSLVAAFFLFALTIIQTIYTV 308
V ++ ++C PW W+A+L + YF NPW+ S+VAA L LT +Q I ++
Sbjct: 367 TVYGNLQAHCNAPWNKWKATLRRDYFHNPWSAASVVAACVLLLLTFVQAICSI 419
>At2g36430 unknown protein (At2g36430)
Length = 448
Score = 105 bits (261), Expect = 4e-23
Identities = 62/184 (33%), Positives = 95/184 (50%), Gaps = 3/184 (1%)
Query: 133 HLNRSAMELKEAGISFKKSK-TRSLKDVSFNRGVLRLPKLVVDDHTECMFLNLIAFELLH 191
H+ S +L+ AGI ++ K S V F G + +P + VDD N +A+E H
Sbjct: 266 HIIHSISKLRRAGIKLRELKDAESFLVVRFRHGTIEMPAITVDDFMSSFLENCVAYEQCH 325
Query: 192 VDGTRKEVISFVCFIDTIIDSAVDVAILIRSGIIINYLESDKAVAKLFNSLAKEIPMD-R 250
V ++ +D + ++ DV L II NY +D +AK NSL +++ D
Sbjct: 326 V-ACSMHFTTYATLLDCLTNTYKDVEYLCDQNIIENYFGTDTELAKFVNSLGRDVAFDIT 384
Query: 251 EGELEEVTKSMISYCKKPWKSWRASLIQTYFRNPWAMVSLVAAFFLFALTIIQTIYTVGQ 310
+ L+++ + + Y K W A+ TYF +PW+ VS +AA L L++IQTIYTV Q
Sbjct: 385 QCYLKDLFEEVNEYYKSSWHVEWATFKFTYFNSPWSFVSALAALVLLVLSVIQTIYTVFQ 444
Query: 311 FYQK 314
YQK
Sbjct: 445 AYQK 448
>At5g22540 unknown protein
Length = 440
Score = 100 bits (250), Expect = 8e-22
Identities = 61/200 (30%), Positives = 104/200 (51%), Gaps = 5/200 (2%)
Query: 118 KTSETSLKVSKEDEDHLN--RSAMELKEAGISFKKSK-TRSLKDVSFNRGVLRLPKLVVD 174
+ + S K S D ++L SA +L GI FK K T S+ D+S++ GVL +P +V+D
Sbjct: 237 RIKDHSSKSSFNDHEYLGFVLSAKKLHLRGIKFKPRKNTDSILDISYSNGVLHIPPVVMD 296
Query: 175 DHTECMFLNLIAFELLHVDGTRKEVISFVCFIDTIIDSAVDVAILIRSGIIINYLESDKA 234
D T +FLN +AFE L+ D + + S+V F+ +I+ D + L I+ NY ++
Sbjct: 297 DFTASIFLNCVAFEQLYADSS-NHITSYVAFMACLINEESDASFLSERRILENYFGTEDE 355
Query: 235 VAKLFNSLAKEIPMDRE-GELEEVTKSMISYCKKPWKSWRASLIQTYFRNPWAMVSLVAA 293
V++ + + K+I +D E L +V + + Y + + A I T+F +PW S AA
Sbjct: 356 VSRFYKRIGKDIALDLEKSYLAKVFEGVNEYTSQGFHVHCAEFIHTHFDSPWTFASSFAA 415
Query: 294 FFLFALTIIQTIYTVGQFYQ 313
L +Q + +++
Sbjct: 416 LLLLLFAALQVFFAAYSYFR 435
>At3g50190 putative protein
Length = 463
Score = 97.4 bits (241), Expect = 9e-21
Identities = 50/124 (40%), Positives = 77/124 (61%), Gaps = 1/124 (0%)
Query: 126 VSKEDEDHLNRSAMELKEAGISFKKSKTRSLKDVSFNRGVLRLPKLVVDDHTECMFLNLI 185
V+ + + L EL+EAGI FK+ K+ L D+ F G L +PKL++ D T+ + NLI
Sbjct: 300 VADKRQQKLLHCVTELREAGIKFKRRKSDRLWDIQFKNGCLEIPKLLIHDGTKSLLSNLI 359
Query: 186 AFELLHVDGTRKEVISFVCFIDTIIDSAVDVAILIRSGIIINYLESDKAVAKLFNSLAKE 245
AFE H+D T K++ S++ F++ +I+S DV L GII ++LESD V LF+ L ++
Sbjct: 360 AFEQCHIDST-KQITSYIIFVENLINSNEDVRYLQYCGIIEHWLESDSEVVDLFHKLCQD 418
Query: 246 IPMD 249
+ D
Sbjct: 419 VVFD 422
>At3g50140 putative protein
Length = 508
Score = 96.7 bits (239), Expect = 1e-20
Identities = 51/138 (36%), Positives = 80/138 (57%), Gaps = 2/138 (1%)
Query: 177 TECMFLNLIAFELLHVDGTRKEVISFVCFIDTIIDSAVDVAILIRSGIIINYLESDKAVA 236
T+ +F NLIA+E H+D T ++ S++ F+D +IDSA D+ L II ++L +D VA
Sbjct: 369 TKSLFSNLIAYEQCHIDSTN-DITSYIIFMDNLIDSAEDIRYLHYYDIIEHWLGNDSEVA 427
Query: 237 KLFNSLAKEIPMDREGE-LEEVTKSMISYCKKPWKSWRASLIQTYFRNPWAMVSLVAAFF 295
+FN L +E+ D E L E++ + Y + W +A+L YF NPWA S AA
Sbjct: 428 DVFNRLCQEVAFDLENTYLSELSNKVDRYYNRKWNVLKATLKHKYFSNPWAYFSFFAAVI 487
Query: 296 LFALTIIQTIYTVGQFYQ 313
L LT+ Q+ +T +++
Sbjct: 488 LLLLTLFQSFFTSYPYFK 505
>At5g22560 putative protein
Length = 517
Score = 92.8 bits (229), Expect = 2e-19
Identities = 56/172 (32%), Positives = 91/172 (52%), Gaps = 3/172 (1%)
Query: 137 SAMELKEAGISFK-KSKTRSLKDVSFNRGVLRLPKLVVDDHTECMFLNLIAFELLHVDGT 195
SA +L+ GI F+ K K + D++ GVL++P L+ DD + +N +AFE +V GT
Sbjct: 337 SARKLRLRGIKFQQKKKFETPLDITLKNGVLKIPPLLFDDFFSSLLINCVAFEQFNVQGT 396
Query: 196 RKEVISFVCFIDTIIDSAVDVAILIRSGIIINYLESDKAVAKLFNSLAKEIPMD-REGEL 254
E+ S+V F+ +I++A D L GII NY + + ++ F + K+I + L
Sbjct: 397 -TEMTSYVTFMGCLINTADDATFLSEKGIIENYFGTGEQLSVFFKNTGKDISFTISKSYL 455
Query: 255 EEVTKSMISYCKKPWKSWRASLIQTYFRNPWAMVSLVAAFFLFALTIIQTIY 306
V + + Y + + A + TYF++PW +S AA L LTI Q +
Sbjct: 456 ANVFEGVNKYTSQGYHVHWAGVKYTYFKSPWTFLSSFAALVLILLTIFQAFF 507
>At3g44710 unknown protein
Length = 504
Score = 91.3 bits (225), Expect = 6e-19
Identities = 60/196 (30%), Positives = 96/196 (48%), Gaps = 10/196 (5%)
Query: 127 SKEDED------HLNR--SAMELKEAGISFK-KSKTRSLKDVSFNRGVLRLPKLVVDDHT 177
S E+ED HL SA +L+ GI FK + K +L D+ +L++P L++DD
Sbjct: 305 SLEEEDSTTGRHHLKMVLSARKLQLKGIKFKARKKAETLMDIRHKGKLLQIPPLILDDFL 364
Query: 178 ECMFLNLIAFELLHVDGTRKEVISFVCFIDTIIDSAVDVAILIRSGIIINYLESDKAVAK 237
+ LN +AFE + T++ + S+V F+ ++ S D L GI+ NY S V++
Sbjct: 365 IAVLLNCVAFEQYYSYCTKQHMTSYVAFMGCLLKSEADAMFLSEVGILENYFGSGDEVSR 424
Query: 238 LFNSLAKEIPMD-REGELEEVTKSMISYCKKPWKSWRASLIQTYFRNPWAMVSLVAAFFL 296
F + K++ D E L + + + Y W A T+F +PW +S AA L
Sbjct: 425 FFKVVGKDVLFDIDESYLAGIFEGVNKYTSSGWHVQWAGFKHTHFDSPWTALSSCAALTL 484
Query: 297 FALTIIQTIYTVGQFY 312
LTIIQ + ++
Sbjct: 485 VILTIIQAFFAAYGYF 500
>At3g60470 putative protein
Length = 540
Score = 89.4 bits (220), Expect = 2e-18
Identities = 59/179 (32%), Positives = 95/179 (52%), Gaps = 3/179 (1%)
Query: 137 SAMELKEAGISFKKSKTRSLKDVSFNRGVLRLPKLVVDDHTECMFLNLIAFELLHVDGTR 196
+A +L+ AG+ FK DV+F G L++P L V D E N++A E H +
Sbjct: 364 NAAKLQSAGVKFKAVTDEFSIDVTFENGCLKIPCLWVPDDAEITLRNIMALEQCHYP-FK 422
Query: 197 KEVISFVCFIDTIIDSAVDVAILIRSGIIINYLE-SDKAVAKLFNSLAKEIPMDREGELE 255
V +FV F+D +ID+ DV +L+ +GI+ N+ E S AVA++ N+L + ++
Sbjct: 423 AYVCNFVSFLDFLIDTDKDVDLLMENGILNNWGEKSSVAVAEMVNTLFSGV-VESRSYYA 481
Query: 256 EVTKSMISYCKKPWKSWRASLIQTYFRNPWAMVSLVAAFFLFALTIIQTIYTVGQFYQK 314
+ + +Y + P R L + YF N W + VAA L +T+IQT+ ++ Q QK
Sbjct: 482 GIASRVNAYYENPVNRSRTILGRQYFGNLWRGTATVAAALLLVMTLIQTVASILQVMQK 540
>At3g47250 unknown protein
Length = 480
Score = 88.6 bits (218), Expect = 4e-18
Identities = 57/198 (28%), Positives = 96/198 (47%), Gaps = 7/198 (3%)
Query: 118 KTSETSLKVSKEDEDHLNRSAMELKEAGISFK-KSKTRSLKDVSFNRGVLRLPKLVVDDH 176
K+ E S S E L SA L+ GI F+ +S S+ D+ + L++P L +D
Sbjct: 283 KSGEVS---SSESTFPLILSAKRLRLQGIKFRLRSDAESILDIKLKKNKLQIPLLRLDGF 339
Query: 177 TECMFLNLIAFELLHVDGTRKEVISFVCFIDTIIDSAVDVAILIRSGIII-NYLESDKAV 235
+FLN +AFE + + T ++ S+V F+ +++ D L II NY ++ V
Sbjct: 340 ISSIFLNCVAFEQFYTESTN-DITSYVVFMGCLLNDQEDATFLNNDKRIIENYFGNENEV 398
Query: 236 AKLFNSLAKEIPMD-REGELEEVTKSMISYCKKPWKSWRASLIQTYFRNPWAMVSLVAAF 294
++ F ++ K++ D R L V + + Y K + A T+F +PW +S A
Sbjct: 399 SQFFKTICKDVVFDTRRSYLRNVFEGVNEYTSKTYNGVWAGFRHTHFESPWTALSSFAVV 458
Query: 295 FLFALTIIQTIYTVGQFY 312
F+ LT+ Q Y + +Y
Sbjct: 459 FVILLTMTQAAYAIRSYY 476
>At5g22550 unknown protein
Length = 443
Score = 88.2 bits (217), Expect = 5e-18
Identities = 53/179 (29%), Positives = 90/179 (49%), Gaps = 3/179 (1%)
Query: 137 SAMELKEAGISF-KKSKTRSLKDVSFNRGVLRLPKLVVDDHTECMFLNLIAFELLHVDGT 195
SA +L+ GI F +K + D+SF G++ +P LV DD + +N +AFE ++ +
Sbjct: 261 SARKLRLRGIKFMRKENVETPLDISFKSGLVEIPLLVFDDFISNLLINCVAFEQFNMSCS 320
Query: 196 RKEVISFVCFIDTIIDSAVDVAILIRSGIIINYLESDKAVAKLFNSLAKEIPMD-REGEL 254
E+ SFV F+ +I++ D LI GI+ NY + + V+ F ++ K+I + L
Sbjct: 321 T-EITSFVIFMGCLINTEDDATFLIEKGILENYFGTGEEVSLFFKNIGKDISFSISKSFL 379
Query: 255 EEVTKSMISYCKKPWKSWRASLIQTYFRNPWAMVSLVAAFFLFALTIIQTIYTVGQFYQ 313
V + + Y + + A T+F PW +S AA L LTI Q + +++
Sbjct: 380 SNVFEGVNEYTSQGYHVHWAGFKYTHFNTPWTFLSSCAALVLLLLTIFQAFFAAYAYFR 438
>At2g44930 unknown protein
Length = 515
Score = 84.7 bits (208), Expect = 6e-17
Identities = 56/201 (27%), Positives = 107/201 (52%), Gaps = 7/201 (3%)
Query: 120 SETSLKVSKEDEDHLNRS---AMELKEAGISFKKSKTRSLKD--VSFNRGVLRLPKLVVD 174
++ +K +K+ + +S A +L AG+ F + + ++ ++F RG+L +P + D
Sbjct: 304 TKEQIKSAKDKPPEIIKSLHNADKLDSAGVDFVRLERKNDLSLVITFERGILEIPCFLAD 363
Query: 175 DHTECMFLNLIAFELLHVDGTRKEVISFVCFIDTIIDSAVDVAILIRSGIIINYLESDKA 234
D+TE + NL+A E H T V +++ F+D +ID+ DV +L++ G+I N+L +
Sbjct: 364 DNTERIMRNLMALEQCHYPLTAY-VCNYIAFLDFLIDTDQDVDLLVKKGVIKNWLGHQAS 422
Query: 235 VAKLFNSLAKEIPMDREGELEEVTKSMISYCKKPWKSWRASLIQTYFRNPWAMVSLVAAF 294
VA++ N L + +D + + + + A+L + YF++ W + +AA
Sbjct: 423 VAEMVNKLCLGL-VDFGSHYYGIADRLNKHYESRRNRSIATLRRVYFKDLWTGTATIAAA 481
Query: 295 FLFALTIIQTIYTVGQFYQKD 315
+ LT+I T+ +V Q QKD
Sbjct: 482 VILVLTLIGTVASVLQVTQKD 502
>At3g47200 unknown protein
Length = 476
Score = 84.0 bits (206), Expect = 1e-16
Identities = 50/176 (28%), Positives = 93/176 (52%), Gaps = 5/176 (2%)
Query: 137 SAMELKEAGISFK--KSKTRSLKDVSFNRGVLRLPKLVVDDHTECMFLNLIAFELLHVDG 194
SA L+ GI F+ +SK S+ +V + L++P+L D FLN +AFE + D
Sbjct: 281 SAKRLRLQGIKFRLRRSKEDSILNVRLKKNKLQIPQLRFDGFISSFFLNCVAFEQFYTDS 340
Query: 195 TRKEVISFVCFIDTIIDSAVDVAILIRSGIII-NYLESDKAVAKLFNSLAKEIPMDREGE 253
+ E+ +++ F+ ++++ DV L +II N+ S+ V++ F +++K++ + +
Sbjct: 341 SN-EITTYIVFMGCLLNNEEDVTFLRNDKLIIENHFGSNNEVSEFFKTISKDVVFEVDTS 399
Query: 254 -LEEVTKSMISYCKKPWKSWRASLIQTYFRNPWAMVSLVAAFFLFALTIIQTIYTV 308
L V K + Y KK + A T+F +PW +S A F+ LT++Q+ +
Sbjct: 400 YLNNVFKGVNEYTKKWYNGLWAGFRHTHFESPWTFLSSCAVLFVILLTMLQSTVAI 455
>At3g02650 hypothetical protein
Length = 1077
Score = 80.5 bits (197), Expect = 1e-15
Identities = 46/162 (28%), Positives = 87/162 (53%), Gaps = 3/162 (1%)
Query: 137 SAMELKEAGISFKKSKTRSLKDVSF--NRGVLRLPKLVVDDHTECMFLNLIAFELLHVDG 194
S +L +AG+ FK + ++ V+F N G LP + +D +TE + NL+A+E + G
Sbjct: 347 SVSDLHKAGVRFKPTAHGNISTVTFDSNSGQFYLPVINLDINTETVLRNLVAYEATNTSG 406
Query: 195 TRKEVISFVCFIDTIIDSAVDVAILIRSGIIINYLESDKAVAKLFNSLAKEIPMDREGEL 254
+ I+ IIDS DV +L G++++ L+SD+ A+++N ++K + + + G L
Sbjct: 407 PLV-FTRYTELINGIIDSEEDVRLLREQGVLVSRLKSDQEAAEMWNGMSKSVRLTKVGFL 465
Query: 255 EEVTKSMISYCKKPWKSWRASLIQTYFRNPWAMVSLVAAFFL 296
++ + + Y WK L++ Y W +++ +AA L
Sbjct: 466 DKTIEDVNRYYTGRWKVKIGRLVEVYVYGSWQILAFLAAVLL 507
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.132 0.358
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,305,583
Number of Sequences: 26719
Number of extensions: 256098
Number of successful extensions: 1557
Number of sequences better than 10.0: 106
Number of HSP's better than 10.0 without gapping: 38
Number of HSP's successfully gapped in prelim test: 68
Number of HSP's that attempted gapping in prelim test: 1375
Number of HSP's gapped (non-prelim): 143
length of query: 315
length of database: 11,318,596
effective HSP length: 99
effective length of query: 216
effective length of database: 8,673,415
effective search space: 1873457640
effective search space used: 1873457640
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC148403.7


