

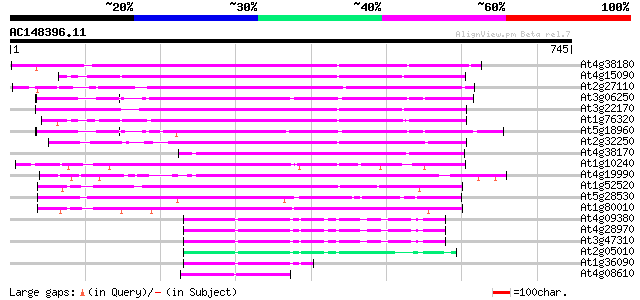
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148396.11 + phase: 0
(745 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g38180 unknown protein (At4g38180) 394 e-110
At4g15090 unknown protein 336 3e-92
At2g27110 Mutator-like transposase 301 9e-82
At3g06250 unknown protein 296 4e-80
At3g22170 far-red impaired response protein, putative 288 8e-78
At1g76320 putative phytochrome A signaling protein 282 4e-76
At5g18960 FAR1 - like protein 270 2e-72
At2g32250 Mutator-like transposase 267 1e-71
At4g38170 hypothetical protein 245 7e-65
At1g10240 unknown protein 230 2e-60
At4g19990 putative protein 229 4e-60
At1g52520 F6D8.26 229 4e-60
At5g28530 far-red impaired response protein (FAR1) - like 228 1e-59
At1g80010 hypothetical protein 226 3e-59
At4g09380 putative protein 81 2e-15
At4g28970 putative protein 81 2e-15
At3g47310 putative protein 77 4e-14
At2g05010 Mutator-like transposase 76 8e-14
At1g36090 hypothetical protein 74 4e-13
At4g08610 predicted transposon protein 67 5e-11
>At4g38180 unknown protein (At4g38180)
Length = 788
Score = 394 bits (1013), Expect = e-110
Identities = 228/635 (35%), Positives = 345/635 (53%), Gaps = 23/635 (3%)
Query: 3 LDDNEVDFSSPSKDGNDPKNTSVEWIPACEDE------LKPVIGKVFDTLVEGGDFYKAY 56
LDD+++ S GN S + P E+E L+P G F++ FY +Y
Sbjct: 33 LDDDDMLDSPIMPCGNGLVGNSGNYFPNQEEEACDLLDLEPYDGLEFESEEAAKAFYNSY 92
Query: 57 AYVAGFSVR-NSIKTKDKDG-VKWKYFLCSKEGFKE--EKKVDKPQLLIAENSLSKSRKR 112
A GFS R +S + +DG + + F+C+KEGF+ EK+ ++ R R
Sbjct: 93 ARRIGFSTRVSSSRRSRRDGAIIQRQFVCAKEGFRNMNEKRTKDREI---------KRPR 143
Query: 113 KLTREGCKARLVLKRTIDGKYEVSNFYEGHSHGLVSPSKRQFLRSARNVTSVHKNILFSC 172
+TR GCKA L +K GK+ VS F + H+H LV P + LRS R ++ K ++ +
Sbjct: 144 TITRVGCKASLSVKMQDSGKWLVSGFVKDHNHELVPPDQVHCLRSHRQISGPAKTLIDTL 203
Query: 173 NRANVGTSKSYQIMKEQVGSYENIGCTQRDLQNYSRNLKELIKDSDADMFIDNFRRKREI 232
A +G + + ++ G +G T+ D +NY RN ++ + + + +D R+
Sbjct: 204 QAAGMGPRRIMSALIKEYGGISKVGFTEVDCRNYMRNNRQKSIEGEIQLLLDYLRQMNAD 263
Query: 233 NPSFFYDYEADNEGKLKHVFWADGICRKNYSLFGDVVSFDTTYRTNKYFMIFAPFTGINH 292
NP+FFY + + + +VFWAD +++ FGD V+FDTTYR+N+Y + FAPFTG+NH
Sbjct: 264 NPNFFYSVQGSEDQSVGNVFWADPKAIMDFTHFGDTVTFDTTYRSNRYRLPFAPFTGVNH 323
Query: 293 HRQSITFGAALLKNEKEESFVWLFETFLKAMGGHKPVMIITDQDGGMKNAIGAVLKGSSH 352
H Q I FG A + NE E SFVWLF T+L AM H PV I TD D ++ AI V G+ H
Sbjct: 324 HGQPILFGCAFIINETEASFVWLFNTWLAAMSAHPPVSITTDHDAVIRAAIMHVFPGARH 383
Query: 353 RFCMWHILKKLSEKVGSSMDENSGFNDRFKSCVWNSESSEEFDLEWNNIISDYSLEGNGW 412
RFC WHILKK EK+ ++ F F CV +ES E+F+ W +++ Y L + W
Sbjct: 384 RFCKWHILKKCQEKLSHVFLKHPSFESDFHKCVNLTESVEDFERCWFSLLDKYELRDHEW 443
Query: 413 LSTMYDLRSMWIPAYFKDTFMAGILRTTSRSESENSFFGNYLNHNLTLVEFWVRFDSALA 472
L +Y R W+P Y +DTF A + T RS+S NS+F Y+N + L +F+ ++ AL
Sbjct: 444 LQAIYSDRRQWVPVYLRDTFFAD-MSLTHRSDSINSYFDGYINASTNLSQFFKLYEKALE 502
Query: 473 AQRHKELFADNNTLHSNPELNMHMNLEKHAREVYTHENFYIFQKELWSACVDCGIEGTKE 532
++ KE+ AD +T++S P L +EK A E+YT + F FQ+EL + +
Sbjct: 503 SRLEKEVKADYDTMNSPPVLKTPSPMEKQASELYTRKLFMRFQEELVGTLTFMASK-ADD 561
Query: 533 EGENLSFSILD-NAVRKHREVVYCLSNNIAHCSCKMFESEGIPCRHILFILKGKGFSEIP 591
+G+ +++ + K V + + A+CSC+MFE GI CRHIL + + +P
Sbjct: 562 DGDLVTYQVAKYGEAHKAHFVKFNVLEMRANCSCQMFEFSGIICRHILAVFRVTNLLTLP 621
Query: 592 SHYIVNRWTKLATSKPIFDCDGNVLEACSKFESES 626
+YI+ RWT+ A S IFD D N+ + ES +
Sbjct: 622 PYYILKRWTRNAKSSVIFD-DYNLHAYANYLESHT 655
>At4g15090 unknown protein
Length = 768
Score = 336 bits (861), Expect = 3e-92
Identities = 194/543 (35%), Positives = 292/543 (53%), Gaps = 18/543 (3%)
Query: 65 RNSIKTKDKDGVKWKYFLCSKEGFKEEKKVDKPQLLIAENSLSKSRKRKLTREGCKARLV 124
R S KTKD K F CS+ G E +E+S S SR+ + + CKA +
Sbjct: 25 RRSKKTKDFIDAK---FACSRYGVTPE----------SESSGSSSRRSTVKKTDCKASMH 71
Query: 125 LKRTIDGKYEVSNFYEGHSHGLVSPSKRQFLRSARNVTSVHKNILFSCNRANVGTSKSYQ 184
+KR DGK+ + F + H+H L+ P+ R RNV KN + + + T K Y
Sbjct: 72 VKRRPDGKWIIHEFVKDHNHELL-PALAYHFRIQRNVKLAEKNNIDILHAVSERTKKMYV 130
Query: 185 IMKEQVGSYENIGCT-QRDLQNYSRNLKEL-IKDSDADMFIDNFRRKREINPSFFYDYEA 242
M Q G Y+NIG Q D+ + + L +++ D+ + ++ F+R ++ NP FFY +
Sbjct: 131 EMSRQSGGYKNIGSLLQTDVSSQVDKGRYLALEEGDSQVLLEYFKRIKKENPKFFYAIDL 190
Query: 243 DNEGKLKHVFWADGICRKNYSLFGDVVSFDTTYRTNKYFMIFAPFTGINHHRQSITFGAA 302
+ + +L+++FWAD R +Y F DVVSFDTTY + A F G+NHH Q + G A
Sbjct: 191 NEDQRLRNLFWADAKSRDDYLSFNDVVSFDTTYVKFNDKLPLALFIGVNHHSQPMLLGCA 250
Query: 303 LLKNEKEESFVWLFETFLKAMGGHKPVMIITDQDGGMKNAIGAVLKGSSHRFCMWHILKK 362
L+ +E E+FVWL +T+L+AMGG P +I+TDQD + +A+ +L + H F +WH+L+K
Sbjct: 251 LVADESMETFVWLIKTWLRAMGGRAPKVILTDQDKFLMSAVSELLPNTRHCFALWHVLEK 310
Query: 363 LSEKVGSSMDENSGFNDRFKSCVWNSESSEEFDLEWNNIISDYSLEGNGWLSTMYDLRSM 422
+ E M + F +F C++ S + +EFD+ W ++S + LE + WL +++ R
Sbjct: 311 IPEYFSHVMKRHENFLLKFNKCIFRSWTDDEFDMRWWKMVSQFGLENDEWLLWLHEHRQK 370
Query: 423 WIPAYFKDTFMAGILRTTSRSESENSFFGNYLNHNLTLVEFWVRFDSALAAQRHKELFAD 482
W+P + D F+AG + T+ RSES NSFF Y++ +TL EF ++ L + +E AD
Sbjct: 371 WVPTFMSDVFLAG-MSTSQRSESVNSFFDKYIHKKITLKEFLRQYGVILQNRYEEESVAD 429
Query: 483 NNTLHSNPELNMHMNLEKHAREVYTHENFYIFQKELWSACVDCGIEGTKEEGENLSFSIL 542
+T H P L EK YTH F FQ E+ V C KE+ +F +
Sbjct: 430 FDTCHKQPALKSPSPWEKQMATTYTHTIFKKFQVEVLGV-VACHPRKEKEDENMATFRVQ 488
Query: 543 DNAVRKHREVVYCLSNNIAHCSCKMFESEGIPCRHILFILKGKGFSEIPSHYIVNRWTKL 602
D V + + + C C+MFE +G CRH L IL+ GF+ IP YI+ RWTK
Sbjct: 489 DCEKDDDFLVTWSKTKSELCCFCRMFEYKGFLCRHALMILQMCGFASIPPQYILKRWTKD 548
Query: 603 ATS 605
A S
Sbjct: 549 AKS 551
>At2g27110 Mutator-like transposase
Length = 851
Score = 301 bits (771), Expect = 9e-82
Identities = 193/622 (31%), Positives = 310/622 (49%), Gaps = 58/622 (9%)
Query: 4 DDNEVDFSSPSKDGNDPKNTSVEWIPACEDEL---KPVIGKVFDTLVEGGDFYKAYAYVA 60
D+ +V+ S S N + V+ DE+ +P +G F++ E FY Y+
Sbjct: 19 DEGDVEPSDCSGQNNMDNSLGVQ------DEIGIAEPCVGMEFNSEKEAKSFYDEYSRQL 72
Query: 61 GFSVRNSIKTKDKDGVKWKYFLCSKEGFKEEKKVDKPQLLIAENSLSKSRKRKLTREGCK 120
GF+ + + + V + F+CS S SK KR+L+ E C
Sbjct: 73 GFT--SKLLPRTDGSVSVREFVCS--------------------SSSKRSKRRLS-ESCD 109
Query: 121 ARLVLKRTIDGKYEVSNFYEGHSHGLVSPSKRQFLRSARNVTSVHKNILFSCNRANVGTS 180
A + ++ K+ V+ F + H+HGL S + LR R+ + K+
Sbjct: 110 AMVRIELQGHEKWVVTKFVKEHTHGLASSNMLHCLRPRRHFANSEKS------------- 156
Query: 181 KSYQ--IMKEQVGSYENIGCTQRDLQNYSR--NLKELIKDSDADMFIDNFRRKREINPSF 236
SYQ + Y ++ R +N S N K I DA ++ F+R + NP F
Sbjct: 157 -SYQEGVNVPSGMMYVSMDANSRGARNASMATNTKRTI-GRDAHNLLEYFKRMQAENPGF 214
Query: 237 FYDYEADNEGKLKHVFWADGICRKNYSLFGDVVSFDTTYRTNKYFMIFAPFTGINHHRQS 296
FY + D + ++ +VFWAD R Y+ FGD V+ DT YR N++ + FAPFTG+NHH Q+
Sbjct: 215 FYAVQLDEDNQMSNVFWADSRSRVAYTHFGDTVTLDTRYRCNQFRVPFAPFTGVNHHGQA 274
Query: 297 ITFGAALLKNEKEESFVWLFETFLKAMGGHKPVMIITDQDGGMKNAIGAVLKGSSHRFCM 356
I FG AL+ +E + SF+WLF+TFL AM PV ++TDQD ++ A G V G+ H
Sbjct: 275 ILFGCALILDESDTSFIWLFKTFLTAMRDQPPVSLVTDQDRAIQIAAGQVFPGARHCINK 334
Query: 357 WHILKKLSEKVGSSMDENSGFNDRFKSCVWNSESSEEFDLEWNNIISDYSLEGNGWLSTM 416
W +L++ EK+ F +C+ +E+ EEF+ W+++I Y L + WL+++
Sbjct: 335 WDVLREGQEKLAHVCLAYPSFQVELYNCINFTETIEEFESSWSSVIDKYDLGRHEWLNSL 394
Query: 417 YDLRSMWIPAYFKDTFMAGILRTTSRSESENSFFGNYLNHNLTLVEFWVRFDSALAAQRH 476
Y+ R+ W+P YF+D+F A + + S SFF Y+N TL F+ ++ A+ +
Sbjct: 395 YNARAQWVPVYFRDSFFAAVFPSQGYS---GSFFDGYVNQQTTLPMFFRLYERAMESWFE 451
Query: 477 KELFADNNTLHSNPELNMHMNLEKHAREVYTHENFYIFQKELWSACVDCGIEGTKEEGEN 536
E+ AD +T+++ P L +E A ++T + F FQ+EL +++G
Sbjct: 452 MEIEADLDTVNTPPVLKTPSPMENQAANLFTRKIFGKFQEELVETFAHTA-NRIEDDGTT 510
Query: 537 LSFSILD-NAVRKHREVVYCLSNNIAHCSCKMFESEGIPCRHILFILKGKGFSEIPSHYI 595
+F + + K V +C A+CSC+MFE GI CRH+L + +P HYI
Sbjct: 511 STFRVANFENDNKAYIVTFCYPEMRANCSCQMFEHSGILCRHVLTVFTVTNILTLPPHYI 570
Query: 596 VNRWTKLATSKPIFDCDGNVLE 617
+ RWT+ +K + + D +V E
Sbjct: 571 LRRWTR--NAKSMVELDEHVSE 590
>At3g06250 unknown protein
Length = 764
Score = 296 bits (757), Expect = 4e-80
Identities = 193/586 (32%), Positives = 287/586 (48%), Gaps = 65/586 (11%)
Query: 36 KPVIGKVFDTLVEGGDFYKAYAYVAGFSVR--NSIKTKDKDGVKWKYFLCSKEGFKEEKK 93
+P G F++ E FY+AYA V GF VR ++K + + F+CSKEGF+
Sbjct: 189 EPYAGLEFNSANEACQFYQAYAEVVGFRVRIGQLFRSKVDGSITSRRFVCSKEGFQHP-- 246
Query: 94 VDKPQLLIAENSLSKSRKRKLTREGCKARLVLKRTIDGKYEVSNFYEGHSHGLVSPSKRQ 153
+R GC A + +KR G + V + H+H L P K+
Sbjct: 247 ---------------------SRMGCGAYMRIKRQDSGGWIVDRLNKDHNHDL-EPGKK- 283
Query: 154 FLRSARNVTSVHKNILFSCNRANVGTSKSYQIMKEQVGSYENIGCTQ-RDLQNYSRNLKE 212
N G K I + G +++ + DL N+ + +E
Sbjct: 284 ----------------------NAGMKK---ITDDVTGGLDSVDLIELNDLSNHISSTRE 318
Query: 213 -LIKDSDADMFIDNFRRKREINPSFFYDYEADNEGKLKHVFWADGICRKNYSLFGDVVSF 271
I + +D F+ K+ + FFY E D+ G +FWAD R S FGD V F
Sbjct: 319 NTIGKEWYPVLLDYFQSKQAEDMGFFYAIELDSNGSCMSIFWADSRSRFACSQFGDAVVF 378
Query: 272 DTTYRTNKYFMIFAPFTGINHHRQSITFGAALLKNEKEESFVWLFETFLKAMGGHKPVMI 331
DT+YR Y + FA F G NHHRQ + G AL+ +E +E+F WLF+T+L+AM G +P +
Sbjct: 379 DTSYRKGDYSVPFATFIGFNHHRQPVLLGGALVADESKEAFSWLFQTWLRAMSGRRPRSM 438
Query: 332 ITDQDGGMKNAIGAVLKGSSHRFCMWHILKKLSEKVGSSMDENSGFNDRFKSCVWNSESS 391
+ DQD ++ A+ V G+ HRF W I K E + S +E F ++ C++ S+++
Sbjct: 439 VADQDLPIQQAVAQVFPGTHHRFSAWQIRSKERENLRSFPNE---FKYEYEKCLYQSQTT 495
Query: 392 EEFDLEWNNIISDYSLEGNGWLSTMYDLRSMWIPAYFKDTFMAGILRTTSRSESENSFFG 451
EFD W+++++ Y L N WL +Y+ R W+PAY + +F GI + + F+G
Sbjct: 496 VEFDTMWSSLVNKYGLRDNMWLREIYEKREKWVPAYLRASFFGGI----HVDGTFDPFYG 551
Query: 452 NYLNHNLTLVEFWVRFDSALAAQRHKELFADNNTLHSNPELNMHMNLEKHAREVYTHENF 511
LN +L EF R++ L +R +E D N+ + P L +E+ R +YT F
Sbjct: 552 TSLNSLTSLREFISRYEQGLEQRREEERKEDFNSYNLQPFLQTKEPVEEQCRRLYTLTIF 611
Query: 512 YIFQKELWSACVDCGIEGTKEEGENLSFSI--LDNAVRKHREVVYCLSNNIAHCSCKMFE 569
IFQ EL + G++ T EEG F + N KH V + SN A CSC+MFE
Sbjct: 612 RIFQSELAQSYNYLGLK-TYEEGAISRFLVRKCGNENEKH-AVTFSASNLNASCSCQMFE 669
Query: 570 SEGIPCRHILFILKGKGFSEIPSHYIVNRWTKLATSKPIFDCDGNV 615
EG+ CRHIL + E+PS YI++RWTK A + D + V
Sbjct: 670 YEGLLCRHILKVFNLLDIRELPSRYILHRWTKNAEFGFVRDVESGV 715
Score = 51.2 bits (121), Expect = 2e-06
Identities = 33/114 (28%), Positives = 51/114 (43%), Gaps = 25/114 (21%)
Query: 35 LKPVIGKVFDTLVEGGDFYKAYAYVAGFSVRNS--IKTKDKDGVKWKYFLCSKEGFKEEK 92
L+P +G FDT E D+Y +YA GF VR +++ V + F+CSKEGF+
Sbjct: 26 LEPYVGLEFDTAEEARDYYNSYATRTGFKVRTGQLYRSRTDGTVSSRRFVCSKEGFQLN- 84
Query: 93 KVDKPQLLIAENSLSKSRKRKLTREGCKARLVLKRTIDGKYEVSNFYEGHSHGL 146
+R GC A + ++R GK+ + + H+H L
Sbjct: 85 ----------------------SRTGCPAFIRVQRRDTGKWVLDQIQKEHNHDL 116
>At3g22170 far-red impaired response protein, putative
Length = 814
Score = 288 bits (737), Expect = 8e-78
Identities = 170/578 (29%), Positives = 290/578 (49%), Gaps = 31/578 (5%)
Query: 35 LKPVIGKVFDTLVEGGDFYKAYAYVAGFS--VRNSIKTKDKDGVKWKYFLCSKEGFKEE- 91
L+P+ G F++ E FY+ Y+ GF+ ++NS ++K F CS+ G K E
Sbjct: 68 LEPLNGMEFESHGEAYSFYQEYSRAMGFNTAIQNSRRSKTTREFIDAKFACSRYGTKREY 127
Query: 92 -KKVDKPQLLIA-ENSLSKSRKRKLTREGCKARLVLKRTIDGKYEVSNFYEGHSHGLVSP 149
K ++P+ + ++ + + +R + CKA + +KR DGK+ + +F H+H L+
Sbjct: 128 DKSFNRPRARQSKQDPENMAGRRTCAKTDCKASMHVKRRPDGKWVIHSFVREHNHELLP- 186
Query: 150 SKRQFLRSARNVTSVHKNILFSCNRANVGTSKSYQIMKEQVGSYENIGCTQRDLQNYSRN 209
+ T K Y M +Q Y+ + + D ++
Sbjct: 187 ----------------------AQAVSEQTRKIYAAMAKQFAEYKTVISLKSDSKSSFEK 224
Query: 210 LKEL-IKDSDADMFIDNFRRKREINPSFFYDYEADNEGKLKHVFWADGICRKNYSLFGDV 268
+ L ++ D + +D R + +N +FFY + ++ ++K+VFW D R NY F DV
Sbjct: 225 GRTLSVETGDFKILLDFLSRMQSLNSNFFYAVDLGDDQRVKNVFWVDAKSRHNYGSFCDV 284
Query: 269 VSFDTTYRTNKYFMIFAPFTGINHHRQSITFGAALLKNEKEESFVWLFETFLKAMGGHKP 328
VS DTTY NKY M A F G+N H Q + G AL+ +E ++ WL ET+L+A+GG P
Sbjct: 285 VSLDTTYVRNKYKMPLAIFVGVNQHYQYMVLGCALISDESAATYSWLMETWLRAIGGQAP 344
Query: 329 VMIITDQDGGMKNAIGAVLKGSSHRFCMWHILKKLSEKVGSSMDENSGFNDRFKSCVWNS 388
++IT+ D M + + + + H +WH+L K+SE +G + ++ F +F+ C++ S
Sbjct: 345 KVLITELDVVMNSIVPEIFPNTRHCLFLWHVLMKVSENLGQVVKQHDNFMPKFEKCIYKS 404
Query: 389 ESSEEFDLEWNNIISDYSLEGNGWLSTMYDLRSMWIPAYFKDTFMAGILRTTSRSESENS 448
E+F +W ++ + L+ + W+ ++Y+ R W P Y D +AG + T+ R++S N+
Sbjct: 405 GKDEDFARKWYKNLARFGLKDDQWMISLYEDRKKWAPTYMTDVLLAG-MSTSQRADSINA 463
Query: 449 FFGNYLNHNLTLVEFWVRFDSALAAQRHKELFADNNTLHSNPELNMHMNLEKHAREVYTH 508
FF Y++ ++ EF +D+ L + +E AD+ + P + EK EVYT
Sbjct: 464 FFDKYMHKKTSVQEFVKVYDTVLQDRCEEEAKADSEMWNKQPAMKSPSPFEKSVSEVYTP 523
Query: 509 ENFYIFQKELWSACVDCGIEGTKEEGENLSFSILDNAVRKHREVVYCLSNNIAHCSCKMF 568
F FQ E+ A + C + +F + D + V + + C C++F
Sbjct: 524 AVFKKFQIEVLGA-IACSPREENRDATCSTFRVQDFENNQDFMVTWNQTKAEVSCICRLF 582
Query: 569 ESEGIPCRHILFILKGKGFSEIPSHYIVNRWTKLATSK 606
E +G CRH L +L+ S IPS YI+ RWTK A S+
Sbjct: 583 EYKGYLCRHTLNVLQCCHLSSIPSQYILKRWTKDAKSR 620
>At1g76320 putative phytochrome A signaling protein
Length = 670
Score = 282 bits (722), Expect = 4e-76
Identities = 178/570 (31%), Positives = 287/570 (50%), Gaps = 32/570 (5%)
Query: 43 FDTLVEGGDFYKAYAYVAGF-----SVRNSIKTKDKDGVKWKYFLCSKEGFKEEKKVDKP 97
F+T + FYK YA GF S R S +K+ K F C + G K++
Sbjct: 3 FETHEDAYLFYKDYAKSVGFGTAKLSSRRSRASKEFIDAK---FSCIRYGSKQQ------ 53
Query: 98 QLLIAENSLSKSRKRKLTREGCKARLVLKRTIDGKYEVSNFYEGHSHGLVSPSKRQFLRS 157
++++++ K+ GCKA + +KR DGK+ V +F + H+H L+ P + + RS
Sbjct: 54 ----SDDAINPRASPKI---GCKASMHVKRRPDGKWYVYSFVKEHNHDLL-PEQAHYFRS 105
Query: 158 ARNVTSVHKNILFSCNRANVGTSKSYQIMKEQVGSYENIGCTQRDLQNYSRNLKELIKDS 217
RN V N + N + + + +Y ++ ++N + L+ D+
Sbjct: 106 HRNTELVKSNDSRLRRKKNTPLTDC-----KHLSAYHDLDFIDGYMRNQHDKGRRLVLDT 160
Query: 218 -DADMFIDNFRRKREINPSFFYDYEADNEGKLKHVFWADGICRKNYSLFGDVVSFDTTYR 276
DA++ ++ R +E NP FF+ + + L++VFW D ++Y F DVVSF+T+Y
Sbjct: 161 GDAEILLEFLMRMQEENPKFFFAVDFSEDHLLRNVFWVDAKGIEDYKSFSDVVSFETSYF 220
Query: 277 TNKYFMIFAPFTGINHHRQSITFGAALLKNEKEESFVWLFETFLKAMGGHKPVMIITDQD 336
+KY + F G+NHH Q + G LL ++ ++VWL +++L AMGG KP +++TDQ+
Sbjct: 221 VSKYKVPLVLFVGVNHHVQPVLLGCGLLADDTVYTYVWLMQSWLVAMGGQKPKVMLTDQN 280
Query: 337 GGMKNAIGAVLKGSSHRFCMWHILKKLSEKVGSSMDENSGFNDRFKSCVWNSESSEEFDL 396
+K AI AVL + H +C+WH+L +L + F + C++ S S EEFD
Sbjct: 281 NAIKAAIAAVLPETRHCYCLWHVLDQLPRNLDYWSMWQDTFMKKLFKCIYRSWSEEEFDR 340
Query: 397 EWNNIISDYSLEGNGWLSTMYDLRSMWIPAYFKDTFMAGILRTTSRSESENSFFGNYLNH 456
W +I + L W+ ++Y+ R W P + + AG L RSES NS F Y++
Sbjct: 341 RWLKLIDKFHLRDVPWMRSLYEERKFWAPTFMRGITFAG-LSMRCRSESVNSLFDRYVHP 399
Query: 457 NLTLVEFWVRFDSALAAQRHKELFADNNTLHSNPELNMHMNLEKHAREVYTHENFYIFQK 516
+L EF + L + +E AD + H PEL EK VY+HE F FQ
Sbjct: 400 ETSLKEFLEGYGLMLEDRYEEEAKADFDAWHEAPELKSPSPFEKQMLLVYSHEIFRRFQL 459
Query: 517 ELWSACVDCGIEGTKEEGENLSFSILDNAVRKHREVVYCLSNNIAHCSCKMFESEGIPCR 576
E+ A C + TKE E ++S+ D + V + + +CSC+ FE +G CR
Sbjct: 460 EVLGAAA-CHL--TKESEEGTTYSVKDFDDEQKYLVDWDEFKSDIYCSCRSFEYKGYLCR 516
Query: 577 HILFILKGKGFSEIPSHYIVNRWTKLATSK 606
H + +L+ G IP +Y++ RWT A ++
Sbjct: 517 HAIVVLQMSGVFTIPINYVLQRWTNAARNR 546
>At5g18960 FAR1 - like protein
Length = 788
Score = 270 bits (690), Expect = 2e-72
Identities = 191/630 (30%), Positives = 299/630 (47%), Gaps = 74/630 (11%)
Query: 36 KPVIGKVFDTLVEGGDFYKAYAYVAGFSVR--NSIKTKDKDGVKWKYFLCSKEGFKEEKK 93
+P G F + E FY+AYA V GF VR ++K + + F+CS+EGF+
Sbjct: 210 EPYAGLEFGSANEACQFYQAYAEVVGFRVRIGQLFRSKVDGSITSRRFVCSREGFQHP-- 267
Query: 94 VDKPQLLIAENSLSKSRKRKLTREGCKARLVLKRTIDGKYEVSNFYEGHSHGLVSPSKRQ 153
+R GC A + +KR G + V + H+H L P K+
Sbjct: 268 ---------------------SRMGCGAYMRIKRQDSGGWIVDRLNKDHNHDL-EPGKKN 305
Query: 154 FLRSARNVTSVHKNILFSCNRANVGTSKSYQIMKEQVGSYENIGCTQRDLQNYSRNLKEL 213
+ G K I + G +++ + L ++ N +
Sbjct: 306 ----------------------DAGMKK---IPDDGTGGLDSVDLIE--LNDFGNNHIKK 338
Query: 214 IKDSDAD-----MFIDNFRRKREINPSFFYDYEAD-NEGKLKHVFWADGICRKNYSLFGD 267
+++ + +D F+ ++ + FFY E D N G +FWAD R S FGD
Sbjct: 339 TRENRIGKEWYPLLLDYFQSRQTEDMGFFYAVELDVNNGSCMSIFWADSRARFACSQFGD 398
Query: 268 VVSFDTTYRTNKYFMIFAPFTGINHHRQSITFGAALLKNEKEESFVWLFETFLKAMGGHK 327
V FDT+YR Y + FA G NHHRQ + G A++ +E +E+F+WLF+T+L+AM G +
Sbjct: 399 SVVFDTSYRKGSYSVPFATIIGFNHHRQPVLLGCAMVADESKEAFLWLFQTWLRAMSGRR 458
Query: 328 PVMIITDQDGGMKNAIGAVLKGSSHRFCMWHILKKLSEKVGSSMDENSGFNDRFKSCVWN 387
P I+ DQD ++ A+ V G+ HR+ W I +K E + + S F ++ C++
Sbjct: 459 PRSIVADQDLPIQQALVQVFPGAHHRYSAWQIREKERENL---IPFPSEFKYEYEKCIYQ 515
Query: 388 SESSEEFDLEWNNIISDYSLEGNGWLSTMYDLRSMWIPAYFKDTFMAGILRTTSRSESEN 447
+++ EFD W+ +I+ Y L + WL +Y+ R W+PAY + +F AGI + +
Sbjct: 516 TQTIVEFDSVWSALINKYGLRDDVWLREIYEQRENWVPAYLRASFFAGI----PINGTIE 571
Query: 448 SFFGNYLNHNLTLVEFWVRFDSALAAQRHKELFADNNTLHSNPELNMHMNLEKHAREVYT 507
FFG L+ L EF R++ AL +R +E D N+ + P L +E+ R +YT
Sbjct: 572 PFFGASLDALTPLREFISRYEQALEQRREEERKEDFNSYNLQPFLQTKEPVEEQCRRLYT 631
Query: 508 HENFYIFQKELWSACVDCGIEGTKEEGENLSFSI--LDNAVRKHREVVYCLSNNIAHCSC 565
F IFQ EL + ++ T EEG F + N KH V + SN + CSC
Sbjct: 632 LTVFRIFQNELVQSYNYLCLK-TYEEGAISRFLVRKCGNESEKH-AVTFSASNLNSSCSC 689
Query: 566 KMFESEGIPCRHILFILKGKGFSEIPSHYIVNRWTKLATSKPIFDCDGNVLEACSKFESE 625
+MFE EG+ CRHIL + E+PS YI++RWTK A + D + V S + +
Sbjct: 690 QMFEHEGLLCRHILKVFNLLDIRELPSRYILHRWTKNAEFGFVRDMESGV----SAQDLK 745
Query: 626 STLVSKTWSQLLKCMHMAGTNKEKLLLIYD 655
+ +V K + ++ EK L Y+
Sbjct: 746 ALMVWSLREAASKYIEFGTSSLEKYKLAYE 775
Score = 50.4 bits (119), Expect = 4e-06
Identities = 33/114 (28%), Positives = 51/114 (43%), Gaps = 25/114 (21%)
Query: 35 LKPVIGKVFDTLVEGGDFYKAYAYVAGFSVRNS--IKTKDKDGVKWKYFLCSKEGFKEEK 92
++P +G FDT E +FY AYA GF VR +++ V + F+CSKEGF+
Sbjct: 41 VEPYVGLEFDTAEEAREFYNAYAARTGFKVRTGQLYRSRTDGTVSSRRFVCSKEGFQLN- 99
Query: 93 KVDKPQLLIAENSLSKSRKRKLTREGCKARLVLKRTIDGKYEVSNFYEGHSHGL 146
+R GC A + ++R GK+ + + H+H L
Sbjct: 100 ----------------------SRTGCTAFIRVQRRDTGKWVLDQIQKEHNHEL 131
>At2g32250 Mutator-like transposase
Length = 684
Score = 267 bits (683), Expect = 1e-71
Identities = 168/559 (30%), Positives = 263/559 (46%), Gaps = 60/559 (10%)
Query: 52 FYKAYAYVAGFSVRNSIKTKDKDGVKW--KYFLCSKEGFKEEKKVDKPQLLIAENSLSKS 109
FY+ YA GF + + K K+ CS+ G K EK +
Sbjct: 53 FYREYARSVGFGITIKASRRSKRSGKFIDVKIACSRFGTKREKA-------------TAI 99
Query: 110 RKRKLTREGCKARLVLKRTIDGKYEVSNFYEGHSHGLVSPSKRQFLRSARNVTSVHKNIL 169
R + GCKA L +KR D K+ + NF + H+H + F S R
Sbjct: 100 NPRSCPKTGCKAGLHMKRKEDEKWVIYNFVKEHNHEICPDD---FYVSVR---------- 146
Query: 170 FSCNRANVGTSKSYQIMKEQVGSYENIGCTQRDLQNYSRNLKELIKDSDADMFIDNFRRK 229
G +K + + G L+ +++ D + +++F
Sbjct: 147 --------GKNKPAGALAIKKG------------------LQLALEEEDLKLLLEHFMEM 180
Query: 230 REINPSFFYDYEADNEGKLKHVFWADGICRKNYSLFGDVVSFDTTYRTNKYFMIFAPFTG 289
++ P FFY + D++ ++++VFW D + +Y F DVV FDT Y N Y + FAPF G
Sbjct: 181 QDKQPGFFYAVDFDSDKRVRNVFWLDAKAKHDYCSFSDVVLFDTFYVRNGYRIPFAPFIG 240
Query: 290 INHHRQSITFGAALLKNEKEESFVWLFETFLKAMGGHKPVMIITDQDGGMKNAIGAVLKG 349
++HHRQ + G AL+ E ++ WLF T+LKA+GG P ++ITDQD + + + V
Sbjct: 241 VSHHRQYVLLGCALIGEVSESTYSWLFRTWLKAVGGQAPGVMITDQDKLLSDIVVEVFPD 300
Query: 350 SSHRFCMWHILKKLSEKVGSSMDENSGFNDRFKSCVWNSESSEEFDLEWNNIISDYSLEG 409
H FC+W +L K+SE + + ++ GF + F +CV +S + E F+ W+N+I + L
Sbjct: 301 VRHIFCLWSVLSKISEMLNPFVSQDDGFMESFGNCVASSWTDEHFERRWSNMIGKFELNE 360
Query: 410 NGWLSTMYDLRSMWIPAYFKDTFMAGILRTTSRSESENSFFGNYLNHNLTLVEFWVRFDS 469
N W+ ++ R W+P YF +AG L RS S S F Y+N T +F+ +
Sbjct: 361 NEWVQLLFRDRKKWVPHYFHGICLAG-LSGPERSGSIASHFDKYMNSEATFKDFFELYMK 419
Query: 470 ALAAQRHKELFADNNTLHSNPELNMHMNLEKHAREVYTHENFYIFQKELWSACVDCGIEG 529
L + E D P L + EK +YT F FQ E+ V C ++
Sbjct: 420 FLQYRCDVEAKDDLEYQSKQPTLRSSLAFEKQLSLIYTDAAFKKFQAEV-PGVVSCQLQK 478
Query: 530 TKEEGENLSFSILDNAVRKHREVVYCLSNNI--AHCSCKMFESEGIPCRHILFILKGKGF 587
+E+G F I D R++ V L+N + A CSC +FE +G C+H + +L+
Sbjct: 479 EREDGTTAIFRIEDFEERQNFFV--ALNNELLDACCSCHLFEYQGFLCKHAILVLQSADV 536
Query: 588 SEIPSHYIVNRWTKLATSK 606
S +PS YI+ RW+K +K
Sbjct: 537 SRVPSQYILKRWSKKGNNK 555
>At4g38170 hypothetical protein
Length = 531
Score = 245 bits (625), Expect = 7e-65
Identities = 137/383 (35%), Positives = 206/383 (53%), Gaps = 11/383 (2%)
Query: 225 NFRRKREI-NPSFFYDYEADNEGKLKHVFWADGICRKNYSLFGDVVSFDTTYRTNKYFMI 283
N+ ++R++ NP F Y E D +VFWAD CR NY+ FGD + FDTTYR K + +
Sbjct: 9 NYLKRRQLENPGFLYAIEDD----CGNVFWADPTCRLNYTYFGDTLVFDTTYRRGKRYQV 64
Query: 284 -FAPFTGINHHRQSITFGAALLKNEKEESFVWLFETFLKAMGGHKPVMIITDQDGGMKNA 342
FA FTG NHH Q + FG AL+ NE E SF WLF+T+L+AM P I + D ++ A
Sbjct: 65 PFAAFTGFNHHGQPVLFGCALILNESESSFAWLFQTWLQAMSAPPPPSITVEPDRLIQVA 124
Query: 343 IGAVLKGSSHRFCMWHILKKLSEKVGSSMDENSGFNDRFKSCVWNSESSEEFDLEWNNII 402
+ V + RF I ++ EK+ + F F +CV +E++ EF+ W++I+
Sbjct: 125 VSRVFSQTRLRFSQPLIFEETEEKLAHVFQAHPTFESEFINCVTETETAAEFEASWDSIV 184
Query: 403 SDYSLEGNGWLSTMYDLRSMWIPAYFKDTFMAGILRTTSRSESENSFFGNYLNHNLTLVE 462
Y +E N WL ++Y+ R W+ + +DTF G L T S NSFF +++ + T+
Sbjct: 185 RRYYMEDNDWLQSIYNARQQWVRVFIRDTFY-GELSTNEGSSILNSFFQGFVDASTTMQM 243
Query: 463 FWVRFDSALAAQRHKELFADNNTLHSNPELNMHMNLEKHAREVYTHENFYIFQKE-LWSA 521
+++ A+ + R KEL AD +S P + +EK A +YT F FQ+E + +
Sbjct: 244 LIKQYEKAIDSWREKELKADYEATNSTPVMKTPSPMEKQAASLYTRAAFIKFQEEFVETL 303
Query: 522 CVDCGIEGTKEEGENLSFSILD-NAVRKHREVVYCLSNNIAHCSCKMFESEGIPCRHILF 580
+ I + G + ++ + V K V + A+CSC+MFE GI CRHIL
Sbjct: 304 AIPANI--ISDSGTHTTYRVAKFGEVHKGHTVSFDSLEVKANCSCQMFEYSGIICRHILA 361
Query: 581 ILKGKGFSEIPSHYIVNRWTKLA 603
+ K +PS Y++ RWTK A
Sbjct: 362 VFSAKNVLALPSRYLLRRWTKEA 384
>At1g10240 unknown protein
Length = 680
Score = 230 bits (587), Expect = 2e-60
Identities = 161/621 (25%), Positives = 300/621 (47%), Gaps = 58/621 (9%)
Query: 8 VDFSSPSKDGNDPKNTSVEWIPACEDELKPVIGKVFDTLVEGGDFYKAYAYVAGFSVRNS 67
+D +S +++ D N S+E + P +G++F T +FY +A GFS+R
Sbjct: 23 LDDASSTEESPDDNNLSLEAV----HNAIPYLGQIFLTHDTAYEFYSTFAKRCGFSIRRH 78
Query: 68 IKTKDKDGV----KWKYFLCSKEGFKEEKKVD--KPQLLIAENSLSKSRKRKLTREGCKA 121
+T+ KDGV +YF+C + G K + KPQ R R+ +R GC+A
Sbjct: 79 -RTEGKDGVGKGLTRRYFVCHRAGNTPIKTLSEGKPQ-----------RNRRSSRCGCQA 126
Query: 122 RLVLKRTID---GKYEVSNFYEGHSHGLVSPSKRQFLRSARNVTSVHKNILFSCNRANVG 178
L + + + ++ V+ F H+H L+ P++ +FL + R+++ K+ + ++ +
Sbjct: 127 YLRISKLTELGSTEWRVTGFANHHNHELLEPNQVRFLPAYRSISDADKSRILMFSKTGIS 186
Query: 179 TSKSYQIMK-EQVGSYENIGCTQRDLQNYSRNLKELIKDSDADMFIDNFRRKREINPSFF 237
+ ++++ E+ + T++D++N ++ K+L + + F+ + +E +P+F
Sbjct: 187 VQQMMRLLELEKCVEPGFLPFTEKDVRNLLQSFKKLDPEDENIDFLRMCQSIKEKDPNFK 246
Query: 238 YDYEADNEGKLKHVFWADGICRKNYSLFGDVVSFDTTYRTNKYFMIFAPFTGINHHRQSI 297
+++ D KL+++ W+ ++Y LFGD V FDTT+R + M + G+N++
Sbjct: 247 FEFTLDANDKLENIAWSYASSIQSYELFGDAVVFDTTHRLSAVEMPLGIWVGVNNYGVPC 306
Query: 298 TFGAALLKNEKEESFVWLFETFLKAMGGHKPVMIITDQDGGMKNAIGAVLKGSSHRFCMW 357
FG LL++E S+ W + F M G P I+TD + +K AI + + H C+W
Sbjct: 307 FFGCVLLRDENLRSWSWALQAFTGFMNGKAPQTILTDHNMCLKEAIAGEMPATKHALCIW 366
Query: 358 HILKKLSEKVGSSMDENSGFNDRFKS---CVWNSESSEEFDLEWNNIISDYSLEGNGWLS 414
++ K + + E +ND +K+ +++ ES EEF+L W ++++ + L N ++
Sbjct: 367 MVVGKFPSWFNAGLGER--YND-WKAEFYRLYHLESVEEFELGWRDMVNSFGLHTNRHIN 423
Query: 415 TMYDLRSMWIPAYFKDTFMAGILRTTSRSESENSFFGNYLNHNLTLVEFWVRFDSALAAQ 474
+Y RS+W Y + F+AG + T RS++ N+F +L+ L F +
Sbjct: 424 NLYASRSLWSLPYLRSHFLAG-MTLTGRSKAINAFIQRFLSAQTRLAHF---VEQVAVVV 479
Query: 475 RHKELFADNNTLHSNPE---LNMHMNLEKHAREVYTHENFYIFQKELWSACVDCGIEGTK 531
K+ + T+ N + L +E HA V T F Q++L A
Sbjct: 480 DFKDQATEQQTMQQNLQNISLKTGAPMESHAASVLTPFAFSKLQEQLVLA---------- 529
Query: 532 EEGENLSFSILDNAVRKH-------REVVYCLSNNIAHCSCKMFESEGIPCRHILFILKG 584
SF + + + +H R+V + I CSC++FE G CRH L +L
Sbjct: 530 --AHYASFQMDEGYLVRHHTKLDGGRKVYWVPQEGIISCSCQLFEFSGFLCRHALRVLST 587
Query: 585 KGFSEIPSHYIVNRWTKLATS 605
++P Y+ RW +++TS
Sbjct: 588 GNCFQVPDRYLPLRWRRISTS 608
>At4g19990 putative protein
Length = 672
Score = 229 bits (584), Expect = 4e-60
Identities = 176/646 (27%), Positives = 290/646 (44%), Gaps = 94/646 (14%)
Query: 40 GKVFDTLVEGGDFYKAYAYVAGFSVRNSIKTKDKDGVKWKY----FLCSKEGFKEEKKVD 95
G+ F++ E +FYK YA GF+ IK + + K+ F+C++ G K+E +D
Sbjct: 24 GREFESKEEAFEFYKEYANSVGFTT--IIKASRRSRMTGKFIDAKFVCTRYGSKKED-ID 80
Query: 96 KPQLLIAENSLSKSRKRKLTREG----CKARLVLKRTIDGKYEVSNFYEGHSHGLVSPSK 151
N ++ ++ R CKA L +KR DG++ V + + H+H + +
Sbjct: 81 TGLGTDGFNIPQARKRGRINRSSSKTDCKAFLHVKRRQDGRWVVRSLVKEHNHEIFTGQA 140
Query: 152 RQFLRSARNVTSVHKNILFSCNRANVGTSKSYQIMKEQVGSYENIGCTQRDLQNYSRNLK 211
S R ++ K L N A V KS ++
Sbjct: 141 D----SLRELSGRRK--LEKLNGAIVKEVKSRKL-------------------------- 168
Query: 212 ELIKDSDADMFIDNFRRKREINPSFFYDYEADNEGKLKHVFWADGICRKNYSLFGDVVSF 271
+D D + ++ FF D ++ L+++FW D R +Y+ F DVVS
Sbjct: 169 ---EDGDVERLLN-----------FFTDMQS-----LRNIFWVDAKGRFDYTCFSDVVSI 209
Query: 272 DTTYRTNKYFMIFAPFTGINHHRQSITFG-AALLKNEKEESFVWLFETFLKAMGGHKPVM 330
DTT+ N+Y + FTG+NHH Q + G LL +E + FVWLF +LKAM G +P +
Sbjct: 210 DTTFIKNEYKLPLVAFTGVNHHGQFLLLGFGLLLTDESKSGFVWLFRAWLKAMHGCRPRV 269
Query: 331 IITDQDGGMKNAIGAVLKGSSHRFCMWHILKKLSEKVGSSMDENSGFNDRFKSCVWNSES 390
I+T D +K A+ V S H F MW L ++ EK+G + D ++ S
Sbjct: 270 ILTKHDQMLKEAVLEVFPSSRHCFYMWDTLGQMPEKLGHVIRLEKKLVDEINDAIYGSCQ 329
Query: 391 SEEFDLEWNNIISDYSLEGNGWLSTMYDLRSMWIPAYFKDTFMAGILRTTSRSESENSFF 450
SE+F+ W ++ + + N WL ++Y+ R W+P Y KD +AG+ T RS+S NS
Sbjct: 330 SEDFEKNWWEVVDRFHMRDNVWLQSLYEDREYWVPVYMKDVSLAGMC-TAQRSDSVNSGL 388
Query: 451 GNYLNHNLTLVEFWVRFDSALAAQRHKELFADNNTLHSNPELNMHMNLEKHAREVYTHEN 510
Y+ T F ++ + + +E ++ TL+ P L K EVYT E
Sbjct: 389 DKYIQRKTTFKAFLEQYKKMIQERYEEEEKSEIETLYKQPGLKSPSPFGKQMAEVYTREM 448
Query: 511 FYIFQKELWSACVDCGIEGTKEEGEN-LSFSILDNAVRKHREVVYCLSNNIAHCSCKMFE 569
F FQ E+ + ++E+G N +F + D + VV+ ++ CSC++FE
Sbjct: 449 FKKFQVEVLGGVACHPKKESEEDGVNKRTFRVQDYEQNRSFVVVWNSESSEVVCSCRLFE 508
Query: 570 SEGIPCRHILFILKGKGFSEIPSHYIVNRWTKLATSKPIFDCDGNVLEACS--------- 620
KG IPS Y++ RWTK A S+ + + D +E+
Sbjct: 509 L--------------KGELSIPSQYVLKRWTKDAKSREVMESDQTDVESTKAQRYKDLCL 554
Query: 621 ---KFESESTLVSKTWSQLLKCMHMA---GTNKEKLLLIYDEGCSI 660
K E++L ++++ ++ ++ A NK L+ +E S+
Sbjct: 555 RSLKLSEEASLSEESYNAVVNVLNEALRKWENKSNLIQNLEESESV 600
>At1g52520 F6D8.26
Length = 703
Score = 229 bits (584), Expect = 4e-60
Identities = 168/581 (28%), Positives = 266/581 (44%), Gaps = 46/581 (7%)
Query: 37 PVIGKVFDTLVEGGDFYKAYAYVAGFSVR--NSI---KTKDKDGVKWKYFLCSKEGFKEE 91
P +G F++ + ++Y YA GF VR NS ++K+K G CS +GFK
Sbjct: 85 PAVGMEFESYDDAYNYYNCYASEVGFRVRVKNSWFKRRSKEKYGA---VLCCSSQGFKRI 141
Query: 92 KKVDKPQLLIAENSLSKSRKRKLTREGCKARLVLKRTIDGKYEVSNFYEGHSHGLVSPSK 151
V+ R RK TR GC A + +++ ++ V H+H L
Sbjct: 142 NDVN--------------RVRKETRTGCPAMIRMRQVDSKRWRVVEVTLDHNHLLGCKLY 187
Query: 152 RQFLRSARNVTSVHKNILFSCNRANVGTSKSYQIMKEQVGSYENIGCTQRDLQNYSRNLK 211
+ R + V+S ++ T K Y+ GS N T S
Sbjct: 188 KSVKRKRKCVSSPV---------SDAKTIKLYRACVVDNGSNVNPNSTLNKKFQNSTGSP 238
Query: 212 ELI--KDSDADMFIDNFRRKREINPSFFYDYEADNEGKLKHVFWADGICRKNYSLFGDVV 269
+L+ K D+ + F R + NP+FFY + ++EG+L++VFWAD + + S FGDV+
Sbjct: 239 DLLNLKRGDSAAIYNYFCRMQLTNPNFFYLMDVNDEGQLRNVFWADAFSKVSCSYFGDVI 298
Query: 270 SFDTTYRTNKYFMIFAPFTGINHHRQSITFGAALLKNEKEESFVWLFETFLKAMGGHKPV 329
D++Y + K+ + FTG+NHH ++ L E ES+ WL + +L M P
Sbjct: 299 FIDSSYISGKFEIPLVTFTGVNHHGKTTLLSCGFLAGETMESYHWLLKVWLSVM-KRSPQ 357
Query: 330 MIITDQDGGMKNAIGAVLKGSSHRFCMWHILKKLSEKVGSSMDENSGFNDRFKSCVWNSE 389
I+TD+ ++ AI V S RF + HI++K+ EK+G + F V+ +
Sbjct: 358 TIVTDRCKPLEAAISQVFPRSHQRFSLTHIMRKIPEKLG-GLHNYDAVRKAFTKAVYETL 416
Query: 390 SSEEFDLEWNNIISDYSLEGNGWLSTMYDLRSMWIPAYFKDTFMAGILRTTSRSESENSF 449
EF+ W ++ ++ + N WL ++Y+ R+ W P Y KDTF AGI E+ F
Sbjct: 417 KVVEFEAAWGFMVHNFGVIENEWLRSLYEERAKWAPVYLKDTFFAGI-AAAHPGETLKPF 475
Query: 450 FGNYLNHNLTLVEFWVRFDSALAAQRHKELFAD--NNTLHSNPELNMHMNLEKHAREVYT 507
F Y++ L EF +++ AL + +E +D + TL++ EL + E +YT
Sbjct: 476 FERYVHKQTPLKEFLDKYELALQKKHREETLSDIESQTLNT-AELKTKCSFETQLSRIYT 534
Query: 508 HENFYIFQKELWSACVDCGIEGTKEEGENLSFSIL-----DNAVRKHR--EVVYCLSNNI 560
+ F FQ E+ +G + F + +++ R+ R EV+Y S
Sbjct: 535 RDMFKKFQIEVEEMYSCFSTTQVHVDGPFVIFLVKERVRGESSRREIRDFEVLYNRSVGE 594
Query: 561 AHCSCKMFESEGIPCRHILFILKGKGFSEIPSHYIVNRWTK 601
C C F G CRH L +L G EIP YI+ RW K
Sbjct: 595 VRCICSCFNFYGYLCRHALCVLNFNGVEEIPLRYILPRWRK 635
>At5g28530 far-red impaired response protein (FAR1) - like
Length = 700
Score = 228 bits (580), Expect = 1e-59
Identities = 162/585 (27%), Positives = 294/585 (49%), Gaps = 45/585 (7%)
Query: 37 PVIGKVFDTLVEGGDFYKAYAYVAGFSVRNSIKTKDKD-GVKWKYFLCSKEGFKEEKKVD 95
P +G++F T E ++Y +A +GFS+R + T+ ++ GV + F+C + GF + +K
Sbjct: 70 PYVGQIFTTDDEAFEYYSTFARKSGFSIRKARSTESQNLGVYRRDFVCYRSGFNQPRK-- 127
Query: 96 KPQLLIAENSLSKSRKRKLTREGCKARLVL-KRTIDG--KYEVSNFYEGHSHGLVSPSKR 152
+ ++ R+RK R GC +L L K +DG + VS F H+H L+ +
Sbjct: 128 -------KANVEHPRERKSVRCGCDGKLYLTKEVVDGVSHWYVSQFSNVHNHELLEDDQV 180
Query: 153 QFLRSARNVTSVHKNILFSCNRANVGTSKSYQIMKEQVGSYEN-IGCTQRDLQNYSRNLK 211
+ L + R + + + ++A ++ ++++ + G + ++D++N+ R K
Sbjct: 181 RLLPAYRKIQQSDQERILLLSKAGFPVNRIVKLLELEKGVVSGQLPFIEKDVRNFVRACK 240
Query: 212 ELIKDSDADM----------FIDNFRRKREINPSFFYDYEADNEGKLKHVFWADGICRKN 261
+ ++++DA M ++ + E + F YD +D K++++ WA G +
Sbjct: 241 KSVQENDAFMTEKRESDTLELLECCKGLAERDMDFVYDCTSDENQKVENIAWAYGDSVRG 300
Query: 262 YSLFGDVVSFDTTYRTNKYFMIFAPFTGINHHRQSITFGAALLKNEKEESFVWLFETFLK 321
YSLFGDVV FDT+YR+ Y ++ F GI+++ +++ G LL++E SF W +TF++
Sbjct: 301 YSLFGDVVVFDTSYRSVPYGLLLGVFFGIDNNGKAMLLGCVLLQDESCRSFTWALQTFVR 360
Query: 322 AMGGHKPVMIITDQDGGMKNAIGAVLKGSSHRFCMWHILKKL----SEKVGSSMDE-NSG 376
M G P I+TD D G+K+AIG + ++H M HI+ KL S+ +GS +E +G
Sbjct: 361 FMRGRHPQTILTDIDTGLKDAIGREMPNTNHVVFMSHIVSKLASWFSQTLGSHYEEFRAG 420
Query: 377 FNDRFKSCVWNSESSEEFDLEWNNIISDYSLEGNGWLSTMYDLRSMWIPAYFKDTFMAGI 436
F+ + + + +EF+ +W+ +++ + L + + +Y R+ W+P ++ F+A
Sbjct: 421 FD-----MLCRAGNVDEFEQQWDLLVTRFGLVPDRHAALLYSCRASWLPCCIREHFVAQT 475
Query: 437 LRTTSRSESENSFFGNYLNHNLTLVEFWVRFDSALAAQRHKELFADNNTLHSNPELNMHM 496
+ T+ + S +SF ++ T ++ + +SAL L + P L M
Sbjct: 476 M-TSEFNLSIDSFLKRVVD-GATCMQLLLE-ESALQVSAAASLAKQILPRFTYPSLKTCM 532
Query: 497 NLEKHAREVYTHENFYIFQKELWSACVDCGIEGTKEEGENLSFSILDNAVRKHRE--VVY 554
+E HAR + T F + Q E+ ++ E N F I+ + + E V++
Sbjct: 533 PMEDHARGILTPYAFSVLQNEM-----VLSVQYAVAEMANGPF-IVHHYKKMEGECCVIW 586
Query: 555 CLSNNIAHCSCKMFESEGIPCRHILFILKGKGFSEIPSHYIVNRW 599
N CSCK FE GI CRH L +L K IP Y + RW
Sbjct: 587 NPENEEIQCSCKEFEHSGILCRHTLRVLTVKNCFHIPEQYFLLRW 631
>At1g80010 hypothetical protein
Length = 696
Score = 226 bits (577), Expect = 3e-59
Identities = 163/583 (27%), Positives = 260/583 (43%), Gaps = 37/583 (6%)
Query: 37 PVIGKVFDTLVEGGDFYKAYAYVAGFSVR-----NSIKTKDKDGVKWKYFLCSKEGFKEE 91
P G F++ + FY +YA GF++R +K+K G C+ +GFK
Sbjct: 66 PTPGMEFESYDDAYSFYNSYARELGFAIRVKSSWTKRNSKEKRGA---VLCCNCQGFKLL 122
Query: 92 KKVDKPQLLIAENSLSKSRKRKLTREGCKARLVLKRTIDGKYEVSNFYEGHSHGLV---S 148
K +RK TR GC+A + L+ +++V H+H +
Sbjct: 123 KDAHS--------------RRKETRTGCQAMIRLRLIHFDRWKVDQVKLDHNHSFDPQRA 168
Query: 149 PSKRQFLRSARNVTSVHKNILFSCNRANVGTSKSYQIMK----EQVGSYENIGCTQRDLQ 204
+ + +S+ + + K V T K Y+ + +G+ + G T
Sbjct: 169 HNSKSHKKSSSSASPATKTNPEPPPHVQVRTIKLYRTLALDTPPALGTSLSSGETSDLSL 228
Query: 205 NYSRNLKELIKDSDADMFIDNFRRKREINPSFFYDYEADNEGKLKHVFWADGICRKNYSL 264
++ ++ + L D F + + +P+F Y + ++G L++VFW D R YS
Sbjct: 229 DHFQSSRRLELRGGFRALQDFFFQIQLSSPNFLYLMDLADDGSLRNVFWIDARARAAYSH 288
Query: 265 FGDVVSFDTTYRTNKYFMIFAPFTGINHHRQSITFGAALLKNEKEESFVWLFETFLKAMG 324
FGDV+ FDTT +N Y + F GINHH +I G LL ++ E++VWLF +L M
Sbjct: 289 FGDVLLFDTTCLSNAYELPLVAFVGINHHGDTILLGCGLLADQSFETYVWLFRAWLTCML 348
Query: 325 GHKPVMIITDQDGGMKNAIGAVLKGSSHRFCMWHILKKLSEKVGSSMDENSGFNDRFKSC 384
G P + IT+Q M+ A+ V + HR + H+L + + V D + F
Sbjct: 349 GRPPQIFITEQCKAMRTAVSEVFPRAHHRLSLTHVLHNICQSVVQLQDSDL-FPMALNRV 407
Query: 385 VWNSESSEEFDLEWNNIISDYSLEGNGWLSTMYDLRSMWIPAYFKDTFMAGILRTTSRSE 444
V+ EEF+ W +I + + N + M+ R +W P Y KDTF+AG L +
Sbjct: 408 VYGCLKVEEFETAWEEMIIRFGMTNNETIRDMFQDRELWAPVYLKDTFLAGALTFPLGNV 467
Query: 445 SENSFFGNYLNHNLTLVEFWVRFDSALAAQRHKELFADNNTLHSNPELNMHMNLEKHARE 504
+ F Y++ N +L EF ++S L + +E D+ +L P+L E +
Sbjct: 468 AAPFIFSGYVHENTSLREFLEGYESFLDKKYTREALCDSESLKLIPKLKTTHPYESQMAK 527
Query: 505 VYTHENFYIFQKELWSACVDC-GIEGTKEEGENLSFSILDNAVRKHR--EVVY---CLSN 558
V+T E F FQ E+ SA C G+ G S+ + + K R EV+Y +
Sbjct: 528 VFTMEIFRRFQDEV-SAMSSCFGVTQVHSNGSASSYVVKEREGDKVRDFEVIYETSAAAQ 586
Query: 559 NIAHCSCKMFESEGIPCRHILFILKGKGFSEIPSHYIVNRWTK 601
C C F G CRH+L +L G E+P YI+ RW K
Sbjct: 587 VRCFCVCGGFSFNGYQCRHVLLLLSHNGLQEVPPQYILQRWRK 629
>At4g09380 putative protein
Length = 960
Score = 81.3 bits (199), Expect = 2e-15
Identities = 73/353 (20%), Positives = 149/353 (41%), Gaps = 42/353 (11%)
Query: 231 EINPSFFYDYEADNEGKLKHVFWADGICRKNYSLFGDVVSFDTTYRTNKY--FMIFAPFT 288
++NP E + E K K++F A G C + + V+ D T+ Y ++ A
Sbjct: 373 KVNPGTVTYVELEGEKKFKYLFIALGACIEGFRAMRKVIVVDATHLKTVYGGMLVIATAQ 432
Query: 289 GINHHRQSITFGAALLKNEKEESFVWLFETFLKAMGGHKPVMIITDQDGGMKNAIGAVLK 348
NHH + FG ++ +EK+ S++W E ++ I+D+ +K A+ V
Sbjct: 433 DPNHHHYPLAFG--IIDSEKDVSWIWFLENLKTVYSDVPGLVFISDRHQSIKKAVKTVYP 490
Query: 349 GSSHRFCMWHILKKLSEKVGSSMDENSGFNDRFKSCVWNSESSEEFDLEWNNIISDYSLE 408
+ H C+WH+ + + ++V D G +F+ C ++ + EF+ E+ + S +
Sbjct: 491 NALHAACIWHLCQNMRDRVKIDKD---GAAVKFRDCA-HAYTESEFEKEFGHFTSLWPKA 546
Query: 409 GNGWLSTMYDLRSMWIPAYFK-DTFMAGILRTTSRSESENSFFGNYLN-HNLTLVEFWVR 466
+ + ++ W +FK D + + T++ +ES N F H L +++ +
Sbjct: 547 ADFLVKVGFE---KWSRCHFKGDKYN---IDTSNSAESINGVFKKARKYHLLPMIDVMI- 599
Query: 467 FDSALAAQRHKELFADNNTLHSNPELNMHMNLEKHAREVYTHENFYIFQKELWSACVDCG 526
+ E F ++ + + A+ V T EN + ++ +
Sbjct: 600 -------SKFSEWFNEHRQASGSCPIT--------AQVVPTVENILHIRCQVAAKLTVFE 644
Query: 527 IEGTKEEGENLSFSILDNAVRKHREVVYCLSNNIAHCSCKMFESEGIPCRHIL 579
+ +E ++++D V + + + CSCK F+ + IPC H +
Sbjct: 645 LNSYNQE-----YNVID-----LNSVSFLVDLKMKSCSCKRFDIDKIPCVHAM 687
>At4g28970 putative protein
Length = 914
Score = 80.9 bits (198), Expect = 2e-15
Identities = 73/353 (20%), Positives = 149/353 (41%), Gaps = 42/353 (11%)
Query: 231 EINPSFFYDYEADNEGKLKHVFWADGICRKNYSLFGDVVSFDTTYRTNKY--FMIFAPFT 288
++NP E + E K K++F A G C + + V+ D T+ Y ++ A
Sbjct: 373 KVNPGTVTYVELEGEKKFKYLFIALGACIEGFRAMRKVIVVDATHLKTVYGGMLVIATAQ 432
Query: 289 GINHHRQSITFGAALLKNEKEESFVWLFETFLKAMGGHKPVMIITDQDGGMKNAIGAVLK 348
NHH + FG ++ +EK+ S++W E ++ I+D+ +K A+ V
Sbjct: 433 DPNHHHYPLAFG--IIDSEKDVSWIWFLEKLKTVYSDVPGLVFISDRHQSIKKAVKTVYP 490
Query: 349 GSSHRFCMWHILKKLSEKVGSSMDENSGFNDRFKSCVWNSESSEEFDLEWNNIISDYSLE 408
+ H C+WH+ + + ++V D G +F+ C ++ + EF+ E+ + S +
Sbjct: 491 NALHAACIWHLCQNMRDRVTIDKD---GAAVKFRDCA-HAYTESEFEKEFGHFTSLWPKA 546
Query: 409 GNGWLSTMYDLRSMWIPAYFK-DTFMAGILRTTSRSESENSFFGNYLN-HNLTLVEFWVR 466
+ + ++ W +FK D + + T++ +ES N F H L +++ +
Sbjct: 547 ADFLVKVGFE---KWSRCHFKGDKYN---IDTSNSAESINGVFKKARKYHLLPMIDVMI- 599
Query: 467 FDSALAAQRHKELFADNNTLHSNPELNMHMNLEKHAREVYTHENFYIFQKELWSACVDCG 526
+ E F ++ + + A+ V T EN + ++ +
Sbjct: 600 -------SKFSEWFNEHRQASGSCPIT--------AQVVPTVENILHIRCQVAAKLTVFE 644
Query: 527 IEGTKEEGENLSFSILDNAVRKHREVVYCLSNNIAHCSCKMFESEGIPCRHIL 579
+ +E ++++D V + + + CSCK F+ + IPC H +
Sbjct: 645 LNSYNQE-----YNVID-----LNSVSFLVDLKMKSCSCKCFDIDKIPCVHAM 687
>At3g47310 putative protein
Length = 735
Score = 77.0 bits (188), Expect = 4e-14
Identities = 72/353 (20%), Positives = 147/353 (41%), Gaps = 42/353 (11%)
Query: 231 EINPSFFYDYEADNEGKLKHVFWADGICRKNYSLFGDVVSFDTTYRTNKY--FMIFAPFT 288
++NP E + E K K++F A G C + + V+ D T+ Y ++ A
Sbjct: 322 KVNPGTVTYVELEGEKKFKYLFIALGACIEGFRTMRKVIVVDATHLKTVYGGMLVIATAQ 381
Query: 289 GINHHRQSITFGAALLKNEKEESFVWLFETFLKAMGGHKPVMIITDQDGGMKNAIGAVLK 348
NHH + FG ++ +E + S++W E ++ I+D+ +K + V
Sbjct: 382 DPNHHHYPLAFG--IIDSENDVSWIWFLEKLKTVYSDVPGLVFISDRHQSIKKVVKTVYP 439
Query: 349 GSSHRFCMWHILKKLSEKVGSSMDENSGFNDRFKSCVWNSESSEEFDLEWNNIISDYSLE 408
+ H C+WH+ + + ++V D G +F+ C ++ + EF+ E+ + S +
Sbjct: 440 NALHAACIWHLCQNMRDRVKIDKD---GAAVKFRDCA-HAYTESEFEKEFGHFTSLWPKA 495
Query: 409 GNGWLSTMYDLRSMWIPAYFK-DTFMAGILRTTSRSESENSFFGNYLN-HNLTLVEFWVR 466
+ + ++ W +FK D + + T++ +ES N F H L +++ +
Sbjct: 496 ADFLVKVGFE---KWSRCHFKGDKYN---IDTSNSAESINGVFKKARKYHLLPMIDVMI- 548
Query: 467 FDSALAAQRHKELFADNNTLHSNPELNMHMNLEKHAREVYTHENFYIFQKELWSACVDCG 526
+ E F ++ S+ + A+ V T EN + + +
Sbjct: 549 -------SKFSEWFNEHRQDSSSCPIT--------AQVVPTVENILHIRCPIAAKLTVFE 593
Query: 527 IEGTKEEGENLSFSILDNAVRKHREVVYCLSNNIAHCSCKMFESEGIPCRHIL 579
+ +E ++++D V + + + CSCK F+ + IPC H +
Sbjct: 594 LNSYNQE-----YNVID-----LNSVSFLVDLKMKSCSCKCFDIDKIPCVHAM 636
>At2g05010 Mutator-like transposase
Length = 731
Score = 75.9 bits (185), Expect = 8e-14
Identities = 76/369 (20%), Positives = 147/369 (39%), Gaps = 54/369 (14%)
Query: 231 EINPSFFYDYEADNEGKLKHVFWADGICRKNYSLFGDVVSFDTTYRTNKY--FMIFAPFT 288
++NP E + E K K++F A G C + + V+ D T+ Y ++ A
Sbjct: 372 KVNPGTVTSVELEGEKKFKYLFIALGACIEGFREMRKVIVVDATHLKTVYGGMLVIATTQ 431
Query: 289 GINHHRQSITFGAALLKNEKEESFVWLFETFLKAMGGHKPVMIITDQDGGMKNAIGAVLK 348
NHH + FG ++ +EK+ S++W E ++ I+D+ +K A+ V
Sbjct: 432 DPNHHHYPLAFG--IIDSEKDVSWIWFLEKLKTVYPNVPGLVFISDRHQNIKKAVKMVYL 489
Query: 349 GSSHRFCMWHILKKLSEKVGSSMDENSGFNDRFKSCVWNSESSEEFDLEWNNIISDYSLE 408
+ H C+WH+ + + + + D G D+F+ C ++ + EF+ E+ + S + +
Sbjct: 490 NALHASCIWHLSQNMRLPLKINKD---GAADKFRDCA-HAYTESEFNKEFGHFTSIWPKD 545
Query: 409 GNGWLSTMYDLRSMWIPAYFK-DTFMAGILRTTSRSESENSFFGNYLN-HNLTLVEFWV- 465
+ ++ W +FK D + + T++ +ES N F H L +++ +
Sbjct: 546 ADFLEKVGFE---KWSRCHFKRDKYN---IDTSNYAESINGVFRKARKYHLLPMIDVMIS 599
Query: 466 RFDSALAAQRHKELFAD-NNTLHSNPELNMHMNLEKHAREVYTHENFYIFQKELWSACVD 524
+F R F + + E +H+ A+ N Y
Sbjct: 600 KFSECFNVHRQDSCFCSITSQVVPTVENILHLRCPVAAKLTVIELNSY------------ 647
Query: 525 CGIEGTKEEGENLSFSILDNAVRKHREVVYCLSNNIAHCSCKMFESEGIPCRHILFILKG 584
N +++++ V + + + SCK F+ + IPC H
Sbjct: 648 -----------NQEYNVIN-----LNSVSFLVDLKMKSVSCKRFDIDKIPCVH------- 684
Query: 585 KGFSEIPSH 593
G S+ P H
Sbjct: 685 -GRSKAPRH 692
>At1g36090 hypothetical protein
Length = 645
Score = 73.6 bits (179), Expect = 4e-13
Identities = 43/175 (24%), Positives = 82/175 (46%), Gaps = 8/175 (4%)
Query: 231 EINPSFFYDYEADNEGKLKHVFWADGICRKNYSLFGDVVSFDTTYRTNKY--FMIFAPFT 288
++NP E + E K K++F A G C + + V+ D T+ Y ++ A
Sbjct: 333 KVNPGTVTYVELEGEKKFKYLFIALGACIEGFRAMRKVIVVDATHLKTVYGGMLVIATAH 392
Query: 289 GINHHRQSITFGAALLKNEKEESFVWLFETFLKAMGGHKPVMIITDQDGGMKNAIGAVLK 348
NHH + FG ++ +EK+ S++W E ++ I+D+ +K A+ V
Sbjct: 393 DPNHHHYPLAFG--IIDSEKDVSWIWFLEKLKTVYSDVPRLVFISDRHQSIKKAVKTVYP 450
Query: 349 GSSHRFCMWHILKKLSEKVGSSMDENSGFNDRFKSCVWNSESSEEFDLEWNNIIS 403
+ H C+WH+ + + ++V D G +F+ C ++ + EF+ E+ + S
Sbjct: 451 NALHAACIWHLCQNMRDRVKIDKD---GAAVKFRDCA-HAYTESEFEKEFGHFTS 501
>At4g08610 predicted transposon protein
Length = 907
Score = 66.6 bits (161), Expect = 5e-11
Identities = 35/149 (23%), Positives = 71/149 (47%), Gaps = 4/149 (2%)
Query: 227 RRKREINPSFFYDYEADNEGKLKHVFWADGICRKNYSLFGDVVSFDTTYRTNKY--FMIF 284
+R+ ++NP E D + K K++F + G+C + + + VV D T+ Y ++F
Sbjct: 371 KRQHDVNPGTISYVEVDAQQKFKYLFVSLGVCIEGFKVMRKVVIVDATFLKTVYGDMLVF 430
Query: 285 APFTGINHHRQSITFGAALLKNEKEESFVWLFETFLKAMGGHKPVMIITDQDGGMKNAIG 344
A NHH I +A++ E + S+ W F + ++ ++D+ + +I
Sbjct: 431 ATAQDPNHHNYIIA--SAVIDRENDASWSWFFNKLKTVIPDELGLVFVSDRHQSIIKSIM 488
Query: 345 AVLKGSSHRFCMWHILKKLSEKVGSSMDE 373
V + H C+WH+ + + +V + +E
Sbjct: 489 HVFPNARHGHCVWHLSQNVKVRVKTEKEE 517
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.134 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,133,692
Number of Sequences: 26719
Number of extensions: 825312
Number of successful extensions: 2349
Number of sequences better than 10.0: 88
Number of HSP's better than 10.0 without gapping: 63
Number of HSP's successfully gapped in prelim test: 25
Number of HSP's that attempted gapping in prelim test: 2149
Number of HSP's gapped (non-prelim): 150
length of query: 745
length of database: 11,318,596
effective HSP length: 107
effective length of query: 638
effective length of database: 8,459,663
effective search space: 5397264994
effective search space used: 5397264994
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 64 (29.3 bits)
Medicago: description of AC148396.11


