

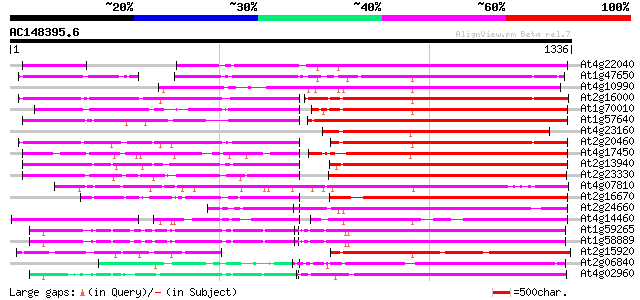
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148395.6 - phase: 0
(1336 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g22040 LTR retrotransposon like protein 678 0.0
At1g47650 hypothetical protein 669 0.0
At4g10990 putative retrotransposon polyprotein 665 0.0
At2g16000 putative retroelement pol polyprotein 568 e-162
At1g70010 hypothetical protein 552 e-157
At1g57640 550 e-156
At4g23160 putative protein 547 e-155
At2g20460 putative retroelement pol polyprotein 538 e-153
At4g17450 retrotransposon like protein 530 e-150
At2g13940 putative retroelement pol polyprotein 528 e-150
At2g23330 putative retroelement pol polyprotein 527 e-149
At4g07810 putative polyprotein 511 e-144
At2g16670 putative retroelement pol polyprotein 508 e-143
At2g24660 putative retroelement pol polyprotein 497 e-140
At4g14460 retrovirus-related like polyprotein 492 e-139
At1g59265 polyprotein, putative 470 e-132
At1g58889 polyprotein, putative 468 e-132
At2g15920 putative retroelement pol polyprotein 465 e-131
At2g06840 putative retroelement pol polyprotein 462 e-130
At4g02960 putative polyprotein of LTR transposon 451 e-126
>At4g22040 LTR retrotransposon like protein
Length = 1109
Score = 678 bits (1750), Expect = 0.0
Identities = 380/966 (39%), Positives = 557/966 (57%), Gaps = 63/966 (6%)
Query: 398 ASDHICSSIKFFDSYAPIIPVHIKLPNGNMAIAKFSGTVNFSPGLIARNVLYVAEFKLNL 457
AS H+ +++ + PV I L +GN +A GTV LI ++V YV E + +L
Sbjct: 169 ASHHMTGNLELLSGMRSMSPVLIILADGNKRVAVSEGTVRLGSHLILKSVFYVKELESDL 228
Query: 458 LSVPKLCMDNNCIVTFDNDKCFIQDKSNLKMTGLGELIEGLYFLNLGSQVFSQFNHQPVI 517
+SV ++ +N+C+V + IQD++ +TG+G+ G + F
Sbjct: 229 ISVGQMMDENHCVVQLADHFLVIQDRTTRMVTGIGKRENGSFC----------FRGMENA 278
Query: 518 ALSQSSSSTTFLPQEALWHFRLGHLSNDRLLRM--QQHFPCIKVDSNSVCDICHYSRHKK 575
A +S F LWH RLGH S D+++ + ++ K +VCD C ++ +
Sbjct: 279 AAVHTSVKAPF----DLWHRRLGHAS-DKIVNLLPRELLSSGKEILENVCDTCMRAKQTR 333
Query: 576 LPFKLSMNKASHCYELIHFDIWGPISTHSIHGHKYFITALDDYSRFTWIMLCKTKSEVSK 635
F L N++ ++LIH D+WGP T S G +YF+T +DDYSR W+ L KSE K
Sbjct: 334 DTFPLRDNRSMDSFQLIHCDVWGPYRTPSYSGARYFLTIVDDYSRGVWVYLMTDKSETQK 393
Query: 636 SVQNFILNIENQFNCKVKTVRTDNGPEFL-MPDFYSSKGIEHQTSCVETPQQNGRVLPYS 694
+++F+ +E QF+ ++KTVR+DNG EFL M +++ KGI H+TSCV TP QNGRV
Sbjct: 394 HLKDFMALVERQFDTEIKTVRSDNGTEFLCMREYFLHKGIAHETSCVGTPHQNGRV---- 449
Query: 695 NSNQYFQWQYHSNHPIIPATEQTSSNSELPLENQSET--------NKEPAMHIDNNTHIQ 746
+ I+ S LP++ E N+ P+M + + +
Sbjct: 450 ---------ERKHRHILNIARALRFQSYLPIQFWGECILSAAYLINRTPSMLLQGKSPYE 500
Query: 747 DPTQIEHEPQQLNTRKSTRVSQKPNHLSDYVCNSSS-----------------DSTKQ-- 787
+ + L S + NH D S D +Q
Sbjct: 501 MLYKTAPKYSHLRVFGSLCYAHNQNHKGDKFAARSRRCVFVGYPHGQKGWRLFDLEEQKF 560
Query: 788 -ASSGILYPIKHYH----SLNNISASHQKFALAITDASEPISYKEASQQDCWVKAMNSEL 842
S +++ + S N ++SH+ F A+T EP +Y EA W +AM++E+
Sbjct: 561 FVSRDVIFQETEFPYSKMSCNRFTSSHKAFLAAVTAGMEPTTYNEAMVDKAWREAMSAEI 620
Query: 843 SALNHNKTWIFVDTPPNIKPIGSKWVYKIKHKSDGTIERYKARLVAKGYTQVEGIDFFDT 902
+L N+T+ V+ PP + +G+KWVYKIK++SDG IERYKARLV G Q EG+D+ +T
Sbjct: 621 ESLRVNQTFSIVNLPPGKRALGNKWVYKIKYRSDGAIERYKARLVVLGNCQKEGVDYDET 680
Query: 903 FSPVAKITTVRILIALASINSWHLHQLDVNNAFLHGDLSENVYMSVPQGVHSPKPNQVCK 962
F+PVAK++TVR+ + +A+ WH+HQ+DV+NAFLHGDL E VYM +PQG P++VC+
Sbjct: 681 FAPVAKMSTVRLFLGVAAARDWHVHQMDVHNAFLHGDLKEEVYMKLPQGFQCDDPSKVCR 740
Query: 963 LLKSLYGLKQASRKWYEKLTGFLLLQGYKQSASDHSLFILHSDTCFTALLVYVDDVILAG 1022
L KSLYGLKQA R W+ KL+ L G+ QS SD+SLF ++D F +LVYVDD+I++G
Sbjct: 741 LHKSLYGLKQAPRCWFSKLSSALKQYGFTQSLSDYSLFSYNNDGVFVHVLVYVDDLIISG 800
Query: 1023 NSMTEIDRIKAVLDVEFKIKDLGKLKYFLGIEVAHSKLGISICQRKYCLDLLKDTGLLGA 1082
+ + + K+ L+ F +KDLG LKYFLGIEV+ + G + QRKY LD++ + GLLGA
Sbjct: 801 SCPDAVAQFKSYLESCFHMKDLGLLKYFLGIEVSRNAQGFYLSQRKYVLDIISEMGLLGA 860
Query: 1083 KPVTTPLDPSIKLHQDNSPPHDDILSYRRLVGKLLYLTTTRPDIAFVVQQLSQFLNAPTI 1142
+P PL+ + KL SP D YRRLVG+L+YL TRP++++ V L+QF+ P
Sbjct: 861 RPSAFPLEQNHKLSLSTSPLLSDSSRYRRLVGRLIYLAVTRPELSYSVHTLAQFMQNPRQ 920
Query: 1143 THYDTACRVVKYLKGTPGQGLLFRRDSQLQLLGFTDADWAGCPDSRRSTSGYCFFLGSSL 1202
H++ A RVV+YLK PGQG+L S LQ+ G+ D+D+A CP +RRS +GY LG +
Sbjct: 921 DHWNAAIRVVRYLKSNPGQGILLSSTSTLQINGWCDSDYAACPLTRRSLTGYFVQLGDTP 980
Query: 1203 ISWRAKKQHTVARSSSEAEYRALSFASCELQWLLYLLKDLRVKCNKLPVLFCDNQSAIHI 1262
ISW+ KKQ T++RSS+EAEYRA++F + EL WL +L DL V + +F D++SAI +
Sbjct: 981 ISWKTKKQPTISRSSAEAEYRAMAFLTQELMWLKRVLYDLGVSHVQAMRIFSDSKSAIAL 1040
Query: 1263 AGNPVFHERTKHLEIDCHFVREKLQQNIFKLLPIKSQSQLADFFTKPLPPKNFHSFLPKL 1322
+ NPV HERTKH+E+DCHF+R+ + I + S QLAD TK L K FL KL
Sbjct: 1041 SVNPVQHERTKHVEVDCHFIRDAILDGIIATSFVPSHKQLADILTKALGEKEVRYFLRKL 1100
Query: 1323 NMIDLY 1328
++D++
Sbjct: 1101 GILDVH 1106
Score = 101 bits (252), Expect = 3e-21
Identities = 44/152 (28%), Positives = 86/152 (55%)
Query: 30 YYVHASDGPSSVAITPVLSHSNYHSWARSMRRALGAKNKFDFVDGSIPVPTDFDPNFKAW 89
Y + A+D +V P+L +NY WA + AL ++ KF F+DG+IP P D P+ + W
Sbjct: 21 YDLTAADNSGAVISHPILKTNNYEEWACGFKTALRSRKKFGFLDGTIPQPLDGSPDLEDW 80
Query: 90 NRCNMLVHSWIMNSVEDSIAQSIVFLENAIDVWNELKERFSQGDFIRISELQCEIFSLKQ 149
N L+ SW+ +++ + +I + A D+W ++++RFS + + +++ ++ + KQ
Sbjct: 81 LTINALLVSWMKMTIDSELLTNISHRDVARDLWEQIRKRFSVSNGPKNQKMKADLATCKQ 140
Query: 150 DSRSVTEFFTALKVLWEELEAYLPTPVCACPH 181
+ +V ++ L +W+ + +Y P +CA H
Sbjct: 141 EGMTVEGYYGKLNKIWDNINSYRPLRICASHH 172
>At1g47650 hypothetical protein
Length = 1409
Score = 669 bits (1725), Expect = 0.0
Identities = 377/955 (39%), Positives = 541/955 (56%), Gaps = 84/955 (8%)
Query: 393 IVDSGASDHICSSIKFFDSYAPIIPVHIKLPNGNMAIAKFSGTVNFSPGLIARNVLYVAE 452
++DSGA H+ F + + + LP G GT+ + ++ +N+L++ E
Sbjct: 333 VIDSGAIHHVSHDKSLFLNLDTSVVSAVNLPAGPTVRISGVGTLRLNDDILLKNILFIPE 392
Query: 453 FKLNLLSVPKLCMDNNCIVTFDNDKCFIQDKSNLKMTGLGELIEGLYFLNLGSQVFSQFN 512
F+LNL+S+ L D V FD C IQD +M G G I LY L++
Sbjct: 393 FRLNLISISSLTDDIGSRVIFDKTSCEIQDPIKARMLGQGRRIANLYVLDI--------- 443
Query: 513 HQPVIALSQSSSSTTFLPQEALWHFRLGHLSNDRLLRMQQHFPCIKVDS--NSVCDICHY 570
P+++++ + ++WH RLGH S RL + K + + C +CH
Sbjct: 444 EDPIVSVNA-------VVDISMWHRRLGHASLQRLDVISDSLGTTKPKNKGSDYCHVCHL 496
Query: 571 SRHKKLPFKLSMNKASHCYELIHFDIWGPISTHSIHGHKYFITALDDYSRFTWIMLCKTK 630
++ +KLPF + ++L+H DIWGP S ++ G+KYF+T +DD+SR TWI L + K
Sbjct: 497 AKQRKLPFPSQNKVCNEIFDLLHIDIWGPFSVETVDGYKYFLTIVDDHSRATWIYLLRNK 556
Query: 631 SEVSKSVQNFILNIENQFNCKVKTVRTDNGPEFLMPDFYSSKGIEHQTSCVETPQQNGRV 690
SEV FI +ENQ+ +VK VR+DN PE Y KGI SC ETP+QN
Sbjct: 557 SEVLTVFPAFIEQVENQYKVRVKAVRSDNAPELKFTSLYQQKGIVSFHSCPETPEQN--- 613
Query: 691 LPYSNSNQYFQWQYHSNHPIIPATEQTSSNSELPLENQSET--------NKEPAMHIDNN 742
L +Q+ I+ + S++PL + N+ P+ + N
Sbjct: 614 LVVERKHQH----------ILNVSRALMFQSQVPLSLWGDCVLTAVFLINRTPSQLLSNK 663
Query: 743 THIQDPTQIEHEPQQLNTRKSTRVSQKPNHLSDYVCNSSSDSTKQASSGILYPI--KHYH 800
T P ++ T K H +A + YP K Y
Sbjct: 664 T-----------PYEILTGTVPFYFAKKRH--------KFQPRSKACVFLGYPAGYKGYK 704
Query: 801 SLN----------NISASHQKFALAITDASEPISYKEASQQDCWVKAMNSELSALNHNKT 850
++ N+ + F LA+ SE S K + + ++SE+ A+ T
Sbjct: 705 LMDLESHTVFISRNVVFHEEVFPLAVDPKSES-SLKLFTP----MVPVSSEIGAMETTHT 759
Query: 851 WIFVDTPPNIKPIGSKWVYKIKHKSDGTIERYKARLVAKGYTQVEGIDFFDTFSPVAKIT 910
W P K +G KWV+ +K +DG++ERYKARLVAKGYTQ EG+D+ DTFSPVAK+T
Sbjct: 760 WEITTLPAGKKAVGCKWVFSLKFLADGSLERYKARLVAKGYTQKEGLDYTDTFSPVAKMT 819
Query: 911 TVRILIALASINSWHLHQLDVNNAFLHGDLSENVYMSVPQGVHSPK-----PNQVCKLLK 965
T+++L+ +++ W L QLDV+NAFL+G+L E +YM +P+G + K PN VC+L +
Sbjct: 820 TIKLLLKISASKKWFLKQLDVSNAFLNGELEEEIYMKLPEGYAARKGITLPPNSVCRLKR 879
Query: 966 SLYGLKQASRKWYEKLTGFLLLQGYKQSASDHSLFILHSDTCFTALLVYVDDVILAGNSM 1025
S+YGLKQASR+W++K + LL G+K++ DH+LFI D F +LVYVDD+ +A S
Sbjct: 880 SIYGLKQASRQWFKKFSASLLDLGFKKTHGDHTLFIKEYDGEFVIVLVYVDDIAIASTSE 939
Query: 1026 TEIDRIKAVLDVEFKIKDLGKLKYFLGIEVAHSKLGISICQRKYCLDLLKDTGLLGAKPV 1085
++ L FK++DLG LKYFLG+E+A ++ GISICQRKY L+LL TG++ KPV
Sbjct: 940 GAAIQLTQDLQQRFKLRDLGDLKYFLGLEIARTEAGISICQRKYALELLASTGMVNCKPV 999
Query: 1086 TTPLDPSIKLHQDNSPPHDDILSYRRLVGKLLYLTTTRPDIAFVVQQLSQFLNAPTITHY 1145
+ P+ P++K+ + + DD YRR+VGKL+YLT TR DI F V +L QF +AP +H
Sbjct: 1000 SVPMIPNVKMMKTDGDLLDDREQYRRIVGKLMYLTITRLDITFAVNKLCQFSSAPRTSHL 1059
Query: 1146 DTACRVVKYLKGTPGQGLLFRRDSQLQLLGFTDADWAGCPDSRRSTSGYCFFLGSSLISW 1205
A RV++Y+KG+ GQGL + + L L GF D+DWA CPDSRRST+G+ F+G SLISW
Sbjct: 1060 QAAYRVLQYIKGSVGQGLFYSASADLTLKGFADSDWASCPDSRRSTTGFSMFVGDSLISW 1119
Query: 1206 RAKKQHTVARSSSEAEYRALSFASCELQWLLYLLKDLRVKCNKLPVLFCDNQSAIHIAGN 1265
R+KKQH V+RSS+EAEYRAL+ +CEL WL LL L+ +PVL+ D+ +AI+IA N
Sbjct: 1120 RSKKQHVVSRSSAEAEYRALALVTCELVWLHTLLMSLKAS-YLVPVLYSDSTTAIYIATN 1178
Query: 1266 PVFHERTKHLEIDCHFVREKLQQNIFKLLPIKSQSQLADFFTKPLPPKNFHSFLP 1320
PVFHERTKH+E+DCH VREKL KLL ++++ Q + T PP N LP
Sbjct: 1179 PVFHERTKHIELDCHTVREKLDNGELKLLHVRTEDQPS---TAAYPPPNLAEPLP 1230
Score = 159 bits (401), Expect = 1e-38
Identities = 94/289 (32%), Positives = 156/289 (53%), Gaps = 27/289 (9%)
Query: 21 DPSQQPGNVYYVHASDGPSSVAITPVLSHSNYHSWARSMRRALGAKNKFDFVDGSIPVPT 80
DP+Q P +++H++D P I+ L +NY W +M +L AKNK F+DG++P P
Sbjct: 59 DPTQSP---FFLHSADHPGLNIISHRLDETNYGDWNVAMLISLDAKNKSGFIDGTLPRPL 115
Query: 81 DFDPNFKAWNRCNMLVHSWIMNSVEDSIAQSIVFLENAIDVWNELKERFSQGDFIRISEL 140
+ D NF+ W+RC +V SW++NSV I +SI+ + +A D+W +L RF+ + R L
Sbjct: 116 ESDKNFRLWSRCKSMVKSWLLNSVSPQIYRSILRMNDASDIWRDLHSRFNVTNLPRTYNL 175
Query: 141 QCEIFSLKQDSRSVTEFFTALKVLWEELEA--YLPTPVCACPHRCMCITG-VMNAKHQHE 197
EI +Q + S++E++T LK LW++LE+ L P CI G M + + E
Sbjct: 176 TQEIQDFRQGTLSLSEYYTRLKTLWDQLESTEELDDP---------CICGKAMRLQQKAE 226
Query: 198 ITRSIRFLTGLNDSFDLVRSQILLMNPLPTINKIFSMVMQHERQFKIS-IPVEDSTILVN 256
+ ++ L GLNDS+ ++R QI+ LP++ +++ ++ Q Q S + + V+
Sbjct: 227 QAKIVKLLAGLNDSYAIIRRQIIAKKALPSLAEVYHILDQDNSQQGFSNVVAPPAAFQVS 286
Query: 257 SAGKSQGRGRGNGNSNSNGKRSCSFCGRDGHTIDICYRKHGFPPNYGNK 305
A + +N++ CS GH + CY+KHGFP + K
Sbjct: 287 EAMMA---------TNNDVNMLCS--EWVGHIAERCYKKHGFPLGFTPK 324
>At4g10990 putative retrotransposon polyprotein
Length = 1203
Score = 665 bits (1715), Expect = 0.0
Identities = 392/1031 (38%), Positives = 555/1031 (53%), Gaps = 138/1031 (13%)
Query: 354 LQSQQASSSKV-----NLSHVTNHVTSGITRTSYTMNHSSFGTWIVDSGASDHICSSIKF 408
L +Q ++S + +L + N++T S N S WI+DSGAS H+CS +
Sbjct: 56 LMAQTSTSGTIPFPSTSLKYENNNLTFQNHTLSSLQNVLSSDAWIIDSGASSHVCSDLTM 115
Query: 409 FDSYAPIIPVHIKLPNGNMAIAKFSGTVNFSPGLIARNVLYVAEFKLNLLSVPKLCMDNN 468
F + V + LPNG +GT+ + LI NVL V +FK NL+SV L +
Sbjct: 116 FRELIHVSGVTVTLPNGTRVAITHTGTICITSTLILHNVLLVPDFKFNLISVCCLVKTLS 175
Query: 469 CIVTFDNDKCFIQDKSNLKMTGLGELIEGLYFLNLGSQVFSQFNHQPVIALSQSSSSTTF 528
F D C+IQ+ + M G G+ LY L FS +L +SS T
Sbjct: 176 YSAHFFADCCYIQELTRGLMIGRGKTYNNLYILETQRTSFSP-------SLPAASSFTGT 228
Query: 529 LPQEAL-WHFRLGHLSNDRLLRMQQHFPCIKVDSNSVCDICHYSRHKKLPFKLSMNKASH 587
+ + L WH RLG I HY ++ L
Sbjct: 229 VQDDCLLWHQRLG--------------------------IRHYLHYRNL----------- 251
Query: 588 CYELIHFDIWGPISTHSIHGHKYFITALDDYSRFTWIMLCKTKSEVSKSVQNFILNIENQ 647
YF+T +DD +R TW+ + K KSEVS F+ I Q
Sbjct: 252 ----------------------YFLTLVDDCTRTTWVYMMKNKSEVSNIFPVFVKLIFTQ 289
Query: 648 FNCKVKTVRTDNGPEFLMPDFYSSKGIEHQTSCVETPQQNGRVLPYSNSNQYFQWQYHSN 707
+N K+K +R+DN E F +G+ HQ SC TPQQN V Y + + ++ +
Sbjct: 290 YNAKIKAIRSDNVKELAFTKFVKEQGMIHQFSCAYTPQQNSVVERYPSGYKGYKVLDLES 349
Query: 708 HPI--------------------IPATEQTSSNSELPL--------------------EN 727
H I + + NS LPL N
Sbjct: 350 HSISITRNVVFHETKFPFKTSKFLKESVDMFPNSILPLPAPLHFVESMPLDDDLRADDNN 409
Query: 728 QSETNKEPAMH----IDNNTHIQDPTQIEHEPQQLNTRKSTRVSQKPNHLSDYVCNS--- 780
S +N + + + + Q+ ++ + + + R ++ P +LS+Y CNS
Sbjct: 410 ASTSNSASSASSIPPLPSTVNTQNTDALDIDTNSVPIARPKRNAKAPAYLSEYHCNSVPF 469
Query: 781 -------SSDSTKQASSGIL-------YPIKHYHSLNNISASHQKFALAITDASEPISYK 826
+S S + SS I YP+ S + ++ + A +EP ++
Sbjct: 470 LSSLSPTTSTSIETPSSSIPPKKITTPYPMSTAISYDKLTPLFHSYICAYNVETEPKAFT 529
Query: 827 EASQQDCWVKAMNSELSALNHNKTWIFVDTPPNIKPIGSKWVYKIKHKSDGTIERYKARL 886
+A + + W +A N EL AL NKTWI +G KWV+ IK+ DG+IERYKARL
Sbjct: 530 QAMKSEKWTRAANEELHALEQNKTWIVESLTEGKNVVGCKWVFTIKYNPDGSIERYKARL 589
Query: 887 VAKGYTQVEGIDFFDTFSPVAKITTVRILIALASINSWHLHQLDVNNAFLHGDLSENVYM 946
VA+G+TQ EGID+ +TFSPVAK +V++L+ LA+ W L Q+DV+NAFLHG+L E +YM
Sbjct: 590 VAQGFTQQEGIDYMETFSPVAKFGSVKLLLGLAAATGWSLTQMDVSNAFLHGELDEEIYM 649
Query: 947 SVPQGVHSPK-----PNQVCKLLKSLYGLKQASRKWYEKLTGFLLLQGYKQSASDHSLFI 1001
S+PQG P VC+LLKSLYGLKQASR+WY++L+ L + QS +D+++F+
Sbjct: 650 SLPQGYTPPTGISLPSKPVCRLLKSLYGLKQASRQWYKRLSSVFLGANFIQSPADNTMFV 709
Query: 1002 LHSDTCFTALLVYVDDVILAGNSMTEIDRIKAVLDVEFKIKDLGKLKYFLGIEVAHSKLG 1061
S T +LVYVDD+++A N + ++ +K +L EFKIKDLG ++FLG+E+A S G
Sbjct: 710 KVSCTSIIVVLVYVDDLMIASNDSSAVENLKELLRSEFKIKDLGPARFFLGLEIARSSEG 769
Query: 1062 ISICQRKYCLDLLKDTGLLGAKPVTTPLDPSIKLHQDNSPPHDDILSYRRLVGKLLYLTT 1121
IS+CQRKY +LL+D GL G KP + P+DP++ L ++ + SYR LVG+LLYL
Sbjct: 770 ISVCQRKYAQNLLEDVGLSGCKPSSIPMDPNLHLTKEMGTLLPNATSYRELVGRLLYLCI 829
Query: 1122 TRPDIAFVVQQLSQFLNAPTITHYDTACRVVKYLKGTPGQGLLFRRDSQLQLLGFTDADW 1181
TRPDI F V LSQFL+APT H A +V++YLKG PGQGL++ S+L L GF+DADW
Sbjct: 830 TRPDITFAVHTLSQFLSAPTDIHMQAAHKVLRYLKGNPGQGLMYSASSELCLNGFSDADW 889
Query: 1182 AGCPDSRRSTSGYCFFLGSSLISWRAKKQHTVARSSSEAEYRALSFASCELQWLLYLLKD 1241
C DSRRS +G+C +LG+SLI+W++KKQ V+RSS+E+EYR+L+ A+CE+ WL LLKD
Sbjct: 890 GTCKDSRRSVTGFCIYLGTSLITWKSKKQSVVSRSSTESEYRSLAQATCEIIWLQQLLKD 949
Query: 1242 LRVKCNKLPVLFCDNQSAIHIAGNPVFHERTKHLEIDCHFVREKLQQNIFKLLPIKSQSQ 1301
L V LFCDN+SA+H+A NPVFHERTKH+EIDCH VR++++ K L + + +Q
Sbjct: 950 LHVTMTCPAKLFCDNKSALHLATNPVFHERTKHIEIDCHTVRDQIKAGKLKTLHVPTGNQ 1009
Query: 1302 LADFFTKPLPP 1312
LAD TKPL P
Sbjct: 1010 LADILTKPLHP 1020
>At2g16000 putative retroelement pol polyprotein
Length = 1454
Score = 568 bits (1464), Expect = e-162
Identities = 289/632 (45%), Positives = 407/632 (63%), Gaps = 18/632 (2%)
Query: 703 QYHSNHPIIPATEQTSSNSELPLENQSETNKEPAMHIDNNTHIQDPTQIEHEPQQLNTRK 762
Q+H + P + S S L L + ++ P+QI P Q++
Sbjct: 833 QFHEE--VFPLAKNPGSESSLKLFTPMVPVSSGIISDTTHSPSSLPSQISDLPPQIS--- 887
Query: 763 STRVSQKPNHLSDYVCNSSSDSTKQASSGILYPIKHYHSLNNISASHQKFALAITDASEP 822
S RV + P HL+DY CN+ K YPI S + IS SH + IT P
Sbjct: 888 SQRVRKPPAHLNDYHCNTMQSDHK-------YPISSTISYSKISPSHMCYINNITKIPIP 940
Query: 823 ISYKEASQQDCWVKAMNSELSALNHNKTWIFVDTPPNIKPIGSKWVYKIKHKSDGTIERY 882
+Y EA W +A+++E+ A+ TW P K +G KWV+ +K +DG +ERY
Sbjct: 941 TNYAEAQDTKEWCEAVDAEIGAMEKTNTWEITTLPKGKKAVGCKWVFTLKFLADGNLERY 1000
Query: 883 KARLVAKGYTQVEGIDFFDTFSPVAKITTVRILIALASINSWHLHQLDVNNAFLHGDLSE 942
KARLVAKGYTQ EG+D+ DTFSPVAK+TT+++L+ +++ W L QLDV+NAFL+G+L E
Sbjct: 1001 KARLVAKGYTQKEGLDYTDTFSPVAKMTTIKLLLKVSASKKWFLKQLDVSNAFLNGELEE 1060
Query: 943 NVYMSVPQGVHSPK-----PNQVCKLLKSLYGLKQASRKWYEKLTGFLLLQGYKQSASDH 997
++M +P+G K N V +L +S+YGLKQASR+W++K + LL G+K++ DH
Sbjct: 1061 EIFMKIPEGYAERKGIVLPSNVVLRLKRSIYGLKQASRQWFKKFSSSLLSLGFKKTHGDH 1120
Query: 998 SLFILHSDTCFTALLVYVDDVILAGNSMTEIDRIKAVLDVEFKIKDLGKLKYFLGIEVAH 1057
+LF+ D F +LVYVDD+++A S ++ LD FK++DLG LKYFLG+EVA
Sbjct: 1121 TLFLKMYDGEFVIVLVYVDDIVIASTSEAAAAQLTEELDQRFKLRDLGDLKYFLGLEVAR 1180
Query: 1058 SKLGISICQRKYCLDLLKDTGLLGAKPVTTPLDPSIKLHQDNSPPHDDILSYRRLVGKLL 1117
+ GISICQRKY L+LL+ TG+L KPV+ P+ P++K+ +D+ +DI YRR+VGKL+
Sbjct: 1181 TTAGISICQRKYALELLQSTGMLACKPVSVPMIPNLKMRKDDGDLIEDIEQYRRIVGKLM 1240
Query: 1118 YLTTTRPDIAFVVQQLSQFLNAPTITHYDTACRVVKYLKGTPGQGLLFRRDSQLQLLGFT 1177
YLT TRPDI F V +L QF +AP TH A RV++Y+KGT GQGL + S L L GF
Sbjct: 1241 YLTITRPDITFAVNKLCQFSSAPRTTHLTAAYRVLQYIKGTVGQGLFYSASSDLTLKGFA 1300
Query: 1178 DADWAGCPDSRRSTSGYCFFLGSSLISWRAKKQHTVARSSSEAEYRALSFASCELQWLLY 1237
D+DWA C DSRRST+ + F+G SLISWR+KKQHTV+RSS+EAEYRAL+ A+CE+ WL
Sbjct: 1301 DSDWASCQDSRRSTTSFTMFVGDSLISWRSKKQHTVSRSSAEAEYRALALATCEMVWLFT 1360
Query: 1238 LLKDLRVKCNKLPVLFCDNQSAIHIAGNPVFHERTKHLEIDCHFVREKLQQNIFKLLPIK 1297
LL L+ +P+L+ D+ +AI+IA NPVFHERTKH+++DCH VRE+L KLL ++
Sbjct: 1361 LLVSLQAS-PPVPILYSDSTAAIYIATNPVFHERTKHIKLDCHTVRERLDNGELKLLHVR 1419
Query: 1298 SQSQLADFFTKPLPPKNFHSFLPKLNMIDLYN 1329
++ Q+AD TKPL P F K++++++++
Sbjct: 1420 TEDQVADILTKPLFPYQFEHLKSKMSILNIFS 1451
Score = 378 bits (970), Expect = e-104
Identities = 226/691 (32%), Positives = 354/691 (50%), Gaps = 52/691 (7%)
Query: 21 DPSQQPGNVYYVHASDGPSSVAITPVLSHSNYHSWARSMRRALGAKNKFDFVDGSIPVPT 80
DP+Q P +++H++D P I+ L +NY W+ +M +L AKNK F+DG++ P
Sbjct: 56 DPTQSP---FFLHSADHPGLNIISHRLDETNYGDWSVAMLISLDAKNKTGFIDGTLSRPL 112
Query: 81 DFDPNFKAWNRCNMLVHSWIMNSVEDSIAQSIVFLENAIDVWNELKERFSQGDFIRISEL 140
+ D NF+ W+RCN +V SW++NSV I +SI+ + +A D+W +L RF+ + R L
Sbjct: 113 ESDLNFRLWSRCNSMVKSWLLNSVSPQIYRSILRMNDASDIWRDLNSRFNVTNLPRTYNL 172
Query: 141 QCEIFSLKQDSRSVTEFFTALKVLWEELEAYLPTPVCACPHRCMCITGVMNAKHQHEITR 200
EI +Q + S++E++T LK LW++L++ A C C M + + E +
Sbjct: 173 TQEIQDFRQGTLSLSEYYTRLKTLWDQLDS-----TEALDEPCTC-GKAMRLQQKAEQAK 226
Query: 201 SIRFLTGLNDSFDLVRSQILLMNPLPTINKIFSMVMQHERQFKIS-IPVEDSTILVNSAG 259
++FL GLN+S+ +VR QI+ LP++ +++ ++ Q Q S + + V+
Sbjct: 227 IVKFLAGLNESYAIVRRQIIAKKALPSLGEVYHILDQDNSQQSFSNVVAPPAAFQVSEIT 286
Query: 260 KSQGRGRGN---GNSNSNGKRSCSFCGRDGHTIDICYRKHGFPPNYGNKNFAKVNNSSIE 316
+S N + G+ CSF R GH + CY+KHGFPP + K A E
Sbjct: 287 QSPSMDPTVCYVQNGPNKGRPICSFYNRVGHIAERCYKKHGFPPGFTPKGKAG------E 340
Query: 317 QNEEREDL--DDSKSCKGNSNTEPSFG-ITKEQYEQLVTLLQSQ-----------QASSS 362
+ ++ + L + ++S + N++ E G ++KEQ +Q + + SQ ++S
Sbjct: 341 KLQKPKPLAANVAESSEVNTSLESMVGNLSKEQLQQFIAMFSSQLQNTPPSTYATASTSQ 400
Query: 363 KVNLSHVTNHVTSGITRTSYTMNHS-SFGTWIVDSGASDHICSSIKFFDSYAPIIPVHIK 421
NL + T H+ S TW++DSGA+ H+ F S + +
Sbjct: 401 SDNLGICFSPSTYSFIGILTVARHTLSSATWVIDSGATHHVSHDRSLFSSLDTSVLSAVN 460
Query: 422 LPNGNMAIAKFSGTVNFSPGLIARNVLYVAEFKLNLLSVPKLCMDNNCIVTFDNDKCFIQ 481
LP G GT+ + ++ +NVL++ EF+LNL+S+ L D V FD + C IQ
Sbjct: 461 LPTGPTVKISGVGTLKLNDDILLKNVLFIPEFRLNLISISSLTDDIGSRVIFDKNSCEIQ 520
Query: 482 DKSNLKMTGLGELIEGLYFLNLGSQVFSQFNHQPVIALSQSSSSTTFLPQEALWHFRLGH 541
D +M G G + LY L++G Q S S + ++WH RLGH
Sbjct: 521 DLIKGRMLGQGRRVANLYLLDVGDQ----------------SISVNAVVDISMWHRRLGH 564
Query: 542 LSNDRLLRMQQHFPCI--KVDSNSVCDICHYSRHKKLPFKLSMNKASHCYELIHFDIWGP 599
S RL + K + C +CH ++ +KL F S ++L+H D+WGP
Sbjct: 565 ASLQRLDAISDSLGTTRHKNKGSDFCHVCHLAKQRKLSFPTSNKVCKEIFDLLHIDVWGP 624
Query: 600 ISTHSIHGHKYFITALDDYSRFTWIMLCKTKSEVSKSVQNFILNIENQFNCKVKTVRTDN 659
S ++ G+KYF+T +DD+SR TW+ L KTKSEV FI +ENQ+ KVK VR+DN
Sbjct: 625 FSVETVEGYKYFLTIVDDHSRATWMYLLKTKSEVLTVFPAFIQQVENQYKVKVKAVRSDN 684
Query: 660 GPEFLMPDFYSSKGIEHQTSCVETPQQNGRV 690
PE FY+ KGI SC ETP+QN V
Sbjct: 685 APELKFTSFYAEKGIVSFHSCPETPEQNSVV 715
>At1g70010 hypothetical protein
Length = 1315
Score = 552 bits (1422), Expect = e-157
Identities = 284/619 (45%), Positives = 406/619 (64%), Gaps = 23/619 (3%)
Query: 720 NSELPLENQSETNKEPAMHIDNNTHIQ-----DPTQIEHEPQQLNTRKSTRVSQKPNHLS 774
N P++ QS + P+ D+++ ++ +PT EP + + S R ++KP +L
Sbjct: 707 NPTPPMQRQSSDHVNPS---DSSSSVEILPSANPTNNVPEP---SVQTSHRKAKKPAYLQ 760
Query: 775 DYVCNSSSDSTKQASSGILYPIKHYHSLNNISASHQKFALAITDASEPISYKEASQQDCW 834
DY C+S ST + I+ + S + I+ + F + EP +Y EA + W
Sbjct: 761 DYYCHSVVSSTP-------HEIRKFLSYDRINDPYLTFLACLDKTKEPSNYTEAEKLQVW 813
Query: 835 VKAMNSELSALNHNKTWIFVDTPPNIKPIGSKWVYKIKHKSDGTIERYKARLVAKGYTQV 894
AM +E L TW P + + IG +W++KIK+ SDG++ERYKARLVA+GYTQ
Sbjct: 814 RDAMGAEFDFLEGTHTWEVCSLPADKRCIGCRWIFKIKYNSDGSVERYKARLVAQGYTQK 873
Query: 895 EGIDFFDTFSPVAKITTVRILIALASINSWHLHQLDVNNAFLHGDLSENVYMSVPQGVHS 954
EGID+ +TFSPVAK+ +V++L+ +A+ L QLD++NAFL+GDL E +YM +PQG S
Sbjct: 874 EGIDYNETFSPVAKLNSVKLLLGVAARFKLSLTQLDISNAFLNGDLDEEIYMRLPQGYAS 933
Query: 955 PK-----PNQVCKLLKSLYGLKQASRKWYEKLTGFLLLQGYKQSASDHSLFILHSDTCFT 1009
+ PN VC+L KSLYGLKQASR+WY K + LL G+ QS DH+ F+ SD F
Sbjct: 934 RQGDSLPPNAVCRLKKSLYGLKQASRQWYLKFSSTLLGLGFIQSYCDHTCFLKISDGIFL 993
Query: 1010 ALLVYVDDVILAGNSMTEIDRIKAVLDVEFKIKDLGKLKYFLGIEVAHSKLGISICQRKY 1069
+LVY+DD+I+A N+ +D +K+ + FK++DLG+LKYFLG+E+ S GI I QRKY
Sbjct: 994 CVLVYIDDIIIASNNDAAVDILKSQMKSFFKLRDLGELKYFLGLEIVRSDKGIHISQRKY 1053
Query: 1070 CLDLLKDTGLLGAKPVTTPLDPSIKLHQDNSPPHDDILSYRRLVGKLLYLTTTRPDIAFV 1129
LDLL +TG LG KP + P+DPS+ D+ ++ YRRL+G+L+YL TRPDI F
Sbjct: 1054 ALDLLDETGQLGCKPSSIPMDPSMVFAHDSGGDFVEVGPYRRLIGRLMYLNITRPDITFA 1113
Query: 1130 VQQLSQFLNAPTITHYDTACRVVKYLKGTPGQGLLFRRDSQLQLLGFTDADWAGCPDSRR 1189
V +L+QF AP H ++++Y+KGT GQGL + S+LQL + +AD+ C DSRR
Sbjct: 1114 VNKLAQFSMAPRKAHLQAVYKILQYIKGTIGQGLFYSATSELQLKVYANADYNSCRDSRR 1173
Query: 1190 STSGYCFFLGSSLISWRAKKQHTVARSSSEAEYRALSFASCELQWLLYLLKDLRVKCNKL 1249
STSGYC FLG SLI W+++KQ V++SS+EAEYR+LS A+ EL WL LK+L+V +K
Sbjct: 1174 STSGYCMFLGDSLICWKSRKQDVVSKSSAEAEYRSLSVATDELVWLTNFLKELQVPLSKP 1233
Query: 1250 PVLFCDNQSAIHIAGNPVFHERTKHLEIDCHFVREKLQQNIFKLLPIKSQSQLADFFTKP 1309
+LFCDN++AIHIA N VFHERTKH+E DCH VRE+L + +F+L I ++ Q+AD FTKP
Sbjct: 1234 TLLFCDNEAAIHIANNHVFHERTKHIESDCHSVRERLLKGLFELYHINTELQIADPFTKP 1293
Query: 1310 LPPKNFHSFLPKLNMIDLY 1328
L P +FH + K+ +++++
Sbjct: 1294 LYPSHFHRLISKMGLLNIF 1312
Score = 345 bits (884), Expect = 1e-94
Identities = 217/637 (34%), Positives = 314/637 (49%), Gaps = 78/637 (12%)
Query: 59 MRRALGAKNKFDFVDGSIPVPTDFDPNFKAWNRCNMLVHSWIMNSVEDSIAQSIVFLENA 118
M ++ AKNK FVDGSIP P D DP K W RCN +V SW++NSV I SI++ A
Sbjct: 1 MTTSIEAKNKLGFVDGSIPKPDDDDPYCKIWRRCNSMVKSWLLNSVSKEIYTSILYFPTA 60
Query: 119 IDVWNELKERFSQGDFIRISELQCEIFSLKQDSRSVTEFFTALKVLWEELEAYLPTPVCA 178
+W +L RF + R+ +L+ +I SL+Q + ++ + T + LWEEL + P
Sbjct: 61 AAIWKDLYTRFHKSSLPRLYKLRQQIHSLRQGNLDLSSYHTRTQTLWEELTSLQAVP--- 117
Query: 179 CPHRCMCITGVMNAKHQHEITRSIRFLTGLNDSFDLVRSQILLMNPLPTINKIFSMVMQH 238
V + + E R I FL GLND +D VRSQIL+ LP+++++F+M+ Q
Sbjct: 118 --------RTVEDLLIERETNRVIDFLMGLNDCYDTVRSQILMKKTLPSLSEVFNMIDQD 169
Query: 239 ERQFKISIPVEDS-TILVNSAGKSQGRGRGNGNSNSNGKRS-CSFCGRDGHTIDICYRKH 296
E Q I T V + NG++ +R CS+C R GH D CY+KH
Sbjct: 170 ETQRSARISTTPGMTSSVFPVSNQSSQSALNGDTYQKKERPVCSYCSRPGHVEDTCYKKH 229
Query: 297 GFPPNYGNKN-FAKVNNSSIEQNEEREDLDDSKSCKGNSNTEPSFGITKEQYEQLVTLLQ 355
G+P ++ +K F K + S+ E ++++ G+ +T Q +QLV+ L
Sbjct: 230 GYPTSFKSKQKFVKPSISANAAIGSEEVVNNTSVSTGD--------LTTSQIQQLVSFLS 281
Query: 356 SQQASSSKVNLSHVTNHVTSGITRTSYTMNHSSFGTWIVDSGASDHICSSIKFFDSYAPI 415
S+ S V HS S +SD
Sbjct: 282 SKLQPPSTPVQPEV----------------HSI-------SVSSD--------------- 303
Query: 416 IPVHIKLPNGNMAIAKFSGTVNFSPGLIARNVLYVAEFKLNLLSVPKLCMDNNCIVTFDN 475
P+ + + SG+V+ LI +VL++ +FK NLLSV L C + FD
Sbjct: 304 -------PSSSSTVCPISGSVHLGRHLILNDVLFIPQFKFNLLSVSSLTKSMGCRIWFDE 356
Query: 476 DKCFIQDKSNLKMTGLGELIEGLYFLNLGSQVFSQFNHQPVIALSQSSSSTTFLPQEALW 535
C +QD + M G+G+ + LY ++L S +H + SS + + LW
Sbjct: 357 TSCVLQDATRELMVGMGKQVANLYIVDLDS-----LSHPG----TDSSITVASVTSHDLW 407
Query: 536 HFRLGHLSNDRLLRMQQ--HFPCIKVDSNSVCDICHYSRHKKLPFKLSMNKASHCYELIH 593
H RLGH S +L M FP K +++ C +CH S+ K LPF NK+S ++LIH
Sbjct: 408 HKRLGHPSVQKLQPMSSLLSFPKQKNNTDFHCRVCHISKQKHLPFVSHNNKSSRPFDLIH 467
Query: 594 FDIWGPISTHSIHGHKYFITALDDYSRFTWIMLCKTKSEVSKSVQNFILNIENQFNCKVK 653
D WGP S + G++YF+T +DDYSR TW+ L + KS+V + F+ +ENQF +K
Sbjct: 468 IDTWGPFSVQTHDGYRYFLTIVDDYSRATWVYLLRNKSDVLTVIPTFVTMVENQFETTIK 527
Query: 654 TVRTDNGPEFLMPDFYSSKGIEHQTSCVETPQQNGRV 690
VR+DN PE FY SKGI SC ETPQQN V
Sbjct: 528 GVRSDNAPELNFTQFYHSKGIVPYHSCPETPQQNSVV 564
>At1g57640
Length = 1444
Score = 550 bits (1418), Expect = e-156
Identities = 286/622 (45%), Positives = 405/622 (64%), Gaps = 6/622 (0%)
Query: 709 PIIPATEQTSSNSELPLENQSETNKEPAMHIDNNTHIQDPTQIEHEPQQLNTRKSTRVSQ 768
PIIP Q SS+ P E S ++ +P + + D P + R+S+R +Q
Sbjct: 824 PIIPEINQESSS---PSEFVSLSSLDPFL-ASSTVQTADLPLSSTTPAPIQLRRSSRQTQ 879
Query: 769 KPNHLSDYVCNSSS--DSTKQASSGILYPIKHYHSLNNISASHQKFALAITDASEPISYK 826
KP L ++V N+ S + +ASS LYPI+ Y + ++SH+ F A+T EP +Y
Sbjct: 880 KPMKLKNFVTNTVSVESISPEASSSSLYPIEKYVDCHRFTSSHKAFLAAVTAGMEPTTYN 939
Query: 827 EASQQDCWVKAMNSELSALNHNKTWIFVDTPPNIKPIGSKWVYKIKHKSDGTIERYKARL 886
EA W +AM++E+ +L N+T+ V+ PP + +G+KWVYKIK++SDG IERYKARL
Sbjct: 940 EAMVDKAWREAMSAEIESLRVNQTFSIVNLPPGKRALGNKWVYKIKYRSDGAIERYKARL 999
Query: 887 VAKGYTQVEGIDFFDTFSPVAKITTVRILIALASINSWHLHQLDVNNAFLHGDLSENVYM 946
V G Q EG+D+ +TF+PVAK++TVR+ + +A+ WH+HQ+DV+NAFLHGDL E VYM
Sbjct: 1000 VVLGNCQKEGVDYDETFAPVAKMSTVRLFLGVAAARDWHVHQMDVHNAFLHGDLKEEVYM 1059
Query: 947 SVPQGVHSPKPNQVCKLLKSLYGLKQASRKWYEKLTGFLLLQGYKQSASDHSLFILHSDT 1006
+PQG P++VC+L KSLYGLKQA R W+ KL+ L G+ QS SD+SLF ++D
Sbjct: 1060 KLPQGFQCDDPSKVCRLHKSLYGLKQAPRCWFSKLSSALKQYGFTQSLSDYSLFSYNNDG 1119
Query: 1007 CFTALLVYVDDVILAGNSMTEIDRIKAVLDVEFKIKDLGKLKYFLGIEVAHSKLGISICQ 1066
F +LVYVDD+I++G+ + + K+ L+ F +KDLG LKYFLGIEV+ + G + Q
Sbjct: 1120 IFVHVLVYVDDLIISGSCPDAVAQFKSYLESCFHMKDLGLLKYFLGIEVSRNAQGFYLSQ 1179
Query: 1067 RKYCLDLLKDTGLLGAKPVTTPLDPSIKLHQDNSPPHDDILSYRRLVGKLLYLTTTRPDI 1126
RKY LD++ + GLLGA+P PL+ + KL SP D YRRLVG+L+YL TRP++
Sbjct: 1180 RKYVLDIISEMGLLGARPSAFPLEQNHKLSLSTSPLLSDSSRYRRLVGRLIYLVVTRPEL 1239
Query: 1127 AFVVQQLSQFLNAPTITHYDTACRVVKYLKGTPGQGLLFRRDSQLQLLGFTDADWAGCPD 1186
++ V L+QF+ P H++ A RVV+YLK PGQG+L S LQ+ G+ D+D+A CP
Sbjct: 1240 SYSVHTLAQFMQNPRQDHWNAAIRVVRYLKSNPGQGILLSSTSTLQINGWCDSDYAACPL 1299
Query: 1187 SRRSTSGYCFFLGSSLISWRAKKQHTVARSSSEAEYRALSFASCELQWLLYLLKDLRVKC 1246
+RRS +GY LG + ISW+ KKQ TV+RSS+EAEYRA++F + EL WL +L DL V
Sbjct: 1300 TRRSLTGYFVQLGDTPISWKTKKQPTVSRSSAEAEYRAMAFLTQELMWLKRVLYDLGVSH 1359
Query: 1247 NKLPVLFCDNQSAIHIAGNPVFHERTKHLEIDCHFVREKLQQNIFKLLPIKSQSQLADFF 1306
+ +F D++SAI ++ NPV HERTKH+E+DCHF+R+ + I + S QLAD
Sbjct: 1360 VQAMRIFSDSKSAIALSVNPVQHERTKHVEVDCHFIRDAILDGIIATSFVPSHKQLADIL 1419
Query: 1307 TKPLPPKNFHSFLPKLNMIDLY 1328
TK L K FL KL ++D++
Sbjct: 1420 TKALGEKEVRYFLRKLGILDVH 1441
Score = 301 bits (770), Expect = 2e-81
Identities = 196/694 (28%), Positives = 336/694 (48%), Gaps = 98/694 (14%)
Query: 30 YYVHASDGPSSVAITPVLSHSNYHSWARSMRRALGAKNKFDFVDGSIPVPTDFDPNFKAW 89
Y + A+D +V P+L +NY WA + AL ++ KF F+DG+IP P D P+ + W
Sbjct: 21 YDLTAADNSGAVISHPILKTNNYEEWACGFKTALRSRKKFGFLDGTIPQPLDGSPDLEDW 80
Query: 90 NRCNMLVHSWIMNSVEDSIAQSIVFLENAIDVWNELKERFSQGDFIRISELQCEIFSLKQ 149
N L+ SW+ +++ + +I + A D+W ++++RFS + + +++ ++ + KQ
Sbjct: 81 LTINALLVSWMKMTIDSELLTNISHRDVARDLWEQIRKRFSVSNGPKNQKMKADLATCKQ 140
Query: 150 DSRSVTEFFTALKVLWEELEAYLPTPVCACPHRCMCITGVMNAKHQHEITRSIRFLTGLN 209
+ +V ++ L +W+ + +Y P +C C RC+C G K++ + ++L GLN
Sbjct: 141 EGMTVEGYYGKLNKIWDNINSYRPLRICKCG-RCICNLGTDQEKYRED-DMVHQYLYGLN 198
Query: 210 DS-FDLVRSQILLMNPLPTINKIFSMVMQHERQFKISIPVEDSTILVNSAGKSQGRGRGN 268
++ F +RS + PLP + +++++V Q E E+ T + A + + R
Sbjct: 199 ETKFHTIRSSLTSRVPLPGLEEVYNIVRQEEDMVNNRSSNEERTDVTAFAVQMRPRSEVI 258
Query: 269 GNSNSNG-----KRSCSFCGRDGHTIDICYRKHGFPPNYGNKNFAKVN-NSSIEQNEER- 321
+N K+ C+ C R GH+ + C+ G+P +G++ K N N S + R
Sbjct: 259 SEKFANSEKLQNKKLCTHCNRGGHSPENCFVLIGYPEWWGDRPRGKSNSNGSTSRGRGRF 318
Query: 322 ----------------------EDLDDSKSCKGNSNTEPSFGITKEQYEQLVTLLQSQQA 359
+ +S+ + G+T EQ+ +V LL + +
Sbjct: 319 GPGFNGGQPRPTYVNVVMTGPFPSSEHVNRVITDSDRDAVSGLTDEQWRGVVKLLNAGR- 377
Query: 360 SSSKVNLSHVTNHVTSGITRTSYTMNHSSFGTWIVDSGASDHICSSIKFFDSYAPIIPVH 419
S +K N +H T T + F +WI+D+GAS H+ +++ + PV
Sbjct: 378 SDNKSN-AHETQSGTCSL-----------FTSWILDTGASHHMTGNLELLSDMRSMSPVL 425
Query: 420 IKLPNGNMAIAKFSGTVNFSPGLIARNVLYVAEFKLNLLSVPKLCMDNNCIVTFDNDKCF 479
I L +GN +A GTV LI ++V YV E + +L+SV ++ +N+C
Sbjct: 426 IILADGNKRVAVSEGTVRLGSHLILKSVFYVKELESDLISVGQMMDENHC---------- 475
Query: 480 IQDKSNLKMTGLGELIEGLYFLNLGSQVFSQFNHQPVIALSQSSSSTTFLPQEALWHFRL 539
++ ++ T+ LWH RL
Sbjct: 476 ---------------------------------------VNAAAVHTSVKAPFDLWHRRL 496
Query: 540 GHLSNDRLLRM--QQHFPCIKVDSNSVCDICHYSRHKKLPFKLSMNKASHCYELIHFDIW 597
GH S D+++ + ++ K +VCD C ++ + F LS N++ ++LIH D+W
Sbjct: 497 GHAS-DKIVNLLPRELLSSGKEILENVCDTCMRAKQTRDTFPLSDNRSMDSFQLIHCDVW 555
Query: 598 GPISTHSIHGHKYFITALDDYSRFTWIMLCKTKSEVSKSVQNFILNIENQFNCKVKTVRT 657
GP S G +YF+T +DDYSR W+ L KSE K +++FI +E QF+ ++K VR+
Sbjct: 556 GPYRAPSYSGARYFLTIVDDYSRGVWVYLMTDKSETQKHLKDFIALVERQFDTEIKIVRS 615
Query: 658 DNGPEFL-MPDFYSSKGIEHQTSCVETPQQNGRV 690
DNG EFL M +++ KGI H+TSCV TP QNGRV
Sbjct: 616 DNGTEFLCMREYFLHKGIAHETSCVGTPHQNGRV 649
>At4g23160 putative protein
Length = 1240
Score = 547 bits (1409), Expect = e-155
Identities = 270/546 (49%), Positives = 371/546 (67%), Gaps = 12/546 (2%)
Query: 745 IQDPTQIEHEPQQLNTRKSTRVSQKPNHLSDYVCNSSSDSTKQASSGILYPIKHYHSLNN 804
I I+++ + + S R ++KP +L DY C+S + T ++ I + S
Sbjct: 16 IMPSANIQNDVPEPSVHTSHRRTRKPAYLQDYYCHSVASLT-------IHDISQFLSYEK 68
Query: 805 ISASHQKFALAITDASEPISYKEASQQDCWVKAMNSELSALNHNKTWIFVDTPPNIKPIG 864
+S + F + I A EP +Y EA + W AM+ E+ A+ TW PPN KPIG
Sbjct: 69 VSPLYHSFLVCIAKAKEPSTYNEAKEFLVWCGAMDDEIGAMETTHTWEICTLPPNKKPIG 128
Query: 865 SKWVYKIKHKSDGTIERYKARLVAKGYTQVEGIDFFDTFSPVAKITTVRILIALASINSW 924
KWVYKIK+ SDGTIERYKARLVAKGYTQ EGIDF +TFSPV K+T+V++++A+++I ++
Sbjct: 129 CKWVYKIKYNSDGTIERYKARLVAKGYTQQEGIDFIETFSPVCKLTSVKLILAISAIYNF 188
Query: 925 HLHQLDVNNAFLHGDLSENVYMSVPQGV-----HSPKPNQVCKLLKSLYGLKQASRKWYE 979
LHQLD++NAFL+GDL E +YM +P G S PN VC L KS+YGLKQASR+W+
Sbjct: 189 TLHQLDISNAFLNGDLDEEIYMKLPPGYAARQGDSLPPNAVCYLKKSIYGLKQASRQWFL 248
Query: 980 KLTGFLLLQGYKQSASDHSLFILHSDTCFTALLVYVDDVILAGNSMTEIDRIKAVLDVEF 1039
K + L+ G+ QS SDH+ F+ + T F +LVYVDD+I+ N+ +D +K+ L F
Sbjct: 249 KFSVTLIGFGFVQSHSDHTYFLKITATLFLCVLVYVDDIIICSNNDAAVDELKSQLKSCF 308
Query: 1040 KIKDLGKLKYFLGIEVAHSKLGISICQRKYCLDLLKDTGLLGAKPVTTPLDPSIKLHQDN 1099
K++DLG LKYFLG+E+A S GI+ICQRKY LDLL +TGLLG KP + P+DPS+ +
Sbjct: 309 KLRDLGPLKYFLGLEIARSAAGINICQRKYALDLLDETGLLGCKPSSVPMDPSVTFSAHS 368
Query: 1100 SPPHDDILSYRRLVGKLLYLTTTRPDIAFVVQQLSQFLNAPTITHYDTACRVVKYLKGTP 1159
D +YRRL+G+L+YL TR DI+F V +LSQF AP + H +++ Y+KGT
Sbjct: 369 GGDFVDAKAYRRLIGRLMYLQITRLDISFAVNKLSQFSEAPRLAHQQAVMKILHYIKGTV 428
Query: 1160 GQGLLFRRDSQLQLLGFTDADWAGCPDSRRSTSGYCFFLGSSLISWRAKKQHTVARSSSE 1219
GQGL + +++QL F+DA + C D+RRST+GYC FLG+SLISW++KKQ V++SS+E
Sbjct: 429 GQGLFYSSQAEMQLQVFSDASFQSCKDTRRSTNGYCMFLGTSLISWKSKKQQVVSKSSAE 488
Query: 1220 AEYRALSFASCELQWLLYLLKDLRVKCNKLPVLFCDNQSAIHIAGNPVFHERTKHLEIDC 1279
AEYRALSFA+ E+ WL ++L++ +K +LFCDN +AIHIA N VFHERTKH+E DC
Sbjct: 489 AEYRALSFATDEMMWLAQFFRELQLPLSKPTLLFCDNTAAIHIATNAVFHERTKHIESDC 548
Query: 1280 HFVREK 1285
H VRE+
Sbjct: 549 HSVRER 554
>At2g20460 putative retroelement pol polyprotein
Length = 1461
Score = 538 bits (1387), Expect = e-153
Identities = 272/571 (47%), Positives = 378/571 (65%), Gaps = 17/571 (2%)
Query: 765 RVSQKPNHLSDYVCN--SSSDSTKQASSGILYPIKHYHSLNNISASHQKFALAITDASEP 822
R+++ P HL DY C + DS +PI S + IS SH + I+ P
Sbjct: 897 RITKFPAHLQDYHCYFVNKDDS---------HPISSSLSYSQISPSHMLYINNISKIPIP 947
Query: 823 ISYKEASQQDCWVKAMNSELSALNHNKTWIFVDTPPNIKPIGSKWVYKIKHKSDGTIERY 882
SY EA W A++ E+ A+ TW PP K +G KWV+ +K +DG++ER+
Sbjct: 948 QSYHEAKDSKEWCGAIDQEIGAMERTDTWEITSLPPGKKAVGCKWVFTVKFHADGSLERF 1007
Query: 883 KARLVAKGYTQVEGIDFFDTFSPVAKITTVRILIALASINSWHLHQLDVNNAFLHGDLSE 942
KAR+VAKGYTQ EG+D+ +TFSPVAK+ TV++L+ +++ W+L+QLD++NAFL+GDL E
Sbjct: 1008 KARIVAKGYTQKEGLDYTETFSPVAKMATVKLLLKVSASKKWYLNQLDISNAFLNGDLEE 1067
Query: 943 NVYMSVPQGVHSPK-----PNQVCKLLKSLYGLKQASRKWYEKLTGFLLLQGYKQSASDH 997
+YM +P G K PN VC+L KS+YGLKQASR+W+ K + LL G+++ DH
Sbjct: 1068 TIYMKLPDGYADIKGTSLPPNVVCRLKKSIYGLKQASRQWFLKFSNSLLALGFEKQHGDH 1127
Query: 998 SLFILHSDTCFTALLVYVDDVILAGNSMTEIDRIKAVLDVEFKIKDLGKLKYFLGIEVAH 1057
+LF+ + F LLVYVDD+++A + + L FK+++LG LKYFLG+EVA
Sbjct: 1128 TLFVRCIGSEFIVLLVYVDDIVIASTTEQAAQSLTEALKASFKLRELGPLKYFLGLEVAR 1187
Query: 1058 SKLGISICQRKYCLDLLKDTGLLGAKPVTTPLDPSIKLHQDNSPPHDDILSYRRLVGKLL 1117
+ GIS+ QRKY L+LL +L KP + P+ P+I+L +++ +D YRRLVGKL+
Sbjct: 1188 TSEGISLSQRKYALELLTSADMLDCKPSSIPMTPNIRLSKNDGLLLEDKEMYRRLVGKLM 1247
Query: 1118 YLTTTRPDIAFVVQQLSQFLNAPTITHYDTACRVVKYLKGTPGQGLLFRRDSQLQLLGFT 1177
YLT TRPDI F V +L QF +AP H +V++Y+KGT GQGL + + L L G+T
Sbjct: 1248 YLTITRPDITFAVNKLCQFSSAPRTAHLAAVYKVLQYIKGTVGQGLFYSAEDDLTLKGYT 1307
Query: 1178 DADWAGCPDSRRSTSGYCFFLGSSLISWRAKKQHTVARSSSEAEYRALSFASCELQWLLY 1237
DADW CPDSRRST+G+ F+GSSLISWR+KKQ TV+RSS+EAEYRAL+ ASCE+ WL
Sbjct: 1308 DADWGTCPDSRRSTTGFTMFVGSSLISWRSKKQPTVSRSSAEAEYRALALASCEMAWLST 1367
Query: 1238 LLKDLRVKCNKLPVLFCDNQSAIHIAGNPVFHERTKHLEIDCHFVREKLQQNIFKLLPIK 1297
LL LRV + +P+L+ D+ +A++IA NPVFHERTKH+EIDCH VREKL KLL +K
Sbjct: 1368 LLLALRVH-SGVPILYSDSTAAVYIATNPVFHERTKHIEIDCHTVREKLDNGQLKLLHVK 1426
Query: 1298 SQSQLADFFTKPLPPKNFHSFLPKLNMIDLY 1328
++ Q+AD TKPL P F L K+++ +++
Sbjct: 1427 TKDQVADILTKPLFPYQFAHLLSKMSIQNIF 1457
Score = 377 bits (969), Expect = e-104
Identities = 235/706 (33%), Positives = 364/706 (51%), Gaps = 75/706 (10%)
Query: 21 DPSQQPGNVYYVHASDGPSSVAITPVLSHSNYHSWARSMRRALGAKNKFDFVDGSIPVPT 80
DP+Q P +++H++D P I+ L + Y W+ +MR +L AKNK FVDGS+P P
Sbjct: 60 DPTQSP---FFLHSADHPGLSIISHRLDETTYGDWSVAMRISLDAKNKLGFVDGSLPRPL 116
Query: 81 DFDPNFKAWNRCNMLVHSWIMNSVEDSIAQSIVFLENAIDVWNELKERFSQGDFIRISEL 140
+ DPNF+ W+RCN +V SW++NSV I +SI+ L +A D+W +L +RF+ + R L
Sbjct: 117 ESDPNFRLWSRCNSMVKSWLLNSVSPQIYRSILRLNDATDIWRDLFDRFNLTNLPRTYNL 176
Query: 141 QCEIFSLKQDSRSVTEFFTALKVLWEELEAYLPTPVCACPHRCMCITGVMNAKHQHEITR 200
EI L+Q + S++E++T LK LW++L++ A C C V + E +
Sbjct: 177 TQEIQDLRQGTMSLSEYYTLLKTLWDQLDS-----TEALDDPCTCGKAV-RLYQKAEKAK 230
Query: 201 SIRFLTGLNDSFDLVRSQILLMNPLPTINKIFSMVMQHERQ------------FKIS--- 245
++FL GLN+S+ +VR QI+ LP++ +++ ++ Q Q F++S
Sbjct: 231 IMKFLAGLNESYAIVRRQIIAKKALPSLAEVYHILDQDNSQKGFFNVVAPPAAFQVSEVS 290
Query: 246 -IPVEDSTILVNSAGKSQGRGRGNGNSNSNGKRSCSFCGRDGHTIDICYRKHGFPPNYGN 304
P+ I+ +G ++GR +CSFC R GH + CY+KHGFPP +
Sbjct: 291 HSPITSPEIMYVQSGPNKGRP------------TCSFCNRVGHIAERCYKKHGFPPGFTP 338
Query: 305 KNFAKVNNSSIEQNEEREDLDDSKSCKGNSNTEPSFGITKEQYEQLVTLLQSQ------- 357
K + + + L K +F + +Q + L+ L SQ
Sbjct: 339 KGKSSDKPPKPQAVAAQVTLSPDKMTGQLETLAGNF--SPDQIQNLIALFSSQLQPQIVS 396
Query: 358 -QASSSKVNLSHVTNHVTSGITRTSYT--------MNHSSFG--TWIVDSGASDHICSSI 406
Q +SS+ S + SGI + T ++H+S TW++DSGA+ H+
Sbjct: 397 PQTASSQHEASSSQSVAPSGILFSPSTYCFIGILAVSHNSLSSDTWVIDSGATHHVSHDR 456
Query: 407 KFFDSYAPIIPVHIKLPNGNMAIAKFSGTVNFSPGLIARNVLYVAEFKLNLLSVPKLCMD 466
K F + I + LP G GTV + +I +NVL++ EF+LNL+S+ L D
Sbjct: 457 KLFQTLDTSIVSFVNLPTGPNVRISGVGTVLINKDIILQNVLFIPEFRLNLISISSLTTD 516
Query: 467 NNCIVTFDNDKCFIQDKSNLKMTGLGELIEGLYFLNLGSQVFSQFNHQPVIALSQSSSST 526
V FD C IQD + G G+ I LY L+ S P I+++
Sbjct: 517 LGTRVIFDPSCCQIQDLTKGLTLGEGKRIGNLYVLDTQS---------PAISVNA----- 562
Query: 527 TFLPQEALWHFRLGHLSNDRLLRMQQHFPCI--KVDSNSVCDICHYSRHKKLPFKLSMNK 584
+ ++WH RLGH S RL + + K ++ C +CH ++ KKL F + N
Sbjct: 563 --VVDVSVWHKRLGHPSFSRLDSLSEVLGTTRHKNKKSAYCHVCHLAKQKKLSFPSANNI 620
Query: 585 ASHCYELIHFDIWGPISTHSIHGHKYFITALDDYSRFTWIMLCKTKSEVSKSVQNFILNI 644
+ +EL+H D+WGP S ++ G+KYF+T +DD+SR TWI L K+KS+V FI +
Sbjct: 621 CNSTFELLHIDVWGPFSVETVEGYKYFLTIVDDHSRATWIYLLKSKSDVLTVFPAFIDLV 680
Query: 645 ENQFNCKVKTVRTDNGPEFLMPDFYSSKGIEHQTSCVETPQQNGRV 690
ENQ++ +VK+VR+DN E +FY +KGI SC ETP+QN V
Sbjct: 681 ENQYDTRVKSVRSDNAKELAFTEFYKAKGIVSFHSCPETPEQNSVV 726
>At4g17450 retrotransposon like protein
Length = 1433
Score = 530 bits (1364), Expect = e-150
Identities = 270/622 (43%), Positives = 382/622 (61%), Gaps = 39/622 (6%)
Query: 712 PATEQTSSNSELPLENQSETNKEPAMHIDNNTHIQDPTQIEHEPQQLNTRKSTRVSQKPN 771
P + + +LPLE S + P + ++ + + +P S R + P
Sbjct: 841 PLLQFPARTDDLPLEQTSIIDTHPHQDVSSSKAL-----VPFDPL------SKRQKKPPK 889
Query: 772 HLSDYVCNSSSDSTKQASSGILYPIKHYHSLNNISASHQKFALAITDASEPISYKEASQQ 831
HL D+ H NN + F IT+A P Y EA
Sbjct: 890 HLQDF-----------------------HCYNNTTEPFHAFINNITNAVIPQRYSEAKDF 926
Query: 832 DCWVKAMNSELSALNHNKTWIFVDTPPNIKPIGSKWVYKIKHKSDGTIERYKARLVAKGY 891
W AM E+ A+ TW V PPN K IG KWV+ IKH +DG+IERYKARLVAKGY
Sbjct: 927 KAWCDAMKEEIGAMVRTNTWSVVSLPPNKKAIGCKWVFTIKHNADGSIERYKARLVAKGY 986
Query: 892 TQVEGIDFFDTFSPVAKITTVRILIALASINSWHLHQLDVNNAFLHGDLSENVYMSVPQG 951
TQ EG+D+ +TFSPVAK+T+VR+++ LA+ W +HQLD++NAFL+GDL E +YM +P G
Sbjct: 987 TQEEGLDYEETFSPVAKLTSVRMMLLLAAKMKWSVHQLDISNAFLNGDLDEEIYMKIPPG 1046
Query: 952 V-----HSPKPNQVCKLLKSLYGLKQASRKWYEKLTGFLLLQGYKQSASDHSLFILHSDT 1006
+ P+ +C+L KS+YGLKQASR+WY KL+ L G+++S +DH+LFI +++
Sbjct: 1047 YADLVGEALPPHAICRLHKSIYGLKQASRQWYLKLSNTLKGMGFQKSNADHTLFIKYANG 1106
Query: 1007 CFTALLVYVDDVILAGNSMTEIDRIKAVLDVEFKIKDLGKLKYFLGIEVAHSKLGISICQ 1066
+LVYVDD+++ NS + + A L FK++DLG KYFLGIE+A S+ GISICQ
Sbjct: 1107 VLMGVLVYVDDIMIVSNSDDAVAQFTAELKSYFKLRDLGAAKYFLGIEIARSEKGISICQ 1166
Query: 1067 RKYCLDLLKDTGLLGAKPVTTPLDPSIKLHQDNSPPHDDILSYRRLVGKLLYLTTTRPDI 1126
RKY L+LL TG LG+KP + PLDPS+KL++++ P D SYR+LVGKL+YL TRPDI
Sbjct: 1167 RKYILELLSTTGFLGSKPSSIPLDPSVKLNKEDGVPLTDSTSYRKLVGKLMYLQITRPDI 1226
Query: 1127 AFVVQQLSQFLNAPTITHYDTACRVVKYLKGTPGQGLLFRRDSQLQLLGFTDADWAGCPD 1186
A+ V L QF +APT H +V++YLKGT GQGL + D + L G+TD+D+ C D
Sbjct: 1227 AYAVNTLCQFSHAPTSVHLSAVHKVLRYLKGTVGQGLFYSADDKFDLRGYTDSDFGSCTD 1286
Query: 1187 SRRSTSGYCFFLGSSLISWRAKKQHTVARSSSEAEYRALSFASCELQWLLYLLKDLRVKC 1246
SRR + YC F+G L+SW++KKQ TV+ S++EAE+RA+S + E+ WL L D +V
Sbjct: 1287 SRRCVAAYCMFIGDYLVSWKSKKQDTVSMSTAEAEFRAMSQGTKEMIWLSRLFDDFKVPF 1346
Query: 1247 NKLPVLFCDNQSAIHIAGNPVFHERTKHLEIDCHFVREKLQQNIFKLLPIKSQSQLADFF 1306
L+CDN +A+HI N VFHERTK +E+DC+ RE ++ K + +++ Q+AD
Sbjct: 1347 IPPAYLYCDNTAALHIVNNSVFHERTKFVELDCYKTREAVESGFLKTMFVETGEQVADPL 1406
Query: 1307 TKPLPPKNFHSFLPKLNMIDLY 1328
TK + P FH + K+ + +++
Sbjct: 1407 TKAIHPAQFHKLIGKMGVCNIF 1428
Score = 285 bits (730), Expect = 9e-77
Identities = 207/706 (29%), Positives = 315/706 (44%), Gaps = 117/706 (16%)
Query: 30 YYVHASDGPSSVAITPVLSHSNYHSWARSMRRALGAKNKFDFVDGSIPVPTDFDPNFKAW 89
Y++H+SD P ++ +L +NY++W+ +MR +L AKNK FVDGS+P P D FK W
Sbjct: 68 YFLHSSDHPGLNIVSHILDGTNYNNWSIAMRMSLDAKNKLSFVDGSLPRPDVSDRMFKIW 127
Query: 90 NRCNMLVHSWIMNSVEDSIAQSIVFLENAIDVWNELKERFSQGDFIRISELQCEIFSLKQ 149
+RCN +V +W++N V ++WN+L RF + R +L+ I +LKQ
Sbjct: 128 SRCNSMVKTWLLNVV--------------TEMWNDLFSRFRVSNLPRKYQLEQSIHTLKQ 173
Query: 150 DSRSVTEFFTALKVLWEEL--EAYLPTPVCACPHRCMCITGVMNAKHQHEITRSIRFLTG 207
+ ++ ++T K LWE+L L C C H V + E +R I+FL G
Sbjct: 174 GNLDLSTYYTKKKTLWEQLANTRVLTVRKCNCEH-------VKELLEEAETSRIIQFLMG 226
Query: 208 LNDSFDLVRSQILLMNPLPTINKIFSMVMQHERQFKISIP----------VEDSTILVNS 257
LND+F +R QIL M P P + +I++M+ Q E Q + P V+ S I+ +
Sbjct: 227 LNDNFAHIRGQILNMKPRPGLTEIYNMLDQDESQRLVGNPTLSNPTAAFQVQASPIIDSQ 286
Query: 258 AGKSQGRGRGNGNSNSNGKRSCSFCGRDGHTIDICYRKHGFPP--------NYGNKNFAK 309
+QG S K CS+C + GH +D CY+KHG+PP G+ N A
Sbjct: 287 VNMAQG---------SYKKPKCSYCNKLGHLVDKCYKKHGYPPGSKWTKGQTIGSTNLAS 337
Query: 310 -----VNNSSIEQNEEREDLDDSKSCKGNSNTEPSFGITKEQYEQLVTLLQSQQASSSKV 364
VN + E+ + E+ + S I T S AS S
Sbjct: 338 TQLQPVNETPNEKTDSYEEFSTDQIQTMISYLSTKLHIASAS-PMPTTSSASISASPSVP 396
Query: 365 NLSHVTNHVTSGITRTSYTMNHSSFGT--------WIVDSGASDHICSSIKFFDSYAPII 416
+S ++ S + Y M SS W++DSGA+ H+ + + ++ +
Sbjct: 397 MISQISGTFLSLFSNAYYDMLISSVSQEPAVSPRGWVIDSGATHHVTHNRDLYLNFRSLE 456
Query: 417 PVHIKLPNGNMAIAKFSGTVNFSPGLIARNVLYVAEFKLNLLSVPKLCMDNNCIVTFDND 476
++LPN G + S + NVLY+ EFK NL+S
Sbjct: 457 NTFVRLPNDCTVKIAGIGFIQLSDAISLHNVLYIPEFKFNLIS----------------- 499
Query: 477 KCFIQDKSNLKMTGLGELIEGLYFLNLGSQVFSQFNHQPVIALSQS-----SSSTTFLPQ 531
+ + M G G + LY L+ F++ NH + + S S ++ +
Sbjct: 500 -----ELTKELMIGRGSQVGNLYVLD-----FNENNHTVSLKGTTSMCPEFSVCSSVVVD 549
Query: 532 EALWHFRLGH--LSNDRLLRMQQHFPCIKVDSN-----SVCDICHYSRHKKLPFKLSMNK 584
WH RLGH S LL + K++ VC +CH S+ K L F+ N
Sbjct: 550 SVTWHKRLGHPAYSKIDLLSDVLNLKVKKINKEHSPVCHVCHVCHLSKQKHLSFQSRQNM 609
Query: 585 ASHCYELIHFDIWGPISTHSIHGHKYFITALDDYSRFTWIMLCKTKSEVSKSVQNFILNI 644
S ++L+H D WGP S + + TWI L K KS+V FI +
Sbjct: 610 CSAAFDLVHIDTWGPFSVPT--------------NDATWIYLLKNKSDVLHVFPAFINMV 655
Query: 645 ENQFNCKVKTVRTDNGPEFLMPDFYSSKGIEHQTSCVETPQQNGRV 690
Q+ K+K+VR+DN E D +++ GI SC ETP+QN V
Sbjct: 656 HTQYQTKLKSVRSDNAHELKFTDLFAAHGIVAYHSCPETPEQNSVV 701
>At2g13940 putative retroelement pol polyprotein
Length = 1501
Score = 528 bits (1361), Expect = e-150
Identities = 278/583 (47%), Positives = 373/583 (63%), Gaps = 17/583 (2%)
Query: 761 RKSTRVSQKPNHLSDYV---------------CNSSSDSTKQASSGILYPIKHYHSLNNI 805
RKS R + P L+DYV + S ST S L+P+ Y S
Sbjct: 918 RKSKRATHPPPKLNDYVLYNAMYTPSSIHALPADPSQSSTVPGKS--LFPLTDYVSDAAF 975
Query: 806 SASHQKFALAITDASEPISYKEASQQDCWVKAMNSELSALNHNKTWIFVDTPPNIKPIGS 865
S+SH+ + AITD EP +KEA Q W AM +E+ AL NKTW VD PP IGS
Sbjct: 976 SSSHRAYLAAITDNVEPKHFKEAVQIKVWNDAMFTEVDALEINKTWDIVDLPPGKVAIGS 1035
Query: 866 KWVYKIKHKSDGTIERYKARLVAKGYTQVEGIDFFDTFSPVAKITTVRILIALASINSWH 925
+WV+K K+ SDGT+ERYKARLV +G QVEG D+ +TF+PV ++TTVR L+ + N W
Sbjct: 1036 QWVFKTKYNSDGTVERYKARLVVQGNKQVEGEDYKETFAPVVRMTTVRTLLRNVAANQWE 1095
Query: 926 LHQLDVNNAFLHGDLSENVYMSVPQGVHSPKPNQVCKLLKSLYGLKQASRKWYEKLTGFL 985
++Q+DV+NAFLHGDL E VYM +P G P++VC+L KSLYGLKQA R W++KL+ L
Sbjct: 1096 VYQMDVHNAFLHGDLEEEVYMKLPPGFRHSHPDKVCRLRKSLYGLKQAPRCWFKKLSDSL 1155
Query: 986 LLQGYKQSASDHSLFILHSDTCFTALLVYVDDVILAGNSMTEIDRIKAVLDVEFKIKDLG 1045
L G+ QS D+SLF + +L+YVDD+++ GN + + K L F +KDLG
Sbjct: 1156 LRFGFVQSYEDYSLFSYTRNNIELRVLIYVDDLLICGNDGYMLQKFKDYLSRCFSMKDLG 1215
Query: 1046 KLKYFLGIEVAHSKLGISICQRKYCLDLLKDTGLLGAKPVTTPLDPSIKLHQDNSPPHDD 1105
KLKYFLGIEV+ GI + QRKY LD++ D+G LG++P TPL+ + L D+ P D
Sbjct: 1216 KLKYFLGIEVSRGPEGIFLSQRKYALDVIADSGNLGSRPAHTPLEQNHHLASDDGPLLSD 1275
Query: 1106 ILSYRRLVGKLLYLTTTRPDIAFVVQQLSQFLNAPTITHYDTACRVVKYLKGTPGQGLLF 1165
YRRLVG+LLYL TRP++++ V L+QF+ P H+D A RVV+YLKG+PGQG+L
Sbjct: 1276 PKPYRRLVGRLLYLLHTRPELSYSVHVLAQFMQNPREAHFDAALRVVRYLKGSPGQGILL 1335
Query: 1166 RRDSQLQLLGFTDADWAGCPDSRRSTSGYCFFLGSSLISWRAKKQHTVARSSSEAEYRAL 1225
D L L + D+DW CP +RRS S Y LG S ISW+ KKQ TV+ SS+EAEYRA+
Sbjct: 1336 NADPDLTLEVYCDSDWQSCPLTRRSISAYVVLLGGSPISWKTKKQDTVSHSSAEAEYRAM 1395
Query: 1226 SFASCELQWLLYLLKDLRVKCNKLPVLFCDNQSAIHIAGNPVFHERTKHLEIDCHFVREK 1285
S+A E++WL LLK+L ++ + L+CD+++AIHIA NPVFHERTKH+E DCH VR+
Sbjct: 1396 SYALKEIKWLRKLLKELGIEQSTPARLYCDSKAAIHIAANPVFHERTKHIESDCHSVRDA 1455
Query: 1286 LQQNIFKLLPIKSQSQLADFFTKPLPPKNFHSFLPKLNMIDLY 1328
++ I +++ QLAD FTK L F + KL + +L+
Sbjct: 1456 VRDGIITTQHVRTTEQLADVFTKALGRNQFLYLMSKLGVQNLH 1498
Score = 347 bits (890), Expect = 3e-95
Identities = 215/686 (31%), Positives = 340/686 (49%), Gaps = 56/686 (8%)
Query: 30 YYVHASDGPSSVAITPVLSHSNYHSWARSMRRALGAKNKFDFVDGSIPVPTDFDPNFKAW 89
Y + +SD P +V + L+ NY+ WA M AL AK K F++G+IP P DPN++ W
Sbjct: 32 YTLASSDNPGAVISSVELNGDNYNQWATEMLNALQAKRKTGFINGTIPRPPPNDPNYENW 91
Query: 90 NRCNMLVHSWIMNSVEDSIAQSIVFLENAIDVWNELKERFSQGDFIRISELQCEIFSLKQ 149
N ++ WI S+E + ++ F+ +A +W +LK+RFS G+ +RI +++ ++ S +Q
Sbjct: 92 TAVNSMIVGWIRTSIEPKVKATVTFISDAHLLWKDLKQRFSVGNKVRIHQIRAQLSSCRQ 151
Query: 150 DSRSVTEFFTALKVLWEELEAYLPTPVCACPHRCMCITGVMNAKHQHEITRSIRFLTGLN 209
D ++V E++ L LWEE Y P VC C C C K + E + +F+ GL+
Sbjct: 152 DGQAVIEYYGRLSNLWEEYNIYKPVTVCTC-GLCRCGATSEPTKEREE-EKIHQFVLGLD 209
Query: 210 DS-FDLVRSQILLMNPLPTINKIFSMVMQHERQFKISIPVEDS----------------- 251
+S F + + ++ M+PLP++ +I+S V++ E++ S+ V +
Sbjct: 210 ESRFGGLCATLINMDPLPSLGEIYSRVIREEQRL-ASVHVREQKEEAVGFLARREQLDHH 268
Query: 252 TILVNSAGKSQGRGRGNGNSNSNGKRSCSFCGRDGHTIDICYRKHGFPPNYGNKNFAKVN 311
+ + S+ +S+ G NS G+ +CS CGR GH C++ GFP + +N + +
Sbjct: 269 SRVDASSSRSEHTGGSRSNSIIKGRVTCSNCGRTGHEKKECWQIVGFPDWWSERNGGRGS 328
Query: 312 NS------SIEQNEEREDLDDSKSCKGNSNTEPSFGITKEQYEQLVTLLQSQQASSSKVN 365
N + + + + NS+ P F T+E L L++ + S S N
Sbjct: 329 NGRGRGGRGSNGGRGQGQVMAAHATSSNSSVFPEF--TEEHMRVLSQLVKEKSNSGSTSN 386
Query: 366 LSHVTNHVTSGITRTSYTMNHSSFGTWIVDSGASDHICSSIKFFDSYAPIIPVHIKLPNG 425
+ + G I+DSGAS H+ ++ + P+ P + +G
Sbjct: 387 ------------NNSDRLSGKTKLGDIILDSGASHHMTGTLSSLTNVVPVPPCPVGFADG 434
Query: 426 NMAIAKFSGTVNFSPGLIARNVLYVAEFKLNLLSVPKLCMDNNCIVTFDNDKCFIQDKSN 485
+ A A G + S + NVL+V L+SV KL C+ TF + CF+QD+S+
Sbjct: 435 SKAFALSVGVLTLSNTVSLTNVLFVPSLNCTLISVSKLLKQTQCLATFTDTLCFLQDRSS 494
Query: 486 LKMTGLGELIEGLYFLNLGSQVFSQFNHQPVIALSQSSSSTTFLPQEALWHFRLGHLSND 545
+ G GE G+Y+L + P + + S +ALWH RLGH S
Sbjct: 495 KTLIGSGEERGGVYYLT---------DVTPAKIHTANVDS-----DQALWHQRLGHPSFS 540
Query: 546 RLLRMQQHFPCIKVDSNSVCDICHYSRHKKLPFKLSMNKASHCYELIHFDIWGPISTHSI 605
L + ++ CD+C ++ + F S+NK C+ LIH D+WGP +
Sbjct: 541 VLSSLPLFSKTSSTVTSHSCDVCFRAKQTREVFPESINKTEECFSLIHCDVWGPYRVPAS 600
Query: 606 HGHKYFITALDDYSRFTWIMLCKTKSEVSKSVQNFILNIENQFNCKVKTVRTDNGPEFL- 664
G YF+T +DDYSR W L KSEV + + NF+ E QF VK VR+DNG EF+
Sbjct: 601 CGAVYFLTIVDDYSRAVWTYLLLEKSEVRQVLTNFLKYAEKQFGKTVKMVRSDNGTEFMC 660
Query: 665 MPDFYSSKGIEHQTSCVETPQQNGRV 690
+ ++ GI HQTSCV TPQQNGRV
Sbjct: 661 LSSYFRENGIIHQTSCVGTPQQNGRV 686
>At2g23330 putative retroelement pol polyprotein
Length = 1496
Score = 527 bits (1357), Expect = e-149
Identities = 273/571 (47%), Positives = 366/571 (63%), Gaps = 3/571 (0%)
Query: 759 NTRKSTRVSQKPNHLSDYVCNS---SSDSTKQASSGILYPIKHYHSLNNISASHQKFALA 815
NT + + H V S +S S S LYP+ + + + SA+H F A
Sbjct: 921 NTTSKKKTASHNIHSPSQVLPSGLPTSLSADSVSGKTLYPLSDFLTNSGYSANHIAFMAA 980
Query: 816 ITDASEPISYKEASQQDCWVKAMNSELSALNHNKTWIFVDTPPNIKPIGSKWVYKIKHKS 875
I D++EP +K+A W +AM+ E+ AL N TW D P K I SKWVYK+K+ S
Sbjct: 981 ILDSNEPKHFKDAILIKEWCEAMSKEIDALEANHTWDITDLPHGKKAISSKWVYKLKYNS 1040
Query: 876 DGTIERYKARLVAKGYTQVEGIDFFDTFSPVAKITTVRILIALASINSWHLHQLDVNNAF 935
DGT+ER+KARLV G Q EG+DF +TF+PVAK+TTVR ++A+A+ W +HQ+DV+NAF
Sbjct: 1041 DGTLERHKARLVVMGNHQKEGVDFKETFAPVAKLTTVRTILAVAAAKDWEVHQMDVHNAF 1100
Query: 936 LHGDLSENVYMSVPQGVHSPKPNQVCKLLKSLYGLKQASRKWYEKLTGFLLLQGYKQSAS 995
LHGDL E VYM +P G P++VC+L KSLYGLKQA R W+ KL+ L G+ QS
Sbjct: 1101 LHGDLEEEVYMRLPPGFKCSDPSKVCRLRKSLYGLKQAPRCWFSKLSTALRNIGFTQSYE 1160
Query: 996 DHSLFILHSDTCFTALLVYVDDVILAGNSMTEIDRIKAVLDVEFKIKDLGKLKYFLGIEV 1055
D+SLF L + +LVYVDD+I+AGN++ IDR K+ L F +KDLGKLKYFLG+EV
Sbjct: 1161 DYSLFSLKNGDTIIHVLVYVDDLIVAGNNLDAIDRFKSQLHKCFHMKDLGKLKYFLGLEV 1220
Query: 1056 AHSKLGISICQRKYCLDLLKDTGLLGAKPVTTPLDPSIKLHQDNSPPHDDILSYRRLVGK 1115
+ G + QRKY LD++K+TGLLG KP P+ + KL P + YRRLVG+
Sbjct: 1221 SRGPDGFCLSQRKYALDIVKETGLLGCKPSAVPIALNHKLASITGPVFTNPEQYRRLVGR 1280
Query: 1116 LLYLTTTRPDIAFVVQQLSQFLNAPTITHYDTACRVVKYLKGTPGQGLLFRRDSQLQLLG 1175
+YLT TRPD+++ V LSQF+ AP + H++ A R+V+YLKG+P QG+ R DS L +
Sbjct: 1281 FIYLTITRPDLSYAVHILSQFMQAPLVAHWEAALRLVRYLKGSPAQGIFLRSDSSLIINA 1340
Query: 1176 FTDADWAGCPDSRRSTSGYCFFLGSSLISWRAKKQHTVARSSSEAEYRALSFASCELQWL 1235
+ D+D+ CP +RRS S Y +LG S ISW+ KKQ TV+ SS+EAEYRA+++ EL+WL
Sbjct: 1341 YCDSDYNACPLTRRSLSAYVVYLGDSPISWKTKKQDTVSYSSAEAEYRAMAYTLKELKWL 1400
Query: 1236 LYLLKDLRVKCNKLPVLFCDNQSAIHIAGNPVFHERTKHLEIDCHFVREKLQQNIFKLLP 1295
LLKDL V + L CD+++AIHIA NPVFHERTKH+E DCH VR+ + +
Sbjct: 1401 KALLKDLGVHHSSPMKLHCDSEAAIHIAANPVFHERTKHIESDCHKVRDAVLDKLITTEH 1460
Query: 1296 IKSQSQLADFFTKPLPPKNFHSFLPKLNMID 1326
I ++ Q+AD TK LP F L L + D
Sbjct: 1461 IYTEDQVADLLTKSLPRPTFERLLSTLGVTD 1491
Score = 334 bits (856), Expect = 2e-91
Identities = 221/690 (32%), Positives = 326/690 (47%), Gaps = 81/690 (11%)
Query: 30 YYVHASDGPSSVAITPVLSHSNYHSWARSMRRALGAKNKFDFVDGSIPVPTDFDPNFKAW 89
Y ++ASD P ++ + VL +NY W+ ++ L AK K F+DGSIP P DP W
Sbjct: 22 YLINASDNPGALISSVVLKENNYAEWSEELQNFLRAKQKLGFIDGSIPKPAA-DPELSLW 80
Query: 90 NRCNMLVHSWIMNSVEDSIAQSIVFLENAIDVWNELKERFSQGDFIRISELQCEIFSLKQ 149
N ++ WI S++ +I ++ F+ A +W L+ RFS G+ +R + L+ EI + Q
Sbjct: 81 IAINSMIVGWIRTSIDPTIRSTVGFVSEASQLWENLRRRFSVGNGVRKTLLKDEIAACTQ 140
Query: 150 DSRSVTEFFTALKVLWEELEAYLPTPVCACPHRCMCITGVMNAKHQHEITRSIRFLTGLN 209
D + V ++ L LWEEL+ Y C C + + + E R +FL GL+
Sbjct: 141 DGQPVLAYYGRLIKLWEELQNYKSGRECKCE-------AASDIEKEREDDRVHKFLLGLD 193
Query: 210 DSFDLVRSQILLMNPLPTINKIFSMVMQHERQFKIS-----IPVEDSTILVNSAGKSQGR 264
F +RS I + PLP + +++S V++ E+ S + E V S+ + R
Sbjct: 194 SRFSSIRSSITDIEPLPDLYQVYSRVVREEQNLNASRTKDVVKTEAIGFSVQSSTTPRFR 253
Query: 265 GRGNGNSNSNGKRSCSFCGRDGHTIDICYRKHGFPPNYGNKNFAKVNNSSIEQNEEREDL 324
+ C+ C R GH + C+ HG+P + +N N +
Sbjct: 254 DKST--------LFCTHCNRKGHEVTQCFLVHGYPDWWLEQN--PQENQPSTRGRGSNGR 303
Query: 325 DDSKSCKGNSNTEPSF-----------------GITKEQYEQLVTLLQSQQASSSKVNLS 367
S GN ++ P+ G +Q QL++LLQ+Q+ SSS LS
Sbjct: 304 GSSSGRGGNRSSAPTTRGRGRANNAQAAAPTVSGDGNDQIAQLISLLQAQRPSSSSERLS 363
Query: 368 HVTNHVTSGITRTSYTMNHSSFGTWIVDSGASDHICSSIKFFDSYAPIIPVHIKLPNGNM 427
T +T G+ +D+GAS H+ I P + P+G
Sbjct: 364 GNTC-LTDGV----------------IDTGASHHMTGDCSILVDVFDITPSPVTKPDGKA 406
Query: 428 AIAKFSGTVNFSPGLIARNVLYVAEFKLNLLSVPKLCMDNNCIVTFDNDKCFIQDKSNLK 487
+ A GT+ +VL+V +F L+SV KL + I F + CF+QD+
Sbjct: 407 SQATKCGTLLLHDSYKLHDVLFVPDFDCTLISVSKLLKQTSSIAIFTDTFCFLQDRFLRT 466
Query: 488 MTGLGELIEGLYFLNLGSQVFSQFNHQPVIALSQSSSSTTFLPQEALWHFRLGHLSNDRL 547
+ G GE EG+Y+ V+A +S+ F LWH RLGH S L
Sbjct: 467 LIGAGEEREGVYYFT------------GVLAPRVHKASSDFAISGDLWHRRLGHPSTSVL 514
Query: 548 L------RMQQHFPCIKVDSNSVCDICHYSRHKKLPFKLSMNKASHCYELIHFDIWGPIS 601
L R Q F K+DS CD C S+ + F +S NK C+ LIH D+WGP
Sbjct: 515 LSLPECNRSSQGFD--KIDS---CDTCFRSKQTREVFPISNNKTMECFSLIHGDVWGPYR 569
Query: 602 THSIHGHKYFITALDDYSRFTWIMLCKTKSEVSKSVQNFILNIENQFNCKVKTVRTDNGP 661
T S G YF+T +DDYSR W L +K+EVS+ ++NF E QF +VK RTDNG
Sbjct: 570 TPSTTGAVYFLTLVDDYSRSVWTYLMSSKTEVSQLIKNFCAMSERQFGKQVKAFRTDNGT 629
Query: 662 EFL-MPDFYSSKGIEHQTSCVETPQQNGRV 690
EF+ + ++ + GI HQTSCV+TPQQNGRV
Sbjct: 630 EFMCLTPYFQTHGILHQTSCVDTPQQNGRV 659
>At4g07810 putative polyprotein
Length = 1366
Score = 511 bits (1316), Expect = e-144
Identities = 393/1350 (29%), Positives = 634/1350 (46%), Gaps = 180/1350 (13%)
Query: 108 IAQSIVFLENAIDVWNELKERFSQGDFIRISELQCEIFSLKQDSRSVTEFFTALKVL--- 164
+ +V+ +A +W +L+ R Q + +I +Q ++ L Q S ++ ++T L V
Sbjct: 67 LGSGMVYFYDAHLLWLKLEGRSRQSNLSKIYSVQNQLDRLHQGSLDLSAYYTRLTVTFIR 126
Query: 165 -----WEELEAYLPTPVCACPHRCMCITGV--MNAKHQHEITRSIRFLTGLNDSFDLVRS 217
WEEL+ + P C C +C C + + +H I +RFL LN+SF R
Sbjct: 127 SQCLSWEELKNFEELPSCTCG-KCTCGSNDRWIQLYEKHNI---VRFLMRLNESFIQARR 182
Query: 218 QILLMNPLPTINKIFSMVMQHERQFKI-SIPVEDSTILVNSAGKSQGRGRGNGNSNSNGK 276
QIL+M+PLP +++ + Q ++Q S+P + + S + + + N +
Sbjct: 183 QILMMDPLPEFTNLYNFISQDDQQRSFNSMPTTEKPVFQASITQQKPK---FFNQQGKSR 239
Query: 277 RSCSFCGRDGHTIDICYRKHGFPPNYGNKNFAKVNNSSIEQNEERED---LDDSKSCKGN 333
C++CG GHT CY+ HG+PP Y NN + + + GN
Sbjct: 240 PLCTYCGLLGHTNARCYKLHGYPPGYKVPVGTCYNNDKSRGQPYPHNGIHMSHLITYNGN 299
Query: 334 S-------NTEPSFGITKEQYEQLVTLLQSQQASSSKVNLSHVTNHVTSGITRTSYTMNH 386
S N + + + Y +Q + + + +S ++ +T S T+NH
Sbjct: 300 SYAPIVQANNNAPYALYNQAYNGNSYAPMAQNFAGNHI-ISDGSSMSAGNVTSESPTVNH 358
Query: 387 S----SFGTWIVDSGASDHICSSIKFF----------DSYAPIIPVHIKLPNGNMAIAKF 432
S + G + G+S H + Y I P + +G+++
Sbjct: 359 SVNMMNSGRGFL--GSSSHGREQVNQMVTQLNTQLQGSPYQVIRPPTVSQNHGSISAQGM 416
Query: 433 SGTVN-----FSPGLIARNVLYVAEFKLN---LLSVPKLC----MDNNCIVTFDNDKCFI 480
S + F P LI + + + + + C +D+ I +N K I
Sbjct: 417 SPIPSHYISAFEPCLIIPQNTWSLDTRASCHICCDLSLFCNVYHIDHTNITLPNNIKISI 476
Query: 481 QDKSNLKMTGLGELIEGLYFLNLGSQVFSQFNHQPVIALSQSSSSTTFLPQE-ALWHFRL 539
+K+ L Y + + S+ H P ++ Q+ SS +PQ+ + +H ++
Sbjct: 477 NIAETVKLNDRLILHLVFYVPSFHFNLISRLGH-PSMSRVQALSSNLHIPQKLSEFHCKI 535
Query: 540 GHLSNDRLLRM-------QQHFPCIKVDSNSVCDICHYSRHKKLPFKLSMN--KASHCYE 590
HLS + L ++ FP I +DS+ + + + F ++ ++ + E
Sbjct: 536 CHLSKQKCLSFVSNNKIYEEPFPLIHIDSDVTTIFPEFLKLVQTQFGCTVKSIRSDNAPE 595
Query: 591 LIHFDI---WGPISTHSI-----------HGHKYFITAL------DDYSRFTWIMLCKTK 630
L D+ +G HS H++ + + W T
Sbjct: 596 LQFKDLLATFGIFHYHSCAYTPQQNYVVERNHQHLLNVARSLYFQSNIPLAYWPECVSTA 655
Query: 631 SEVSKSVQNFILNIENQFNCKVKTVRTDNGPEFLMPDFYSS-------KGIEHQTSCVET 683
+ + L ++ + K + N Y+S K E TSCV
Sbjct: 656 AFLINRTPTPNLEHKSPYEVLYKKLPDYNSLRVFCCLCYASTHQHERHKFTERATSCVFI 715
Query: 684 PQQNG----RVLPYSNSNQYFQWQYHSNHPIIPATEQTSS-------NSELPLENQSETN 732
++G ++L ++ + I P ++ S+ +S LP+ + + N
Sbjct: 716 GYESGFKGYKILDLESNTVSVTRNVVFHETIFPFIDKHSTQNVSFFDDSVLPISEKQKEN 775
Query: 733 KEPAMHIDNNTHIQ-----DPTQIEHEPQQLNTRK----STRVS--------------QK 769
+ N +++ +PT + P +TR ST V+ +K
Sbjct: 776 RFQIYDYFNVLNLEVCPVIEPTTV---PAHTHTRSLAPLSTTVTNDQFGNDMDNTLMPRK 832
Query: 770 PNHLSDYVCNSSSDSTKQASSGILYPIKH----YHSLNNISASHQKFALAITDASEPISY 825
Y+ + + S L+ H + S + +S ++ F AI EP ++
Sbjct: 833 ETRAPSYLSQYHCSNVLKEPSSSLHGTAHSLSSHLSYDKLSNEYRLFCFAIIAEKEPTTF 892
Query: 826 KEASQQDCWVKAMNSELSALNHNKTWIFVDTPPNIKPIGSKWVYKIKHKSDGTIERYKAR 885
KEA+ W+ AMN EL AL T + IG KWV+KIK+KSDGTIERYKAR
Sbjct: 893 KEAALLQKWLDAMNVELDALVSTSTREICSLHDGKRAIGCKWVFKIKYKSDGTIERYKAR 952
Query: 886 LVAKGYTQVEGIDFFDTFSPVAKITTVRILIALASINSWHLHQLDVNNAFLHGDLSENVY 945
LVA GYTQ EG+D+ DTFSP+AK+T+VR+++ALA+I++W + Q+DV NAFLHGD E +Y
Sbjct: 953 LVANGYTQQEGVDYIDTFSPIAKLTSVRLILALAAIHNWSISQMDVTNAFLHGDFEEEIY 1012
Query: 946 MSVPQGVHSPKPNQ------VCKLLKSLYGLKQASRKWYEKLTGFLLLQGYKQSASDHSL 999
M +PQG ++P+ + VC+L+KSLYGLKQASR+W+ K +G L+ G+ QS D +L
Sbjct: 1013 MQLPQG-YTPRKGELLPKRPVCRLVKSLYGLKQASRQWFHKFSGVLIQNGFMQSLFDPTL 1071
Query: 1000 FILHSDTCFTALLVYVDDVILAGNSMTEIDRIKAVLDVEFKIKDLGKLKYFLGIEVAHSK 1059
F+ + F ALLVYVDD++L N + + +K +L EFK+KDLG+ +YFLG+E+A SK
Sbjct: 1072 FVRVREDTFLALLVYVDDIMLVSNKDSAVIEVKQILAKEFKLKDLGQKRYFLGLEIARSK 1131
Query: 1060 LGISICQRKYCLDLLKDTGLLGAKPVTTPLDPSIKLHQDNSPPHDDILSYRRLVGKLLYL 1119
GISI QRKY L+LL++ G LG KPV TP++ ++KL Q++ D YR+L+G+L+YL
Sbjct: 1132 EGISISQRKYALELLEEFGFLGCKPVPTPMELNLKLSQEDGALLLDASHYRKLIGRLVYL 1191
Query: 1120 TTTRPDIAFVVQQLSQFLNAPTITHYDTACRVVKYLKGTPGQGLLFRRDSQLQLLGFTDA 1179
T TRPDI F V +L+Q+++AP H A R+++YLK PGQG+ + S L F DA
Sbjct: 1192 TVTRPDICFAVNKLNQYMSAPREPHLMAARRILRYLKNDPGQGVFYPASSTLTFRAFADA 1251
Query: 1180 DWAGCPDSRRSTSGYCFFLGSSLISWRAKKQHTVARSSSEAEYRALSFASCELQWLLYLL 1239
DW+ CP+S + S++ W + S+EA WL+ L
Sbjct: 1252 DWSNCPESS---------ISISIVFW--------LKLSTEA-------------WLVLSL 1281
Query: 1240 KDLRVKCNKLPVLFCDNQSAIHIAGNPVFHERTKHLEIDCHFVREKLQQNIFKLLPIKSQ 1299
D ++ D++SA+HIA N VFHE TK+ D H VREK+ K L + ++
Sbjct: 1282 PD-------TIFVYYDDESALHIAKNSVFHESTKNFLHDIHVVREKVAVGFIKTLHVDTE 1334
Query: 1300 SQLADFFTKPLPPKNFHSFLPKLNMIDLYN 1329
+ D TKPL F+ L K+ + LY+
Sbjct: 1335 HNIVDLLTKPLTALRFNYLLSKMGLHHLYS 1364
>At2g16670 putative retroelement pol polyprotein
Length = 1333
Score = 508 bits (1307), Expect = e-143
Identities = 271/569 (47%), Positives = 354/569 (61%), Gaps = 28/569 (4%)
Query: 761 RKSTRVSQKPNHLSDYVCNSS-SDSTKQASSGILYPIKHYHSLNNISASHQKFALAITDA 819
R+STR P L D+V N++ + + ILY ++ SASH +
Sbjct: 789 RQSTRNKAPPVTLKDFVVNTTVCQESPSKLNSILYQLQKRDDTRRFSASHTTYV------ 842
Query: 820 SEPISYKEASQQDCWVKAMNSELSALNHNKTWIFVDTPPNIKPIGSKWVYKIKHKSDGTI 879
+ A N TW D PP + IGS+WVYK+KH SDG++
Sbjct: 843 ---------------------AIDAQEENHTWTIEDLPPGKRAIGSQWVYKVKHNSDGSV 881
Query: 880 ERYKARLVAKGYTQVEGIDFFDTFSPVAKITTVRILIALASINSWHLHQLDVNNAFLHGD 939
ERYKARLVA G Q EG D+ +TF+PVAK+ TVR+ + +A +W +HQ+DV+NAFLHGD
Sbjct: 882 ERYKARLVALGNKQKEGEDYGETFAPVAKMATVRLFLDVAVKRNWEIHQMDVHNAFLHGD 941
Query: 940 LSENVYMSVPQGVHSPKPNQVCKLLKSLYGLKQASRKWYEKLTGFLLLQGYKQSASDHSL 999
L E VYM +P G + PN+VC+L K+LYGLKQA R W+EKLT L G++QS +D+SL
Sbjct: 942 LREEVYMKLPPGFEASHPNKVCRLRKALYGLKQAPRCWFEKLTTALKRYGFQQSLADYSL 1001
Query: 1000 FILHSDTCFTALLVYVDDVILAGNSMTEIDRIKAVLDVEFKIKDLGKLKYFLGIEVAHSK 1059
F L + +L+YVDD+I+ GNS + K L F +KDLG LKYFLGIEVA S
Sbjct: 1002 FTLVKGSVRIKILIYVDDLIITGNSQRATQQFKEYLASCFHMKDLGPLKYFLGIEVARST 1061
Query: 1060 LGISICQRKYCLDLLKDTGLLGAKPVTTPLDPSIKLHQDNSPPHDDILSYRRLVGKLLYL 1119
GI ICQRKY LD++ +TGLLG KP PL+ + KL SP D YRRLVG+L+YL
Sbjct: 1062 TGIYICQRKYALDIISETGLLGVKPANFPLEQNHKLGLSTSPLLTDPQRYRRLVGRLIYL 1121
Query: 1120 TTTRPDIAFVVQQLSQFLNAPTITHYDTACRVVKYLKGTPGQGLLFRRDSQLQLLGFTDA 1179
TR D+AF V L++F+ P H+ A RVV+YLK PGQG+ RR Q+ G+ D+
Sbjct: 1122 AVTRLDLAFSVHILARFMQEPREDHWAAALRVVRYLKADPGQGVFLRRSGDFQITGWCDS 1181
Query: 1180 DWAGCPDSRRSTSGYCFFLGSSLISWRAKKQHTVARSSSEAEYRALSFASCELQWLLYLL 1239
DWAG P SRRS +GY G S ISW+ KKQ TV++SS+EAEYRA+SF + EL WL LL
Sbjct: 1182 DWAGDPMSRRSVTGYFVQFGDSPISWKTKKQDTVSKSSAEAEYRAMSFLASELLWLKQLL 1241
Query: 1240 KDLRVKCNKLPVLFCDNQSAIHIAGNPVFHERTKHLEIDCHFVREKLQQNIFKLLPIKSQ 1299
L V + ++ CD++SAI+IA NPVFHERTKH+EID HFVR++ + + + +
Sbjct: 1242 FSLGVSHVQPMIMCCDSKSAIYIATNPVFHERTKHIEIDYHFVRDEFVKGVITPRHVGTT 1301
Query: 1300 SQLADFFTKPLPPKNFHSFLPKLNMIDLY 1328
SQLAD FTKPL F +F KL + +LY
Sbjct: 1302 SQLADIFTKPLGRDCFSAFRIKLGIRNLY 1330
Score = 212 bits (540), Expect = 1e-54
Identities = 152/534 (28%), Positives = 251/534 (46%), Gaps = 65/534 (12%)
Query: 170 AYLPTPVCACPHRCMCITGVMNAKHQHEITRSIRFLTGLNDS-FDLVRSQILLMNPLPTI 228
A+ P +C C C+C G + K + E + FL+GL+D+ F VRS ++ P+ +
Sbjct: 68 AFKPVRICKCGG-CVCDLGALQEKDREE-DKVHEFLSGLDDALFRTVRSSLVSRIPVQPL 125
Query: 229 NKIFSMVMQHERQFKISIPVEDSTILVNSAGKSQG----RGRGNGNSNSNGKRSCSFCGR 284
+++++V Q E + V D VN+ +GRG+ S C C R
Sbjct: 126 EEVYNIVRQEEDLLRNGANVLDDQREVNAFAAQMRPKLYQGRGDEKDKS---MVCKHCNR 182
Query: 285 DGHTIDICYRKHGFPPNYGNKNFAKVNNSSIEQNEEREDLDDSKSCKGNSNTEPSFGITK 344
GH + CY G+P +G++ ++ S++ +G +N+ G
Sbjct: 183 SGHASESCYAVIGYPEWWGDRPRSR----SLQTRG-----------RGGTNSSGGRGRGA 227
Query: 345 EQYEQLVTLLQSQQASSSKVNLSHVTNHVTSGITRTSYTMNHSSFGTWIVDSGASDHICS 404
Y VT+ + L+ +G+T + + S I++SG D
Sbjct: 228 AAYANRVTVPNHDTYEQANYALTDEDRDGVNGLTDSQWRTIKS-----ILNSG-KDAATE 281
Query: 405 SIKFFDSYAPIIPVHIKLPNGNMAIAKFSGTVNFSPGLIARNVLYVAEFKLNLLSVPKLC 464
+ D P+ I L +G I+ GTV L+ +V YV EF+ +L+S+ +L
Sbjct: 282 KLTDMD------PILIVLADGRERISVKEGTVRLGSNLVMISVFYVEEFQSDLISIGQLM 335
Query: 465 MDNNCIVTFDNDKCFIQDKSNLKMTGLGELIEGLYFLNLGSQVFSQFNHQPVIALSQSSS 524
+N C++ + +QD+++ + G G + G + ++ ++
Sbjct: 336 DENRCVLQMSDRFLVVQDRTSRMVMGAGRRVGGTFHFRS----------------TEIAA 379
Query: 525 STTFLPQE--ALWHFRLGHLSNDRLLRMQQHFP--CIKVDS---NSVCDICHYSRHKKLP 577
S T ++ LWH R+GH + R+ P + V S N CD+CH ++ +
Sbjct: 380 SVTVKEEKNYELWHSRMGHPA----ARVVSLIPESSVSVSSTHLNKACDVCHRAKQTRNS 435
Query: 578 FKLSMNKASHCYELIHFDIWGPISTHSIHGHKYFITALDDYSRFTWIMLCKTKSEVSKSV 637
F LS+NK +ELI+ D+WGP T S G +YF+T +DDYSR W+ L KSE +
Sbjct: 436 FPLSINKTLRIFELIYCDLWGPYRTPSHTGARYFLTIIDDYSRGVWLYLLNDKSEAPCHL 495
Query: 638 QNFILNIENQFNCKVKTVRTDNGPEFL-MPDFYSSKGIEHQTSCVETPQQNGRV 690
+NF + QFN K+KTVR+DNG EFL + F+ +G+ H+ SCV TP++N RV
Sbjct: 496 KNFFAMTDRQFNVKIKTVRSDNGTEFLCLTKFFQEQGVIHERSCVATPERNDRV 549
Score = 33.1 bits (74), Expect = 1.1
Identities = 18/58 (31%), Positives = 30/58 (51%), Gaps = 3/58 (5%)
Query: 9 NGGVNGGAAPVQDPSQQPGNV---YYVHASDGPSSVAITPVLSHSNYHSWARSMRRAL 63
N N P+ +P+ + Y + +++ P +V P+L+ SNY WA SM+ AL
Sbjct: 7 NVNTNTNTNPIPNPTVEVRRTILPYDLTSANNPCAVISHPLLTGSNYDEWACSMKTAL 64
>At2g24660 putative retroelement pol polyprotein
Length = 1156
Score = 497 bits (1280), Expect = e-140
Identities = 284/667 (42%), Positives = 386/667 (57%), Gaps = 41/667 (6%)
Query: 677 QTSCVETPQQNGRVLPYSNSNQYFQWQYHSNHPIIPATEQTSSNSELPLENQSETNKEPA 736
QT V P + V P S ++ + S+ P S ++ +++ + P
Sbjct: 484 QTDSVFEPDSDSDVAPASIASDLQNSRVDSSDSSTPDKNLASGDTLAQIDDSPDIVSTPN 543
Query: 737 MHIDNNTHIQDPTQIEHEPQQLNTRKSTRVSQKPNHLSDYVCNSSS----------DSTK 786
+ + P P+Q RK R ++ L DYV +++ DS+
Sbjct: 544 RNNQPLFVVDSPFVEATSPRQ---RK--RQIRQSVRLQDYVLYNATVSPINPHALPDSSS 598
Query: 787 QASSGI-----LYPIKHYHSLNNISASHQKFALAITDASEPISYKEASQQDCWVKAMNSE 841
Q+SS + LYP+ Y S + SA H+ F AIT EP +KEA + W AM E
Sbjct: 599 QSSSMVQGTSSLYPLSDYVSDDCFSAGHKAFLAAITANDEPKHFKEAVRIKVWNDAMFKE 658
Query: 842 LSALNHNKTWIFVDTPPNIKPIGSKWVYKIKHKSDGTIERYKARLVAKGYTQVEGIDFFD 901
+ AL NKTW VD PP IGS+WVYK K+ +DG+IERYKARLV +G QVEG D+ +
Sbjct: 659 VDALEINKTWDIVDLPPGKVAIGSQWVYKTKYNADGSIERYKARLVVQGNKQVEGEDYNE 718
Query: 902 TFSPVAKITTVRILIALASINSWHLHQLDVNNAFLHGDLSENVYMSVPQGVHSPKPNQVC 961
TF+PV K+TTVR L+ L + N W ++Q+DVNNAFLHGDL E VYM +P G P++VC
Sbjct: 719 TFAPVVKMTTVRTLLRLVAANQWEVYQMDVNNAFLHGDLDEEVYMKLPPGFRHSHPDKVC 778
Query: 962 KLLKSLYGLKQASRKWYEKLTGFLLLQGYKQSASDHSLFILHSDTCFTALLVYVDDVILA 1021
+L KSLYGLKQA R W++KL+ LL G+ Q D+S F + +LVYVDD+++
Sbjct: 779 RLRKSLYGLKQAPRCWFKKLSDALLRFGFVQGHEDYSFFSYTRNGIELRVLVYVDDLLIC 838
Query: 1022 GNSMTEIDRIKAVLDVEFKIKDLGKLKYFLGIEVAHSKLGISICQRKYCLDLLKDTGLLG 1081
GN + + K L F +KDLGKLKYFLGIEV+ GI + QRKY LD++ D+G LG
Sbjct: 839 GNDGYMLQKFKEYLGRCFSMKDLGKLKYFLGIEVSRGSEGIFLSQRKYALDIITDSGNLG 898
Query: 1082 AKPVTTPLDPSIKLHQDNSPPHDDILSYRRLVGKLLYLTTTRPDIAFVVQQLSQFLNAPT 1141
+P TPL+ + L D+ P D YRRLVG+LLYL TRP++++ V LSQF+ P
Sbjct: 899 CRPALTPLEQNHHLATDDGPLLADAKPYRRLVGRLLYLLHTRPELSYSVHVLSQFMQTPR 958
Query: 1142 ITHYDTACRVVKYLKGTPGQGLLFRRDSQLQLLGFTDADWAGCPDSRRSTSGYCFFLGSS 1201
H A R+V++LKG+PGQG+L D++LQL + D+D+ CP +RRS S Y LG S
Sbjct: 959 EAHLAAAMRIVRFLKGSPGQGILLNADTELQLDVYCDSDFQTCPKTRRSLSAYVVLLGGS 1018
Query: 1202 LISWRAKKQHTVARSSSEAEYRALSFASCELQWLLYLLKDLRVKCNKLPVLFCDNQSAIH 1261
ISW+ KKQ TV+ SS+EAEYRA+S A E++WL LLK+L
Sbjct: 1019 PISWKTKKQDTVSHSSAEAEYRAMSVALREIKWLRKLLKEL------------------- 1059
Query: 1262 IAGNPVFHERTKHLEIDCHFVREKLQQNIFKLLPIKSQSQLADFFTKPLPPKNFHSFLPK 1321
NPVFHERTKH+E DCH VR+ ++ I +++ QLAD FTK L F + K
Sbjct: 1060 --ANPVFHERTKHIESDCHSVRDAVRDGIITTHHVRTNEQLADIFTKALGRSQFLYLMSK 1117
Query: 1322 LNMIDLY 1328
L + DL+
Sbjct: 1118 LGIQDLH 1124
Score = 142 bits (357), Expect = 2e-33
Identities = 86/225 (38%), Positives = 120/225 (53%), Gaps = 23/225 (10%)
Query: 471 VTFDNDKCFIQDKSNLKMTGLGELIEGLYFLNLGSQVFSQFNHQPVIALSQSSSSTTFLP 530
+T +N K D+ + + G GE +G+Y+L + V + H ++ Q
Sbjct: 41 LTHENLKACKLDRFSRTLIGSGEERDGVYYL---TDVATAKIHTAKVSSDQ--------- 88
Query: 531 QEALWHFRLGHLSNDRLLRMQQHFPCIKVDSNSV----CDICHYSRHKKLPFKLSMNKAS 586
ALWH RLGH S L + P + S SV CD+C ++ + F +S NK+
Sbjct: 89 --ALWHQRLGHPSFSVLSSL----PVLTSSSLSVGSRSCDVCFRAKQTREVFPVSTNKSI 142
Query: 587 HCYELIHFDIWGPISTHSIHGHKYFITALDDYSRFTWIMLCKTKSEVSKSVQNFILNIEN 646
C+ LIH D+WGP S G YF+T +DD+SR W L KSEV + NF++ E
Sbjct: 143 ECFSLIHCDVWGPYRVPSSCGAVYFLTIVDDFSRAVWTYLLLAKSEVRTVLTNFLVYTEK 202
Query: 647 QFNCKVKTVRTDNGPEFL-MPDFYSSKGIEHQTSCVETPQQNGRV 690
QF VK +R+DNG EF+ + ++ GI HQTSCV TPQQNGRV
Sbjct: 203 QFGKSVKVLRSDNGTEFMCLASYFREHGIVHQTSCVGTPQQNGRV 247
>At4g14460 retrovirus-related like polyprotein
Length = 1489
Score = 492 bits (1267), Expect = e-139
Identities = 263/637 (41%), Positives = 380/637 (59%), Gaps = 93/637 (14%)
Query: 717 TSSNSELPLENQSETNKEPAMHIDNNTHIQDPTQIEHEPQQLNTRKSTRVSQKPNHLSDY 776
+SS+S LP +N E ID+N + +S R ++ P++LS+Y
Sbjct: 913 SSSSSALPSIIPPSSNTE-TQDIDSNA--------------VPITRSKRTTRAPSYLSEY 957
Query: 777 VCNS-SSDSTKQASSGIL-------------------YPIKHYHSLNNISASHQKFALAI 816
C+ S ST + + YPI S + + Q + A
Sbjct: 958 HCSLVPSISTLPPTDSSIPIHPLPEIFTASSPKKTTPYPISTVVSYDKYTPLCQSYIFAY 1017
Query: 817 TDASEPISYKEASQQDCWVKAMNSELSALNHNKTWIFVDTPPNIKPIGSKWVYKIKHKSD 876
+EP ++ +A + + W++ EL A+ NKTW PP+ +G KWV+ IK+ D
Sbjct: 1018 NTETEPKTFSQAMKSEKWIRVAVEELQAMELNKTWSVESLPPDKNVVGCKWVFTIKYNPD 1077
Query: 877 GTIERYKARLVAKGYTQVEGIDFFDTFSPVAKITTVRILIALASINSWHLHQLDVNNAFL 936
GT+ERYKARLVA+G+TQ EGIDF DTFSPVAK+T+ ++++ LA+I W L Q+DV++AFL
Sbjct: 1078 GTVERYKARLVAQGFTQQEGIDFLDTFSPVAKLTSAKMMLGLAAITGWTLTQMDVSDAFL 1137
Query: 937 HGDLSENVYMSVPQGVHSPK-----PNQVCKLLKSLYGLKQASRKWYEKLTGFLLLQGYK 991
HGDL E ++MS+PQG P PN VC+LLKS+YGLKQASR+WY++
Sbjct: 1138 HGDLDEEIFMSLPQGYTPPAGTILPPNPVCRLLKSIYGLKQASRQWYKR----------- 1186
Query: 992 QSASDHSLFILHSDTCFTALLVYVDDVILAGNSMTEIDRIKAVLDVEFKIKDLGKLKYFL 1051
F A LVY+DD+++A N+ E++ +KA+L EFKIKDLG ++FL
Sbjct: 1187 ----------------FVAALVYIDDIMIASNNDAEVENLKALLRSEFKIKDLGPARFFL 1230
Query: 1052 GIEVAHSKLGISICQRKYCLDLLKDTGLLGAKPVTTPLDPSIKLHQDNSPPHDDILSYRR 1111
G LLG KP + P+DP++ L +D P + +YR+
Sbjct: 1231 G--------------------------LLGCKPSSIPMDPTLHLVRDMGTPLPNPTAYRK 1264
Query: 1112 LVGKLLYLTTTRPDIAFVVQQLSQFLNAPTITHYDTACRVVKYLKGTPGQGLLFRRDSQL 1171
L+G+LLYLT TRPDI + V QLSQF++AP+ H A +V++Y+K PGQGL++ D ++
Sbjct: 1265 LIGRLLYLTITRPDITYAVHQLSQFISAPSDIHLQAAHKVLRYIKANPGQGLMYSADYEI 1324
Query: 1172 QLLGFTDADWAGCPDSRRSTSGYCFFLGSSLISWRAKKQHTVARSSSEAEYRALSFASCE 1231
L GF+DADWA C D+RRS SG+C +LG+SLISW++KKQ +RSS+E+EYR+++ A+CE
Sbjct: 1325 CLNGFSDADWAACKDTRRSISGFCIYLGTSLISWKSKKQAVASRSSTESEYRSMAQATCE 1384
Query: 1232 LQWLLYLLKDLRVKCNKLPVLFCDNQSAIHIAGNPVFHERTKHLEIDCHFVREKLQQNIF 1291
+ WL LLKDL + LFCDN+SA+H + NPVFHERTKH+EIDCH VR++++
Sbjct: 1385 IIWLQQLLKDLHIPLTCPAKLFCDNKSALHSSLNPVFHERTKHIEIDCHTVRDQIKAGNL 1444
Query: 1292 KLLPIKSQSQLADFFTKPLPPKNFHSFLPKLNMIDLY 1328
K L + +++Q AD TK L P FH L ++++ L+
Sbjct: 1445 KALHVPTENQHADILTKALHPGPFHHLLRQMSLSSLF 1481
Score = 204 bits (519), Expect = 3e-52
Identities = 110/307 (35%), Positives = 172/307 (55%), Gaps = 8/307 (2%)
Query: 5 NTVVNGGVNGGAAPVQDPSQQPGNVYYVHASDGPSSVAITPVLSH-SNYHSWARSMRRAL 63
N V +N G ++ P Q N YY+H++D + ++ L+ S++HSW RS+ AL
Sbjct: 10 NPVTRPPINNGDQ-IRYPVDQYENPYYLHSADHAGLILVSDRLTTASDFHSWRRSILMAL 68
Query: 64 GAKNKFDFVDGSIPVPTDFDPNFKAWNRCNMLVHSWIMNSVEDSIAQSIVFLENAIDVWN 123
+NK F++G+I P + +F AW+RCN +V +W+MNSV+ I QS++++ +WN
Sbjct: 69 NVRNKLGFINGTITKPPEDHRDFGAWSRCNDIVSTWLMNSVDKKIGQSLLYIATVQGIWN 128
Query: 124 ELKERFSQGDFIRISELQCEIFSLKQDSRSVTEFFTALKVLWEELEAYLPTPVCACPHRC 183
L RF Q D RI +++ ++ ++Q S ++ ++TAL LWEE Y+ PVC C RC
Sbjct: 129 NLLSRFKQDDAPRIFDIEQKLSKIEQGSMDISTYYTALLTLWEEHRNYVELPVCTC-GRC 187
Query: 184 MCITGVMNAKHQHEITRSIRFLTGLNDSFDLVRSQILLMNPLPTINKIFSMVMQHERQFK 243
C V +H + +R +FL LN+ FD R IL++ P+PTI + F+MV Q ERQ
Sbjct: 188 ECDAAV-KWEHLQQRSRVTKFLKELNEGFDQTRRHILMLKPIPTIKEAFNMVTQDERQRN 246
Query: 244 IS-IPVEDSTILVNSAGKSQGRGRGNGNSNS---NGKRSCSFCGRDGHTIDICYRKHGFP 299
+ + DS N++ ++ N+ N K C+ CG+ GHTI CY+ HG+P
Sbjct: 247 VKPLTRVDSVAFQNTSMINEDENAYVAAYNTVRPNQKPICTHCGKVGHTIQKCYKVHGYP 306
Query: 300 PNYGNKN 306
P N
Sbjct: 307 PGMKTGN 313
Score = 175 bits (444), Expect = 1e-43
Identities = 122/381 (32%), Positives = 170/381 (44%), Gaps = 79/381 (20%)
Query: 343 TKEQYEQLVTLLQSQ-------QASSSKVNLSHVTNHVTSGITRTSYTM----------- 384
T +Q EQ+++ Q+Q +SS+ L+ V+ H +T TS T+
Sbjct: 400 TPQQIEQMISQFQAQVQVPEPAASSSNPSPLATVSEHGFMALTSTSGTIIPFPSTSLKYE 459
Query: 385 -------NHSSFG--------TWIVDSGASDHICSSIKFFDSYAPIIPVHIKLPNGNMAI 429
NH+ WI+DSGAS H+CS + F +
Sbjct: 460 NNDLKFQNHTLSALQKFLPSDAWIIDSGASSHVCSDLAMFRELKSV-------------- 505
Query: 430 AKFSGTVNFSPGLIARNVLYVAEFKLNLLSVPKLCMDNNCIVTFDNDKCFIQDKSNLKMT 489
SGTV+ + LI NVL+V +FK NL+SV L +C F D C IQ+ S M
Sbjct: 506 ---SGTVHITQKLILHNVLHVPDFKFNLMSVSSLVKTISCSAHFYVDCCLIQELSQGLMI 562
Query: 490 GLGELIEGLYFLNLGSQVFSQFNHQPVIALSQSSSSTTFLPQEALWHFRLGHLSNDRLLR 549
G G L LY L ++ S P L S L LWH RLGH S+ L +
Sbjct: 563 GRGRLYHNLYILE--TENTSPSTSTPAACLFTGS----VLNDGHLWHQRLGHPSSVVLQK 616
Query: 550 MQQHFPCIKVDSNSVCDICHYSRHKKLPFKLSMNKASHCYELIHFDIWGPISTHSIHGHK 609
+ K+L + N AS+ ++L+H DIWGP S SI G +
Sbjct: 617 L-----------------------KRLAYISHNNLASNPFDLVHLDIWGPFSIESIEGFR 653
Query: 610 YFITALDDYSRFTWIMLCKTKSEVSKSVQNFILNIENQFNCKVKTVRTDNGPEFLMPDFY 669
YF+T +DD +R TW+ + + K +VS FI + QFN K+K +R+DN PE +
Sbjct: 654 YFLTVVDDCTRTTWVYMLRNKKDVSSVFPEFIKLVSTQFNAKIKAIRSDNAPELGFTEIV 713
Query: 670 SSKGIEHQTSCVETPQQNGRV 690
G+ H SC TPQQN V
Sbjct: 714 KEHGMLHHFSCAYTPQQNSVV 734
>At1g59265 polyprotein, putative
Length = 1466
Score = 470 bits (1209), Expect = e-132
Identities = 266/669 (39%), Positives = 379/669 (55%), Gaps = 32/669 (4%)
Query: 679 SCVETPQQNGRVLPYSNSNQYFQWQYHSNHPIIPATEQTSSNSELPLENQSETNKEPAMH 738
S P +N +V S+SN + S+ P P E T+ P T + H
Sbjct: 801 SSPSAPFRNSQV---SSSN--LDSSFSSSFPSSP--EPTAPRQNGPQPTTQPTQTQTQTH 853
Query: 739 IDNNTHIQDPTQIEHEPQQLNTRKSTRVSQKPNHLSDYVCNSSSDSTKQASSGILYPIK- 797
NT +PT P QL ST + S SSS ++ S +++P
Sbjct: 854 SSQNTSQNNPTN--ESPSQLAQSLSTPAQSSSSSPSPTTSASSSSTSPTPPSILIHPPPP 911
Query: 798 -------------HYHSLNN-----ISASHQKFALAITDA--SEPISYKEASQQDCWVKA 837
+ HS+ I + K++LA++ A SEP + +A + + W A
Sbjct: 912 LAQIVNNNNQAPLNTHSMGTRAKAGIIKPNPKYSLAVSLAAESEPRTAIQALKDERWRNA 971
Query: 838 MNSELSALNHNKTWIFVDTPPN-IKPIGSKWVYKIKHKSDGTIERYKARLVAKGYTQVEG 896
M SE++A N TW V PP+ + +G +W++ K+ SDG++ RYKARLVAKGY Q G
Sbjct: 972 MGSEINAQIGNHTWDLVPPPPSHVTIVGCRWIFTKKYNSDGSLNRYKARLVAKGYNQRPG 1031
Query: 897 IDFFDTFSPVAKITTVRILIALASINSWHLHQLDVNNAFLHGDLSENVYMSVPQG-VHSP 955
+D+ +TFSPV K T++RI++ +A SW + QLDVNNAFL G L+++VYMS P G +
Sbjct: 1032 LDYAETFSPVIKSTSIRIVLGVAVDRSWPIRQLDVNNAFLQGTLTDDVYMSQPPGFIDKD 1091
Query: 956 KPNQVCKLLKSLYGLKQASRKWYEKLTGFLLLQGYKQSASDHSLFILHSDTCFTALLVYV 1015
+PN VCKL K+LYGLKQA R WY +L +LL G+ S SD SLF+L +LVYV
Sbjct: 1092 RPNYVCKLRKALYGLKQAPRAWYVELRNYLLTIGFVNSVSDTSLFVLQRGKSIVYMLVYV 1151
Query: 1016 DDVILAGNSMTEIDRIKAVLDVEFKIKDLGKLKYFLGIEVAHSKLGISICQRKYCLDLLK 1075
DD+++ GN T + L F +KD +L YFLGIE G+ + QR+Y LDLL
Sbjct: 1152 DDILITGNDPTLLHNTLDNLSQRFSVKDHEELHYFLGIEAKRVPTGLHLSQRRYILDLLA 1211
Query: 1076 DTGLLGAKPVTTPLDPSIKLHQDNSPPHDDILSYRRLVGKLLYLTTTRPDIAFVVQQLSQ 1135
T ++ AKPVTTP+ PS KL + D YR +VG L YL TRPDI++ V +LSQ
Sbjct: 1212 RTNMITAKPVTTPMAPSPKLSLYSGTKLTDPTEYRGIVGSLQYLAFTRPDISYAVNRLSQ 1271
Query: 1136 FLNAPTITHYDTACRVVKYLKGTPGQGLLFRRDSQLQLLGFTDADWAGCPDSRRSTSGYC 1195
F++ PT H R+++YL GTP G+ ++ + L L ++DADWAG D ST+GY
Sbjct: 1272 FMHMPTEEHLQALKRILRYLAGTPNHGIFLKKGNTLSLHAYSDADWAGDKDDYVSTNGYI 1331
Query: 1196 FFLGSSLISWRAKKQHTVARSSSEAEYRALSFASCELQWLLYLLKDLRVKCNKLPVLFCD 1255
+LG ISW +KKQ V RSS+EAEYR+++ S E+QW+ LL +L ++ + PV++CD
Sbjct: 1332 VYLGHHPISWSSKKQKGVVRSSTEAEYRSVANTSSEMQWICSLLTELGIRLTRPPVIYCD 1391
Query: 1256 NQSAIHIAGNPVFHERTKHLEIDCHFVREKLQQNIFKLLPIKSQSQLADFFTKPLPPKNF 1315
N A ++ NPVFH R KH+ ID HF+R ++Q +++ + + QLAD TKPL F
Sbjct: 1392 NVGATYLCANPVFHSRMKHIAIDYHFIRNQVQSGALRVVHVSTHDQLADTLTKPLSRTAF 1451
Query: 1316 HSFLPKLNM 1324
+F K+ +
Sbjct: 1452 QNFASKIGV 1460
Score = 171 bits (433), Expect = 3e-42
Identities = 154/662 (23%), Positives = 290/662 (43%), Gaps = 84/662 (12%)
Query: 47 LSHSNYHSWARSMRRALGAKNKFDFVDGSIPVP-----TD----FDPNFKAWNRCNMLVH 97
L+ +NY W+R + F+DGS +P TD +P++ W R + L++
Sbjct: 26 LTSTNYLMWSRQVHALFDGYELAGFLDGSTTMPPATIGTDAAPRVNPDYTRWKRQDKLIY 85
Query: 98 SWIMNSVEDSIAQSIVFLENAIDVWNELKERFSQGDFIRISELQCEIFSLKQDSRSVTEF 157
S ++ ++ S+ ++ A +W L++ ++ + +++L+ ++ + ++++ ++
Sbjct: 86 SAVLGAISMSVQPAVSRATTAAQIWETLRKIYANPSYGHVTQLRTQLKQWTKGTKTIDDY 145
Query: 158 FTALKVLWEELEAYLPTPVCACPHRCMCITGVMNAKHQHEITRSIRFLTGLNDSFDLVRS 217
L +++L A L P+ H ++ R L L + + V
Sbjct: 146 MQGLVTRFDQL-ALLGKPM----------------DHDEQVER---VLENLPEEYKPVID 185
Query: 218 QILLMNPLPTINKIFSMVMQHERQFKISIPVEDSTILVNSAGKSQGRGRGNGNSNSNGKR 277
QI + PT+ +I ++ HE + + V +T++ +A R N+N+NG R
Sbjct: 186 QIAAKDTPPTLTEIHERLLNHESKI---LAVSSATVIPITANAVSHRNTTTTNNNNNGNR 242
Query: 278 SCSFCGRDGHTIDICYRKHGFPPNYGNKNFAKVNNSSIEQNEEREDLDDSKSC--KGNSN 335
+ + R+ + + P + NF NN S + L + C +G+S
Sbjct: 243 NNRYDNRNNNN-------NSKPWQQSSTNFHPNNNQS------KPYLGKCQICGVQGHSA 289
Query: 336 TEPSFGITKEQYEQLVTLLQSQQASSSKVNLSHVTNHVTSGITRTSYTMNHSSFGTWIVD 395
S Q + ++ + SQQ S N + Y+ N+ W++D
Sbjct: 290 KRCS------QLQHFLSSVNSQQPPSPFTPWQPRANLALG----SPYSSNN-----WLLD 334
Query: 396 SGASDHICSSIKFFDSYAPIIPVH-IKLPNGNMAIAKFSGTVNFSPG---LIARNVLYVA 451
SGA+ HI S + P + + +G+ +G+ + S L N+LYV
Sbjct: 335 SGATHHITSDFNNLSLHQPYTGGDDVMVADGSTIPISHTGSTSLSTKSRPLNLHNILYVP 394
Query: 452 EFKLNLLSVPKLCMDNNCIVTFDNDKCFIQDKSNLKMTGLGELIEGLYFLNLGSQVFSQF 511
NL+SV +LC N V F ++D + G+ + LY + S
Sbjct: 395 NIHKNLISVYRLCNANGVSVEFFPASFQVKDLNTGVPLLQGKTKDELYEWPIASS----- 449
Query: 512 NHQPVIALSQSSSSTTFLPQEALWHFRLGHLSNDRLLRMQQHFPCIKVDSNSV---CDIC 568
QPV + SS T + WH RLGH + L + ++ ++ + C C
Sbjct: 450 --QPVSLFASPSSKAT----HSSWHARLGHPAPSILNSVISNYSLSVLNPSHKFLSCSDC 503
Query: 569 HYSRHKKLPFKLSMNKASHCYELIHFDIWG-PISTHSIHGHKYFITALDDYSRFTWIMLC 627
++ K+PF S ++ E I+ D+W PI +H ++Y++ +D ++R+TW+
Sbjct: 504 LINKSNKVPFSQSTINSTRPLEYIYSDVWSSPILSHD--NYRYYVIFVDHFTRYTWLYPL 561
Query: 628 KTKSEVSKSVQNFILNIENQFNCKVKTVRTDNGPEFL-MPDFYSSKGIEHQTSCVETPQQ 686
K KS+V ++ F +EN+F ++ T +DNG EF+ + +++S GI H TS TP+
Sbjct: 562 KQKSQVKETFITFKNLLENRFQTRIGTFYSDNGGEFVALWEYFSQHGISHLTSPPHTPEH 621
Query: 687 NG 688
NG
Sbjct: 622 NG 623
>At1g58889 polyprotein, putative
Length = 1466
Score = 468 bits (1205), Expect = e-132
Identities = 265/669 (39%), Positives = 378/669 (55%), Gaps = 32/669 (4%)
Query: 679 SCVETPQQNGRVLPYSNSNQYFQWQYHSNHPIIPATEQTSSNSELPLENQSETNKEPAMH 738
S P +N +V S+SN + S+ P P E T+ P T + H
Sbjct: 801 SSPSAPFRNSQV---SSSN--LDSSFSSSFPSSP--EPTAPRQNGPQPTTQPTQTQTQTH 853
Query: 739 IDNNTHIQDPTQIEHEPQQLNTRKSTRVSQKPNHLSDYVCNSSSDSTKQASSGILYPIK- 797
NT +PT P QL ST + S SSS ++ S +++P
Sbjct: 854 SSQNTSQNNPTN--ESPSQLAQSLSTPAQSSSSSPSPTTSASSSSTSPTPPSILIHPPPP 911
Query: 798 -------------HYHSLNN-----ISASHQKFALAITDA--SEPISYKEASQQDCWVKA 837
+ HS+ I + K++LA++ A SEP + +A + + W A
Sbjct: 912 LAQIVNNNNQAPLNTHSMGTRAKAGIIKPNPKYSLAVSLAAESEPRTAIQALKDERWRNA 971
Query: 838 MNSELSALNHNKTWIFVDTPPN-IKPIGSKWVYKIKHKSDGTIERYKARLVAKGYTQVEG 896
M SE++A N TW V PP+ + +G +W++ K+ SDG++ RYKAR VAKGY Q G
Sbjct: 972 MGSEINAQIGNHTWDLVPPPPSHVTIVGCRWIFTKKYNSDGSLNRYKARFVAKGYNQRPG 1031
Query: 897 IDFFDTFSPVAKITTVRILIALASINSWHLHQLDVNNAFLHGDLSENVYMSVPQG-VHSP 955
+D+ +TFSPV K T++RI++ +A SW + QLDVNNAFL G L+++VYMS P G +
Sbjct: 1032 LDYAETFSPVIKSTSIRIVLGVAVDRSWPIRQLDVNNAFLQGTLTDDVYMSQPPGFIDKD 1091
Query: 956 KPNQVCKLLKSLYGLKQASRKWYEKLTGFLLLQGYKQSASDHSLFILHSDTCFTALLVYV 1015
+PN VCKL K+LYGLKQA R WY +L +LL G+ S SD SLF+L +LVYV
Sbjct: 1092 RPNYVCKLRKALYGLKQAPRAWYVELRNYLLTIGFVNSVSDTSLFVLQRGKSIVYMLVYV 1151
Query: 1016 DDVILAGNSMTEIDRIKAVLDVEFKIKDLGKLKYFLGIEVAHSKLGISICQRKYCLDLLK 1075
DD+++ GN T + L F +KD +L YFLGIE G+ + QR+Y LDLL
Sbjct: 1152 DDILITGNDPTLLHNTLDNLSQRFSVKDHEELHYFLGIEAKRVPTGLHLSQRRYILDLLA 1211
Query: 1076 DTGLLGAKPVTTPLDPSIKLHQDNSPPHDDILSYRRLVGKLLYLTTTRPDIAFVVQQLSQ 1135
T ++ AKPVTTP+ PS KL + D YR +VG L YL TRPDI++ V +LSQ
Sbjct: 1212 RTNMITAKPVTTPMAPSPKLSLYSGTKLTDPTEYRGIVGSLQYLAFTRPDISYAVNRLSQ 1271
Query: 1136 FLNAPTITHYDTACRVVKYLKGTPGQGLLFRRDSQLQLLGFTDADWAGCPDSRRSTSGYC 1195
F++ PT H R+++YL GTP G+ ++ + L L ++DADWAG D ST+GY
Sbjct: 1272 FMHMPTEEHLQALKRILRYLAGTPNHGIFLKKGNTLSLHAYSDADWAGDKDDYVSTNGYI 1331
Query: 1196 FFLGSSLISWRAKKQHTVARSSSEAEYRALSFASCELQWLLYLLKDLRVKCNKLPVLFCD 1255
+LG ISW +KKQ V RSS+EAEYR+++ S E+QW+ LL +L ++ + PV++CD
Sbjct: 1332 VYLGHHPISWSSKKQKGVVRSSTEAEYRSVANTSSEMQWICSLLTELGIRLTRPPVIYCD 1391
Query: 1256 NQSAIHIAGNPVFHERTKHLEIDCHFVREKLQQNIFKLLPIKSQSQLADFFTKPLPPKNF 1315
N A ++ NPVFH R KH+ ID HF+R ++Q +++ + + QLAD TKPL F
Sbjct: 1392 NVGATYLCANPVFHSRMKHIAIDYHFIRNQVQSGALRVVHVSTHDQLADTLTKPLSRTAF 1451
Query: 1316 HSFLPKLNM 1324
+F K+ +
Sbjct: 1452 QNFASKIGV 1460
Score = 171 bits (433), Expect = 3e-42
Identities = 154/662 (23%), Positives = 290/662 (43%), Gaps = 84/662 (12%)
Query: 47 LSHSNYHSWARSMRRALGAKNKFDFVDGSIPVP-----TD----FDPNFKAWNRCNMLVH 97
L+ +NY W+R + F+DGS +P TD +P++ W R + L++
Sbjct: 26 LTSTNYLMWSRQVHALFDGYELAGFLDGSTTMPPATIGTDAAPRVNPDYTRWKRQDKLIY 85
Query: 98 SWIMNSVEDSIAQSIVFLENAIDVWNELKERFSQGDFIRISELQCEIFSLKQDSRSVTEF 157
S ++ ++ S+ ++ A +W L++ ++ + +++L+ ++ + ++++ ++
Sbjct: 86 SAVLGAISMSVQPAVSRATTAAQIWETLRKIYANPSYGHVTQLRTQLKQWTKGTKTIDDY 145
Query: 158 FTALKVLWEELEAYLPTPVCACPHRCMCITGVMNAKHQHEITRSIRFLTGLNDSFDLVRS 217
L +++L A L P+ H ++ R L L + + V
Sbjct: 146 MQGLVTRFDQL-ALLGKPM----------------DHDEQVER---VLENLPEEYKPVID 185
Query: 218 QILLMNPLPTINKIFSMVMQHERQFKISIPVEDSTILVNSAGKSQGRGRGNGNSNSNGKR 277
QI + PT+ +I ++ HE + + V +T++ +A R N+N+NG R
Sbjct: 186 QIAAKDTPPTLTEIHERLLNHESKI---LAVSSATVIPITANAVSHRNTTTTNNNNNGNR 242
Query: 278 SCSFCGRDGHTIDICYRKHGFPPNYGNKNFAKVNNSSIEQNEEREDLDDSKSC--KGNSN 335
+ + R+ + + P + NF NN S + L + C +G+S
Sbjct: 243 NNRYDNRNNNN-------NSKPWQQSSTNFHPNNNQS------KPYLGKCQICGVQGHSA 289
Query: 336 TEPSFGITKEQYEQLVTLLQSQQASSSKVNLSHVTNHVTSGITRTSYTMNHSSFGTWIVD 395
S Q + ++ + SQQ S N + Y+ N+ W++D
Sbjct: 290 KRCS------QLQHFLSSVNSQQPPSPFTPWQPRANLALG----SPYSSNN-----WLLD 334
Query: 396 SGASDHICSSIKFFDSYAPIIPVH-IKLPNGNMAIAKFSGTVNFSPG---LIARNVLYVA 451
SGA+ HI S + P + + +G+ +G+ + S L N+LYV
Sbjct: 335 SGATHHITSDFNNLSLHQPYTGGDDVMVADGSTIPISHTGSTSLSTKSRPLNLHNILYVP 394
Query: 452 EFKLNLLSVPKLCMDNNCIVTFDNDKCFIQDKSNLKMTGLGELIEGLYFLNLGSQVFSQF 511
NL+SV +LC N V F ++D + G+ + LY + S
Sbjct: 395 NIHKNLISVYRLCNANGVSVEFFPASFQVKDLNTGVPLLQGKTKDELYEWPIASS----- 449
Query: 512 NHQPVIALSQSSSSTTFLPQEALWHFRLGHLSNDRLLRMQQHFPCIKVDSNSV---CDIC 568
QPV + SS T + WH RLGH + L + ++ ++ + C C
Sbjct: 450 --QPVSLFASPSSKAT----HSSWHARLGHPAPSILNSVISNYSLSVLNPSHKFLSCSDC 503
Query: 569 HYSRHKKLPFKLSMNKASHCYELIHFDIWG-PISTHSIHGHKYFITALDDYSRFTWIMLC 627
++ K+PF S ++ E I+ D+W PI +H ++Y++ +D ++R+TW+
Sbjct: 504 LINKSNKVPFSQSTINSTRPLEYIYSDVWSSPILSHD--NYRYYVIFVDHFTRYTWLYPL 561
Query: 628 KTKSEVSKSVQNFILNIENQFNCKVKTVRTDNGPEFL-MPDFYSSKGIEHQTSCVETPQQ 686
K KS+V ++ F +EN+F ++ T +DNG EF+ + +++S GI H TS TP+
Sbjct: 562 KQKSQVKETFITFKNLLENRFQTRIGTFYSDNGGEFVALWEYFSQHGISHLTSPPHTPEH 621
Query: 687 NG 688
NG
Sbjct: 622 NG 623
>At2g15920 putative retroelement pol polyprotein
Length = 1100
Score = 465 bits (1197), Expect = e-131
Identities = 248/578 (42%), Positives = 357/578 (60%), Gaps = 35/578 (6%)
Query: 765 RVSQKPNHLSDYVCNSSSDSTKQASSGILYPIKHYHSLNNISASHQKFALAITDASEPIS 824
RV + HLSDYVC ++ K YPI S + IS SH + IT +
Sbjct: 550 RVRKPSTHLSDYVCKNTQSDQK-------YPISSTISYSQISPSHMCYIDNITKILISTN 602
Query: 825 YKEASQQDCWVKAMNSELSALNHNKTWIFVDTPPNIKPIGSKWVYKIKHKSDGTIERYKA 884
+ EA W +A++ E+ A+ W P + K +G KWV+ +K +DG++ERYKA
Sbjct: 603 FPEAQGTKEWCEAVDVEIGAMESTNKWEITTLPKDKKAVGCKWVFTLKILADGSLERYKA 662
Query: 885 RLVAKGYTQVEGIDFFDTFSPVAKITTVRILIALASINSWHLHQLDVNNAFLHGDLSENV 944
RLVAKGYTQ EG+D+ +TFSPVAK+TT+++L+ + + W L QLDV+N+FL+G+L E +
Sbjct: 663 RLVAKGYTQKEGLDYTNTFSPVAKMTTIKLLLKIYASKKWFLKQLDVSNSFLNGELEEEI 722
Query: 945 YMSVPQGVHSPK-----PNQVCKLLKSLYGLKQASRKWYEKLTGFLLLQGYKQSASDHSL 999
YM +P+G K N VC+L +S+YGLKQASR+W++K + LL G+ + DH+L
Sbjct: 723 YMKLPEGSAERKGIILPSNAVCRLKRSIYGLKQASRQWFKKFSASLLSLGFFKIHGDHTL 782
Query: 1000 FILH--SDTCFTALLVYVDDVILAGNSMTEIDRIKAVLDVEFKIKDLGKLKYFLGIEVAH 1057
F++ + +LVYVDD+++A + L FK++DLG LKYFL +E++
Sbjct: 783 FLIDCGGEYVVVLVLVYVDDIVIASTNED--------LHNLFKLRDLGDLKYFLCLEISR 834
Query: 1058 SKLGISICQRKYCLDLLKDTGLLGAKPVTTPLDPSIKLHQDNSPPHDDILSYRRLVGKLL 1117
+++GIS+C RK +LL G++ KPV+ P+ ++KL + + +D YRR+VGKL+
Sbjct: 835 TEVGISLCHRKNAWELLASIGMINCKPVSVPMITNVKLMKADGELLEDREQYRRIVGKLM 894
Query: 1118 YLTTTRPDIAFVVQQLSQFLNAPTITHYDTACRVVKYLKGTPGQGLLFRRDSQLQLLGFT 1177
LT TRPDI TH A RV++Y+KG GQGL + S L L GF
Sbjct: 895 DLTITRPDITL------------RTTHLQAAHRVLQYIKGIVGQGLFYSASSDLTLKGFA 942
Query: 1178 DADWAGCPDSRRSTSGYCFFLGSSLISWRAKKQHTVARSSSEAEYRALSFASCELQWLLY 1237
D+DW C DSRRST+G+ F+G SLIS R+KKQ V+RSS+EAEYRAL+ A+CEL L
Sbjct: 943 DSDWVSCADSRRSTTGFTMFVGDSLISLRSKKQRVVSRSSAEAEYRALALATCELVSLHT 1002
Query: 1238 LLKDLRVKCNKLPVLFCDNQSAIHIAGNPVFHERTKHLEIDCHFVREKLQQNIFKLLPIK 1297
LL L +P+LF ++ +AI+IA NPVFHERTKH EIDC+ VREK+ KLL ++
Sbjct: 1003 LLVSL-TAATSVPILFSNSTTAIYIAINPVFHERTKHTEIDCNTVREKVDDGELKLLHVR 1061
Query: 1298 SQSQLADFFTKPLPPKNFHSFLPKLNMIDLYNAKLEGG 1335
++ Q+AD TKPL P F K+++++++ + GG
Sbjct: 1062 TEDQVADIMTKPLSPHQFEHLKFKMSILNIFESSSSGG 1099
Score = 195 bits (495), Expect = 2e-49
Identities = 147/517 (28%), Positives = 236/517 (45%), Gaps = 78/517 (15%)
Query: 17 APVQDPSQQPGNVYYVHASDGPSSVAITPVLSHSNYHSWARSMRRALGAKNKFDFVDGSI 76
A QDP V +D P I+ L +NY W +M +L KNK F+DG++
Sbjct: 9 AQAQDPMVTVARV----TADHPGLNIISHRLDETNYGDWNVAMLISLDVKNKSGFIDGTL 64
Query: 77 PVPTDFDPNFKAWNRCNMLVHSWIMNSVEDSIAQSIVFLENAIDVWNELKERFSQGDFIR 136
P P + D NF W+RCN + I +SI+ + +A D+W +L RF+ + R
Sbjct: 65 PRPLETDKNFCLWSRCNSM------------ICRSILRMNDASDIWRDLNSRFNMTNLPR 112
Query: 137 ISELQCEIFSLKQDSRSVTEFFTALKVLWEELEA--YLPTPVCACPHRCMCITGVMNAKH 194
L EI L+Q + S++E++T LK LW++L++ L P C R
Sbjct: 113 TYNLTQEIQDLRQGTLSLSEYYTRLKTLWDQLDSTEELDDP-CTRQKR------------ 159
Query: 195 QHEITRSIRFLTGLNDSFDLVRSQILLMNPLPTINKIFSMVMQHERQFKISIPVE----- 249
+ ++FL GLN+S+ ++R QI+ LP++ +++ +V Q Q S V
Sbjct: 160 ----AKIVKFLAGLNESYAIIRRQIIAKKILPSLAEVYHIVDQDNSQQGFSNVVARPAAF 215
Query: 250 --DSTILVNSAGKSQGRGRGNGNSNSNGKRSCSFCGRDGHTIDICYRKHGFPPNYGNKNF 307
+ NS + + N + G+ CSFC R GH + CY+KHGFPP + K
Sbjct: 216 QVSEVTITNSIDPTIYYVQ---NGPNKGRSMCSFCNRVGHIAERCYKKHGFPPRFTPKGK 272
Query: 308 A---------KVNNSSIEQNEEREDLDDSKSCKGNSNTEPSFGITKEQYEQLVTL----L 354
+N ++ E + KS GN ++KEQ +Q + + L
Sbjct: 273 VGDKTQKPKPVASNVALATTESNDTHSGLKSLVGN--------LSKEQLQQFIAMFSSQL 324
Query: 355 QSQQASSSKVNLSHVTNHVTSGITRTSYTM-------NHS-SFGTWIVDSGASDHICSSI 406
Q Q S+S V S +++ + ++Y+ H+ S TW++DSGA H+ +
Sbjct: 325 QPQPHSNSAVASSSQADNIGISFSPSTYSFIGILTVAQHTLSSKTWVIDSGAIHHVNLLL 384
Query: 407 KFFDSYAPIIPVHIKLPNGNMAIAKFSGTVNFSPGLIARNVLYVAEFKLNLLSVPKLCMD 466
S + + LP G GT+ + + +NVL++ EF LNL+S+ L D
Sbjct: 385 TLNTS----VLSSVNLPAGPTVKISGVGTLRLNDDNLLKNVLFIPEFCLNLMSISSLTDD 440
Query: 467 NNCIVTFDNDKCFIQDKSNLKMTGLGELIEGLYFLNL 503
V F+ C IQD +M G G + LY +++
Sbjct: 441 IGSRVIFNQHACEIQDLIKGRMLGHGRRVANLYVMDV 477
>At2g06840 putative retroelement pol polyprotein
Length = 1102
Score = 462 bits (1190), Expect = e-130
Identities = 271/656 (41%), Positives = 369/656 (55%), Gaps = 53/656 (8%)
Query: 673 GIEHQTSCVETPQQNGRVLPYSNSNQYFQWQYHSNHPIIPATEQTSSNSELPLENQSETN 732
G H+T+ TP + + P S S+ +N P P T Q +E P + S +
Sbjct: 485 GYPHETA---TPNTHDSIDPTSTSSD------ENNTPPEPVTPQ----AEQP-HSPSSIS 530
Query: 733 KEPAMHIDNNTHIQDPTQIEHE------PQQLNTRKSTRVSQKPNHLSDYVCNSSSDSTK 786
+H + H + + +H+ P+ L K R P +L DYV + S
Sbjct: 531 SPHIVHNKGSVHSRHLNE-DHDSSSPGLPELLG--KGHRPKHPPVYLKDYVAHKVHSSPH 587
Query: 787 QASSGILYPIKHYHSLNNISASHQKFALAITDASEPISYKEASQQDCWVKAMNSELSALN 846
+S G+ + +SA+H F AI D++E +K+ W AM E+ AL
Sbjct: 588 TSSPGL----SDSNVSPTVSANHIAFMAAILDSNEQNHFKDDVLIKEWCDAMQKEIEALE 643
Query: 847 HNKTWIFVDTPPNIKPIGSKWVYKIKHKSDGTIERYKARLVAKGYTQVEGIDFFDTFSPV 906
N TW D P K I SKWVYK+K SDGT+ER+KARLV G Q EGIDF +TF+PV
Sbjct: 644 ANHTWDVTDLPHGKKAISSKWVYKLKFNSDGTLERHKARLVVMGNHQKEGIDFKETFAPV 703
Query: 907 AKITTVRILIALASINSWHLHQLDVNNAFLHGDLSENVYMSVPQGVHSPKPNQVCKLLKS 966
AK+TTVR+L+A+A+ W + Q+DV+NAFLHGDL + +S
Sbjct: 704 AKMTTVRLLLAVAAAKDWDVFQMDVHNAFLHGDLEQ----------------------ES 741
Query: 967 LYGLKQASRKWYEKLTGFLLLQGYKQSASDHSLFILHSDTCFTALLVYVDDVILAGNSMT 1026
LYGLKQA R W+ KL+ L G+ QS D+SLF L+ D LVYVDD I+ GN++
Sbjct: 742 LYGLKQAPRCWFAKLSTALRKLGFTQSYEDYSLFSLNRDGTVIHFLVYVDDFIIVGNNLK 801
Query: 1027 EIDRIKAVLDVEFKIKDLGKLKYFLGIEVAHSKLGISICQRKYCLDLLKDTGLLGAKPVT 1086
ID K L F +KDLGKLKYFLG+EV+ G + Q+KY LD++ + GLLG KP
Sbjct: 802 AIDHFKEHLHKCFHMKDLGKLKYFLGLEVSRGADGFCLSQQKYALDIINEAGLLGYKPSA 861
Query: 1087 TPLDPSIKLHQDNSPPHDDILSYRRLVGKLLYLTTTRPDIAFVVQQLSQFLNAPTITHYD 1146
P++ KL +SP D+ YRRLV + +YLT TRPD+++ V LSQF+ P H+
Sbjct: 862 VPMELHHKLGSISSPVFDNPAQYRRLVDRFIYLTITRPDLSYAVHILSQFMQTPLEAHWH 921
Query: 1147 TACRVVKYLKGTPGQGLLFRRDSQLQLLGFTDADWAGCPDSRRSTSGYCFFLGSSLISWR 1206
R+V+YLKG+P QG+L R D L L + D+D+ CP +RRS S Y +LG + ISW+
Sbjct: 922 ATLRLVRYLKGSPDQGILLRSDRALSLTAYCDSDYNPCPRTRRSLSAYVLYLGDTPISWK 981
Query: 1207 AKKQHTVARSSSEAEYRALSFASCELQWLLYLLKDLRVKCNKLPVLFCDNQSAIHIAGNP 1266
KKQ TV+ SS+EAEYRA+++ E++WL L+ L V + +LFCD+Q+AIHIA NP
Sbjct: 982 TKKQDTVSSSSAEAEYRAMAYTLKEIKWLKALMTTLGVDHTQPILLFCDSQAAIHIAANP 1041
Query: 1267 VFHERTKHLEIDCHFVREKLQQNIFKLLPIKSQSQLADFFTKPLPPKNFHSFLPKL 1322
VFHERTKH+E DCH VR+ + + I + D TK LP F L L
Sbjct: 1042 VFHERTKHIEKDCHQVRDAVTDKVISTPHIST----TDLLTKALPRPTFERLLSTL 1093
Score = 125 bits (315), Expect = 1e-28
Identities = 120/495 (24%), Positives = 192/495 (38%), Gaps = 116/495 (23%)
Query: 212 FDLVRSQILLMNPLPTINKIFSMVMQHERQFKISIPVEDSTI-LVNSAGKSQGRGRGNGN 270
F +RS+I +PLP+ N+++S V++ ++ ++ E + +N + K+ +
Sbjct: 15 FAPIRSKITDEDPLPSHNRVYSRVIRGQQNLDVARSKETTNSEAINFSVKTPSAPQVAAV 74
Query: 271 SNSNGK-RSCSFCGRDGHTIDICYRKHGFPPNY----GNKNFAKVNNSSIEQNEEREDLD 325
+ RSC+ C R GH + C+ HGFP Y G + N + + E +
Sbjct: 75 YAPKPRDRSCTHCHRQGHDVTDCFLVHGFPEWYYEQKGGSRVSSDNREVVSRLENKPAKR 134
Query: 326 DSKSCKGNSNTEPSFGITK---------EQYEQLVTLLQSQQASSSKVNLSHVTNHVTSG 376
+ +S KGN + +Q QL++LLQ+Q+ S+ LS
Sbjct: 135 EGRSSKGNGRGRGRVNSARAPLSSSNGSDQITQLISLLQAQRPKSTSERLS--------- 185
Query: 377 ITRTSYTMNHSSFGTWIVDSGASDHICSSIKFFDSYAPIIPVHIKLPNGNMAIAKFSGTV 436
++ I+DSGAS H+ IIP + P+G + A T+
Sbjct: 186 --------GNTCLTDVIIDSGASHHMTGDCSILVDVFDIIPSAVTKPDGKASCATKCVTL 237
Query: 437 NFSPGLIARNVLYVAEFKLNLLSVPKLCMDNNCIVTFDNDKCFIQDKSNLKMTGLGELIE 496
S ++VL+V +F L+SV KL + + G GE+ E
Sbjct: 238 LLSSSYKLQDVLFVPDFDCTLISVSKLLKQTG--------------PFSRTLIGAGEVRE 283
Query: 497 GLYFLNLGSQVFSQFNHQPVIALSQSSSSTTFLPQEALWHFRLGHLSNDRLLRMQQHFPC 556
+Y+ V+ S +S+ ALWH RLGH S LL + +
Sbjct: 284 RVYYFT------------GVLVASAHKTSSDSTSSGALWHRRLGHPSTSFLLSLPE---- 327
Query: 557 IKVDSNSVCDICHYSRHKKLPFKLSMNKASHCYELIHFDIWGPISTHSIHGHKYFITALD 616
CH S + K C D
Sbjct: 328 -----------CHQS-------SKDLGKIDSC---------------------------D 342
Query: 617 DYSRFTWIMLCKTKSEVSKSVQNFILNIENQFNCKVKTVRTDNGPEFLM-PDFYSSKGIE 675
SR K + ++NF + QF V+++R+DNG EF+ ++ GI
Sbjct: 343 TCSR--------AKQTLPSLIRNFCAMADRQFRKPVRSIRSDNGTEFMCHTSYFQEHGIL 394
Query: 676 HQTSCVETPQQNGRV 690
H+TSCV+TPQQN RV
Sbjct: 395 HETSCVDTPQQNARV 409
>At4g02960 putative polyprotein of LTR transposon
Length = 1456
Score = 451 bits (1160), Expect = e-126
Identities = 255/653 (39%), Positives = 368/653 (56%), Gaps = 32/653 (4%)
Query: 684 PQQNGRVLPYSNSNQYFQWQYHSNHPIIPATEQTSSNSELPLENQSETNKEPAMHIDNNT 743
P NG P + + +SN PI+ + N P N N ++
Sbjct: 813 PSHNG---PQPTAQPHQTQNSNSNSPIL-----NNPNPNSPSPNSPNQNSPLPQSPISSP 864
Query: 744 HIQDPTQIEHEPQQLNTRKSTRVSQKPNHL---------SDYVCNSSSDSTKQASSGILY 794
HI P+ EP ++ ST P L + N+ S +T+ A GI
Sbjct: 865 HIPTPSTSISEPNSPSS-SSTSTPPLPPVLPAPPIIQVNAQAPVNTHSMATR-AKDGIRK 922
Query: 795 PIKHYHSLNNISASHQKFALAITDASEPISYKEASQQDCWVKAMNSELSALNHNKTWIFV 854
P + Y +++A+ SEP + +A + D W +AM SE++A N TW V
Sbjct: 923 PNQKYSYATSLAAN-----------SEPRTAIQAMKDDRWRQAMGSEINAQIGNHTWDLV 971
Query: 855 DTPP-NIKPIGSKWVYKIKHKSDGTIERYKARLVAKGYTQVEGIDFFDTFSPVAKITTVR 913
PP ++ +G +W++ K SDG++ RYKARLVAKGY Q G+D+ +TFSPV K T++R
Sbjct: 972 PPPPPSVTIVGCRWIFTKKFNSDGSLNRYKARLVAKGYNQRPGLDYAETFSPVIKSTSIR 1031
Query: 914 ILIALASINSWHLHQLDVNNAFLHGDLSENVYMSVPQG-VHSPKPNQVCKLLKSLYGLKQ 972
I++ +A SW + QLDVNNAFL G L++ VYMS P G V +P+ VC+L K++YGLKQ
Sbjct: 1032 IVLGVAVDRSWPIRQLDVNNAFLQGTLTDEVYMSQPPGFVDKDRPDYVCRLRKAIYGLKQ 1091
Query: 973 ASRKWYEKLTGFLLLQGYKQSASDHSLFILHSDTCFTALLVYVDDVILAGNSMTEIDRIK 1032
A R WY +L +LL G+ S SD SLF+L +LVYVDD+++ GN +
Sbjct: 1092 APRAWYVELRTYLLTVGFVNSISDTSLFVLQRGRSIIYMLVYVDDILITGNDTVLLKHTL 1151
Query: 1033 AVLDVEFKIKDLGKLKYFLGIEVAHSKLGISICQRKYCLDLLKDTGLLGAKPVTTPLDPS 1092
L F +K+ L YFLGIE G+ + QR+Y LDLL T +L AKPV TP+ S
Sbjct: 1152 DALSQRFSVKEHEDLHYFLGIEAKRVPQGLHLSQRRYTLDLLARTNMLTAKPVATPMATS 1211
Query: 1093 IKLHQDNSPPHDDILSYRRLVGKLLYLTTTRPDIAFVVQQLSQFLNAPTITHYDTACRVV 1152
KL + D YR +VG L YL TRPD+++ V +LSQ+++ PT H++ RV+
Sbjct: 1212 PKLTLHSGTKLPDPTEYRGIVGSLQYLAFTRPDLSYAVNRLSQYMHMPTDDHWNALKRVL 1271
Query: 1153 KYLKGTPGQGLLFRRDSQLQLLGFTDADWAGCPDSRRSTSGYCFFLGSSLISWRAKKQHT 1212
+YL GTP G+ ++ + L L ++DADWAG D ST+GY +LG ISW +KKQ
Sbjct: 1272 RYLAGTPDHGIFLKKGNTLSLHAYSDADWAGDTDDYVSTNGYIVYLGHHPISWSSKKQKG 1331
Query: 1213 VARSSSEAEYRALSFASCELQWLLYLLKDLRVKCNKLPVLFCDNQSAIHIAGNPVFHERT 1272
V RSS+EAEYR+++ S ELQW+ LL +L ++ + PV++CDN A ++ NPVFH R
Sbjct: 1332 VVRSSTEAEYRSVANTSSELQWICSLLTELGIQLSHPPVIYCDNVGATYLCANPVFHSRM 1391
Query: 1273 KHLEIDCHFVREKLQQNIFKLLPIKSQSQLADFFTKPLPPKNFHSFLPKLNMI 1325
KH+ +D HF+R ++Q +++ + + QLAD TKPL F +F K+ +I
Sbjct: 1392 KHIALDYHFIRNQVQSGALRVVHVSTHDQLADTLTKPLSRVAFQNFSRKIGVI 1444
Score = 139 bits (351), Expect = 8e-33
Identities = 155/661 (23%), Positives = 263/661 (39%), Gaps = 103/661 (15%)
Query: 47 LSHSNYHSWARSMRRALGAKNKFDFVDGSIPVP-----TD----FDPNFKAWNRCNMLVH 97
L+ +NY W+R + F+DGS P+P TD +P++ W R + L++
Sbjct: 26 LTSTNYLMWSRQVHALFDGYELAGFLDGSTPMPPATIGTDAVPRVNPDYTRWRRQDKLIY 85
Query: 98 SWIMNSVEDSIAQSIVFLENAIDVWNELKERFSQGDFIRISELQCEIFSLKQDSRSVTEF 157
S I+ ++ S+ ++ A +W L++ ++ + +++L R +T F
Sbjct: 86 SAILGAISMSVQPAVSRATTAAQIWETLRKIYANPSYGHVTQL-----------RFITRF 134
Query: 158 FTALKVLWEELEAYLPTPVCACPHRCMCITGVMNAKHQHEITRSIRFLTGLNDSFDLVRS 217
+ A L P+ H ++ R L L D + V
Sbjct: 135 ---------DQLALLGKPM----------------DHDEQVER---VLENLPDDYKPVID 166
Query: 218 QILLMNPLPTINKIFSMVMQHERQFKISIPVEDSTILVNSAGKSQGRGRGNGNSNSNGKR 277
QI + P++ +I ++ E + + + + ++ +A R N N N N +
Sbjct: 167 QIAAKDTPPSLTEIHERLINRESKL---LALNSAEVVPITANVVTHRNT-NTNRNQNNR- 221
Query: 278 SCSFCGRDGHTIDICYRKHGFPPNYGNKNFAKVNNSSIEQNEEREDLDDSKSCKGNSNTE 337
G NY N N + N+ + R D K G
Sbjct: 222 -------------------GDNRNYNNNN-NRSNSWQPSSSGSRSDNRQPKPYLGRCQIC 261
Query: 338 PSFGITKEQYEQLVTLLQSQQASSSKVNLSHVTNHVTSGITRTSYTMNHS-SFGTWIVDS 396
G + ++ QL S N T+ T R + +N + W++DS
Sbjct: 262 SVQGHSAKRCPQL-------HQFQSTTNQQQSTSPFTPWQPRANLAVNSPYNANNWLLDS 314
Query: 397 GASDHICSSIKFFDSYAPIIPVH-IKLPNGNMAIAKFSGTVNF---SPGLIARNVLYVAE 452
GA+ HI S + P + + +G+ +G+ + S L VLYV
Sbjct: 315 GATHHITSDFNNLSFHQPYTGGDDVMIADGSTIPITHTGSASLPTSSRSLDLNKVLYVPN 374
Query: 453 FKLNLLSVPKLCMDNNCIVTFDNDKCFIQDKSNLKMTGLGELIEGLYFLNLGSQVFSQFN 512
NL+SV +LC N V F ++D + G+ + LY + S
Sbjct: 375 IHKNLISVYRLCNTNRVSVEFFPASFQVKDLNTGVPLLQGKTKDELYEWPIASS------ 428
Query: 513 HQPVIALSQSSSSTTFLPQEALWHFRLGHLSNDRLLRM--QQHFPCIKVDSNSV-CDICH 569
Q V + S T + WH RLGH S L + P + + C C
Sbjct: 429 -QAVSMFASPCSKAT----HSSWHSRLGHPSLAILNSVISNHSLPVLNPSHKLLSCSDCF 483
Query: 570 YSRHKKLPFKLSMNKASHCYELIHFDIWG-PISTHSIHGHKYFITALDDYSRFTWIMLCK 628
++ K+PF S +S E I+ D+W PI SI ++Y++ +D ++R+TW+ K
Sbjct: 484 INKSHKVPFSNSTITSSKPLEYIYSDVWSSPIL--SIDNYRYYVIFVDHFTRYTWLYPLK 541
Query: 629 TKSEVSKSVQNFILNIENQFNCKVKTVRTDNGPEF-LMPDFYSSKGIEHQTSCVETPQQN 687
KS+V + F +EN+F ++ T+ +DNG EF ++ D+ S GI H TS TP+ N
Sbjct: 542 QKSQVKDTFIIFKSLVENRFQTRIGTLYSDNGGEFVVLRDYLSQHGISHFTSPPHTPEHN 601
Query: 688 G 688
G
Sbjct: 602 G 602
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.134 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 31,309,019
Number of Sequences: 26719
Number of extensions: 1389732
Number of successful extensions: 4971
Number of sequences better than 10.0: 165
Number of HSP's better than 10.0 without gapping: 129
Number of HSP's successfully gapped in prelim test: 36
Number of HSP's that attempted gapping in prelim test: 4069
Number of HSP's gapped (non-prelim): 402
length of query: 1336
length of database: 11,318,596
effective HSP length: 111
effective length of query: 1225
effective length of database: 8,352,787
effective search space: 10232164075
effective search space used: 10232164075
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 66 (30.0 bits)
Medicago: description of AC148395.6


