

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148395.10 - phase: 0
(247 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
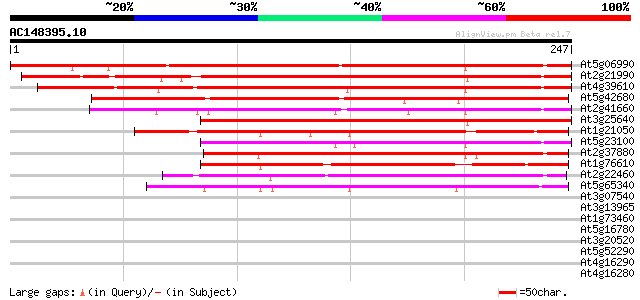
Score E
Sequences producing significant alignments: (bits) Value
At5g06990 putative protein 282 1e-76
At2g21990 unknown protein 230 5e-61
At4g39610 unknown protein 221 4e-58
At5g42680 putative protein 162 2e-40
At2g41660 unknown protein 160 8e-40
At3g25640 unknown protein (At3g25640) 146 1e-35
At1g21050 unknown protein 144 6e-35
At5g23100 putative protein 141 4e-34
At2g37880 unknown protein 132 2e-31
At1g76610 hypothetical protein 130 5e-31
At2g22460 unknown protein 126 9e-30
At5g65340 putative protein 114 6e-26
At3g07540 putative protein 30 1.2
At3g13965 unknown protein 30 1.6
At1g73460 29 2.0
At5g16780 putative protein 28 3.5
At3g20520 Glycerophosphodiesterase-like (GPDL4) 28 3.5
At5g52290 unknown protein 28 5.9
At4g16290 FCA delta protein 28 5.9
At4g16280 FCA gamma protein 28 5.9
>At5g06990 putative protein
Length = 261
Score = 282 bits (721), Expect = 1e-76
Identities = 154/264 (58%), Positives = 184/264 (69%), Gaps = 20/264 (7%)
Query: 1 MAPPHAAQTMTLPTAQRPPPAITSPK----VPMQISLQPANSKSKR--SSTNKFFGKFRS 54
MA P M PT P + S K V +I+L+ + ++K S + K F + RS
Sbjct: 1 MATPPLTPRMMTPTTMSPLGSPKSKKSTATVRPEITLEQPSGRNKTTGSKSTKLFRRVRS 60
Query: 55 MFRSFPIIVPSCKLPTMNGNHRTSETIIHGGTRITGTLFGYRKARVNLAFQEDSKCHPFL 114
+FRS PI+ P CK P + G R E +HGGTR+TGTLFGYRK RVNLA QE+ + P L
Sbjct: 61 VFRSLPIMSPMCKFP-VGGGGRLHENHVHGGTRVTGTLFGYRKTRVNLAVQENPRSLPIL 119
Query: 115 LLELAIPTGKLLQDMGMGLNRIALECEKHSSNDKTKIVDEPIWTLFCNGKKMGYGVKRDP 174
LLELAIPTGKLLQD+G+GL RIALECEK S +KTKI+DEPIW L+CNGKK GYGVKR P
Sbjct: 120 LLELAIPTGKLLQDLGVGLVRIALECEKKPS-EKTKIIDEPIWALYCNGKKSGYGVKRQP 178
Query: 175 TDDDLYVIQMLHAVSVAVGVLPSDM-----------SDPQDGELSYMRAHFERVIGSKDS 223
T++DL V+QMLHAVS+ GVLP Q+G+L+YMRAHFERVIGS+DS
Sbjct: 179 TEEDLVVMQMLHAVSMGAGVLPVSSGAITEQSGGGGGGQQEGDLTYMRAHFERVIGSRDS 238
Query: 224 ETYYMMMPDGNSNGPELSVFFVRV 247
ETYYMM PDGNS GPELS+FFVRV
Sbjct: 239 ETYYMMNPDGNS-GPELSIFFVRV 261
>At2g21990 unknown protein
Length = 252
Score = 230 bits (587), Expect = 5e-61
Identities = 122/249 (48%), Positives = 172/249 (68%), Gaps = 15/249 (6%)
Query: 6 AAQTMTLPTAQRPPPAITSPKVPMQISLQPANSKSKRSSTNKFFGKFRSMFRSFPIIVPS 65
+ + + P+ P +P+ + + L+P++ K+S K F FRS+FRSFPII P+
Sbjct: 12 STDSYSTPSPSASPSPSPAPRQHVTL-LEPSHQHKKKSK--KVFRVFRSVFRSFPIITPA 68
Query: 66 -CKLPTMNGN-----HRTSETIIHGGTRITGTLFGYRKARVNLAFQEDSKCHPFLLLELA 119
CK+P + G HR+ + G+R+TGTLFGYRK RV+L+ QE +C P L++ELA
Sbjct: 69 ACKIPVLPGGSLPDQHRSGSS----GSRVTGTLFGYRKGRVSLSIQESPRCLPSLVVELA 124
Query: 120 IPTGKLLQDMGMGLNRIALECEKHSSNDKTKIVDEPIWTLFCNGKKMGYGVKRDPTDDDL 179
+ T L +++ G+ RIALE EK +K KI+DEP+WT+F NGKK GYGVKRD T++DL
Sbjct: 125 MQTMVLQKELSGGMVRIALETEKRGDKEKIKIMDEPLWTMFSNGKKTGYGVKRDATEEDL 184
Query: 180 YVIQMLHAVSVAVGVLPSDMS-DPQDGELSYMRAHFERVIGSKDSETYYMMMPDGNSNGP 238
V+++L VS+ GVLP + + D E++YMRA+FERV+GSKDSET+YM+ P+GN NGP
Sbjct: 185 NVMELLRPVSMGAGVLPGNTEFEGPDSEMAYMRAYFERVVGSKDSETFYMLSPEGN-NGP 243
Query: 239 ELSVFFVRV 247
ELS+FFVRV
Sbjct: 244 ELSIFFVRV 252
>At4g39610 unknown protein
Length = 264
Score = 221 bits (562), Expect = 4e-58
Identities = 121/247 (48%), Positives = 166/247 (66%), Gaps = 15/247 (6%)
Query: 13 PTAQRPPPAITSPKVPMQISLQPANSKSKRSSTNKFFGKFRSMFRSFPIIVP---SCKLP 69
P++ P P T+ LQP +SK K++ TN F R++FRSFPI +CK+P
Sbjct: 21 PSSPTPSPTSTTTPPRQHPLLQPPSSKKKKNRTN-VFRVLRTVFRSFPIFTTPSVACKIP 79
Query: 70 TMNGNHRTSETIIHGGTRITGTLFGYRKARVNLAFQEDSKCHPFLLLELAIPTGKLLQDM 129
++ + H +RITGTLFGYRK RV+L+ QE+ KC P L++ELA+ T L +++
Sbjct: 80 VIHPGLGLPDPH-HNTSRITGTLFGYRKGRVSLSIQENPKCLPSLVVELAMQTTTLQKEL 138
Query: 130 GMGLNRIALECEKHSSND--------KTKIVDEPIWTLFCNGKKMGYGVKRDPTDDDLYV 181
G+ RIALE EK D KT I++EP+WT++C G+K GYGVKR+ T++DL V
Sbjct: 139 STGMVRIALETEKQPRADNNNSKTEKKTDILEEPLWTMYCKGEKTGYGVKREATEEDLNV 198
Query: 182 IQMLHAVSVAVGVLPSDM-SDPQDGELSYMRAHFERVIGSKDSETYYMMMPDGNSNGPEL 240
+++L VS+ GVLP + S+ DGE++YMRA+FERVIGSKDSET+YM+ P+GN NGPEL
Sbjct: 199 MELLRPVSMGAGVLPGNSESEGPDGEMAYMRAYFERVIGSKDSETFYMLSPEGN-NGPEL 257
Query: 241 SVFFVRV 247
S FFVRV
Sbjct: 258 SFFFVRV 264
>At5g42680 putative protein
Length = 238
Score = 162 bits (410), Expect = 2e-40
Identities = 85/213 (39%), Positives = 131/213 (60%), Gaps = 7/213 (3%)
Query: 37 NSKSKRSSTNKFFGKFRSMFRSFPIIVPSCKLPTMNGNHRTSETIIHGGTRITGTLFGYR 96
+SKS + S+ G MF+ P++ CK+ + + T TGT+FG+R
Sbjct: 28 DSKSSKKSSGSIGGGVLKMFKLIPMLSSGCKMVNLLSRGHRRPLLKDYAT--TGTIFGFR 85
Query: 97 KARVNLAFQEDSKCHPFLLLELAIPTGKLLQDMGMGLNRIALECEKHSSNDKTKIVDEPI 156
K RV LA QED C P ++EL + T L ++M RIALE E +S + K+++E +
Sbjct: 86 KGRVFLAIQEDPHCLPIFIIELPMLTSALQKEMASETVRIALESETKTS--RKKVLEEYV 143
Query: 157 WTLFCNGKKMGYGVKR-DPTDDDLYVIQMLHAVSVAVGVLP--SDMSDPQDGELSYMRAH 213
W ++CNG+K+GY ++R + +++++YVI L VS+ GVLP + +GE++YMRA
Sbjct: 144 WGIYCNGRKIGYSIRRKNMSEEEMYVIDALRGVSMGAGVLPCKNQYDQETEGEMTYMRAR 203
Query: 214 FERVIGSKDSETYYMMMPDGNSNGPELSVFFVR 246
F+RVIGSKDSE YM+ P+G+ G ELS++F+R
Sbjct: 204 FDRVIGSKDSEALYMINPEGSGQGTELSIYFLR 236
>At2g41660 unknown protein
Length = 297
Score = 160 bits (404), Expect = 8e-40
Identities = 97/238 (40%), Positives = 140/238 (58%), Gaps = 29/238 (12%)
Query: 36 ANSKSKRSSTNKFFGKFRSMFRSFPIIV--PSCKLPTMNGNHRTSETI-------IHGGT 86
++S S + KF KF + RSF I+ P+CK+ ++ +S ++ + GG+
Sbjct: 63 SSSSSASHNIGKFHLKFSLLLRSFINIINIPACKMLSLPSPPSSSSSVSNQLISLVTGGS 122
Query: 87 -----RITGTLFGYRKARVNLAFQEDSKCHPFLLLELAIPTGKLLQDMGMGLNRIALECE 141
R+TGTL+G+++ V + Q + + P LLL+LA+ T L+++M GL RIALECE
Sbjct: 123 SSLGRRVTGTLYGHKRGHVTFSVQYNQRSDPVLLLDLAMSTATLVKEMSSGLVRIALECE 182
Query: 142 K-HSSNDKTKIVDEPIWTLFCNGKKMGYGVKRDP--TDDDLYVIQMLHAVSVAVGVLPSD 198
K H S TK+ EP WT++CNG+K GY V R TD D V+ + V+V GV+P+
Sbjct: 183 KRHRSG--TKLFQEPKWTMYCNGRKCGYAVSRGGACTDTDWRVLNTVSRVTVGAGVIPTP 240
Query: 199 M---------SDPQDGELSYMRAHFERVIGSKDSETYYMMMPDGNSNGPELSVFFVRV 247
S + GEL YMR FERV+GS+DSE +YMM PD N GPELS+F +R+
Sbjct: 241 KTIDDVSGVGSGTELGELLYMRGKFERVVGSRDSEAFYMMNPDKN-GGPELSIFLLRI 297
>At3g25640 unknown protein (At3g25640)
Length = 267
Score = 146 bits (368), Expect = 1e-35
Identities = 73/173 (42%), Positives = 109/173 (62%), Gaps = 10/173 (5%)
Query: 85 GTRITGTLFGYRKARVNLAFQEDSKCHPFLLLELAIPTGKLLQDMGMGLNRIALECEKHS 144
G R+ GTLFG R+ V A Q+D P +L++L PT L+++M GL RIALE +
Sbjct: 95 GFRVVGTLFGNRRGHVYFAVQDDPTRLPAVLIQLPTPTSVLVREMASGLVRIALETAAYK 154
Query: 145 SNDKTKIVDEPIWTLFCNGKKMGYGVKRDPTDDDLYVIQMLHAVSVAVGVLPSDMS---- 200
++ K K+++E W +CNGKK GY +++ + + V++ + +++ GVLP+ +
Sbjct: 155 TDSKKKLLEESTWRTYCNGKKCGYAARKECGEAEWKVLKAVGPITMGAGVLPATTTTVDE 214
Query: 201 ------DPQDGELSYMRAHFERVIGSKDSETYYMMMPDGNSNGPELSVFFVRV 247
+ GEL YMRA FERV+GS+DSE +YMM PD +S GPELSV+F+RV
Sbjct: 215 EGNGAVGSEKGELMYMRARFERVVGSRDSEAFYMMNPDVSSGGPELSVYFLRV 267
>At1g21050 unknown protein
Length = 245
Score = 144 bits (362), Expect = 6e-35
Identities = 78/195 (40%), Positives = 125/195 (64%), Gaps = 12/195 (6%)
Query: 56 FRSFPIIVPSCKLPTMNGNHRTSETIIHGGTRITGTLFGYRKARVNLAFQEDSK-CHPFL 114
+++ + V S + + +H +S + ++GT FG+R+ RV+ Q+ + P L
Sbjct: 58 YQNDTLSVSSTSSSSSSSDHSSSS---QSSSTVSGTFFGHRRGRVSFCLQDAAVGSSPLL 114
Query: 115 LLELAIPTGKLLQDMGM-GLNRIALECEKHSSNDK--TKIVDEPIWTLFCNGKKMGYGVK 171
LLELA+PT L ++M G+ RIALEC++ S++ + I D P+W++FCNG+KMG+ V+
Sbjct: 115 LLELAVPTAALAKEMDEEGVLRIALECDRRRSSNSRSSSIFDVPVWSMFCNGRKMGFAVR 174
Query: 172 RDPTDDDLYVIQMLHAVSVAVGVLPSDMSDPQDGELSYMRAHFERVIGSKDSETYYMMMP 231
R T++D ++M+ +VSV GV+PS+ D ++ Y+RA FERV GS DSE+++MM P
Sbjct: 175 RKVTENDAVFLRMMQSVSVGAGVVPSEEED----QMLYLRARFERVTGSSDSESFHMMNP 230
Query: 232 DGNSNGPELSVFFVR 246
G S G ELS+F +R
Sbjct: 231 -GGSYGQELSIFLLR 244
>At5g23100 putative protein
Length = 277
Score = 141 bits (355), Expect = 4e-34
Identities = 76/192 (39%), Positives = 109/192 (56%), Gaps = 30/192 (15%)
Query: 85 GTRITGTLFGYRKARVNLAFQEDSKCHPFLLLELAIPTGKLLQDMGMGLNRIALECEK-- 142
G+R+ GTLFG R+ V+ + Q+D P L+ELA P L+++M GL RIALEC+K
Sbjct: 87 GSRVVGTLFGSRRGHVHFSIQKDPNSPPAFLIELATPISGLVKEMASGLVRIALECDKGK 146
Query: 143 -------------HSSNDKTK----------IVDEPIWTLFCNGKKMGYGVKRDPTDDDL 179
H DKTK +V+EP+W +CNGKK G+ +R+ + +
Sbjct: 147 EEEEGEEKNGTLRHGGGDKTKTTTTAAVSRRLVEEPMWRTYCNGKKCGFATRRECGEKEK 206
Query: 180 YVIQMLHAVSVAVGVLPSDM----SDPQDGELSYMRAHFERVIGSKDSETYYMMMPDGNS 235
V++ L VS+ GVLP G++ YMRA FER++GS+DSE +YMM PD N
Sbjct: 207 KVLKALEMVSMGAGVLPETEEIGGGGGGGGDIMYMRAKFERIVGSRDSEAFYMMNPDSN- 265
Query: 236 NGPELSVFFVRV 247
PELS++ +R+
Sbjct: 266 GAPELSIYLLRI 277
>At2g37880 unknown protein
Length = 247
Score = 132 bits (332), Expect = 2e-31
Identities = 70/167 (41%), Positives = 103/167 (60%), Gaps = 7/167 (4%)
Query: 86 TRITGTLFGYRKARVNLAFQEDS-KCHPFLLLELAIPTGKLLQDMGMGLNRIALECEKHS 144
T + GT+FG RK V Q D P LLLEL+I T +L+ +MG GL R+ALEC
Sbjct: 79 TMVIGTIFGRRKGHVWFCVQHDRLSVKPILLLELSIATSQLVHEMGSGLVRVALECPTRP 138
Query: 145 SNDKTKIVDEPIWTLFCNGKKMGYGVKRDPTDDDLYVIQMLHAVSVAVGVLPSDM----S 200
+ P+WT+FCNG+K+G+ V+R ++ +++ L +++V GVLPS S
Sbjct: 139 ELKSCLLRSVPVWTMFCNGRKLGFAVRRSANEETRMMLKRLESMTVGAGVLPSGSGLGGS 198
Query: 201 DPQD-GELSYMRAHFERVIGSKDSETYYMMMPDGNSNGPELSVFFVR 246
D D E+ YMRA++E V+GS DSE+++++ PD NS ELS+F +R
Sbjct: 199 DESDTDEVMYMRANYEHVVGSSDSESFHLINPDANS-AQELSIFLLR 244
>At1g76610 hypothetical protein
Length = 226
Score = 130 bits (328), Expect = 5e-31
Identities = 67/164 (40%), Positives = 106/164 (63%), Gaps = 13/164 (7%)
Query: 85 GTRITGTLFGYRKARVNLAFQEDSK--CHPFLLLELAIPTGKLLQDMGMGLNRIALECEK 142
G +TGT +G+R+ V+ Q+D++ P LLLELA+PT L ++M G RIAL +
Sbjct: 73 GVVVTGTFYGHRRGHVSFCLQDDTRPSSPPLLLLELAVPTAALAREMEEGFLRIAL---R 129
Query: 143 HSSNDKTKIVDEPIWTLFCNGKKMGYGVKRDPTDDDLYVIQMLHAVSVAVGVLPSDMSDP 202
SN ++ I + P+W+++CNG+K G+ V+R+ T++D+ ++++ +VSV GV+P
Sbjct: 130 SKSNRRSSIFNVPVWSMYCNGRKFGFAVRRETTENDVGFLRLMQSVSVGAGVIP------ 183
Query: 203 QDGELSYMRAHFERVIGSKDSETYYMMMPDGNSNGPELSVFFVR 246
+GE Y+RA FERV GS DSE+++ M+ G G ELS+F R
Sbjct: 184 -NGETLYLRAKFERVTGSSDSESFH-MVNQGGGYGQELSIFLSR 225
>At2g22460 unknown protein
Length = 245
Score = 126 bits (317), Expect = 9e-30
Identities = 70/179 (39%), Positives = 103/179 (57%), Gaps = 5/179 (2%)
Query: 68 LPTMNGNHRTSETIIHGGTRITGTLFGYRKARVNLAFQEDSKCHPF-LLLELAIPTGKLL 126
+ + + R+S ++ + +TGT+FGYRK ++N Q K LLLELA+PT L
Sbjct: 67 IKSRTSSFRSSSSL--SSSTVTGTIFGYRKGKINFCIQTPRKSTNLDLLLELAVPTTVLA 124
Query: 127 QDMGMGLNRIALECEKHSSNDKTKIVDEPIWTLFCNGKKMGYGVKRDPTDDDLYVIQMLH 186
++M G RI LE + D + +P W ++CNGK++GY KR P+ DD+ + L
Sbjct: 125 REMREGALRIVLE-RNNEKQDDDSFLSKPFWNMYCNGKRVGYARKRSPSQDDMTALTALS 183
Query: 187 AVSVAVGVLPSDMSDPQDGELSYMRAHFERVIGSKDSETYYMMMPDGNSNGPELSVFFV 245
V V GV+ D EL Y+RA F RV GSK+SE+++++ P GN G ELS+F V
Sbjct: 184 KVVVGAGVVTGKELGRFDDELMYLRASFRRVNGSKESESFHLIDPAGNI-GQELSIFIV 241
>At5g65340 putative protein
Length = 253
Score = 114 bits (284), Expect = 6e-26
Identities = 65/209 (31%), Positives = 116/209 (55%), Gaps = 24/209 (11%)
Query: 61 IIVPSCKLPTMNGNHRTSETIIHG--------------GTRITGTLFGYRKARVNLAFQE 106
+++PSC ++ N + + H + TGT+FG+R+ +VN Q
Sbjct: 42 LLIPSCYCTIVDPNDSQEDKLSHRQIKPRTSSASSTATNSTFTGTIFGFRRGKVNFCIQA 101
Query: 107 DSK--CHPFL-LLELAIPTGKLLQDMGMGLNRIALECEKHSSNDK-----TKIVDEPIWT 158
+ +P + LLEL +PT L ++M G+ RIALE + D + ++ P+W
Sbjct: 102 TNSKTLNPIIVLLELTVPTEVLAREMQGGVLRIALESNNNDGYDSHEDSSSSLLTTPLWN 161
Query: 159 LFCNGKKMGYGVKRDPTDDDLYVIQMLHAVSVAVGVL-PSDMSDPQDGELSYMRAHFERV 217
++CNG+K+G+ +KR+P+ +L +++L V+ GV+ +++ + + Y+RA F+RV
Sbjct: 162 MYCNGRKVGFAIKREPSKSELAALKVLTPVAEGAGVVNGEEINREKSDHMMYLRASFKRV 221
Query: 218 IGSKDSETYYMMMPDGNSNGPELSVFFVR 246
GS DSE+++++ P G G ELS+FF R
Sbjct: 222 FGSFDSESFHLVDPRG-IIGQELSIFFSR 249
>At3g07540 putative protein
Length = 841
Score = 30.0 bits (66), Expect = 1.2
Identities = 18/40 (45%), Positives = 20/40 (50%), Gaps = 2/40 (5%)
Query: 6 AAQTMTLPTAQRPPPAITSPK--VPMQISLQPANSKSKRS 43
AA +TLP QRPPPA+ P VP S S K S
Sbjct: 420 AAAAVTLPPPQRPPPAMPEPPPLVPPSQSFMVQKSGKKLS 459
>At3g13965 unknown protein
Length = 209
Score = 29.6 bits (65), Expect = 1.6
Identities = 12/35 (34%), Positives = 18/35 (51%)
Query: 13 PTAQRPPPAITSPKVPMQISLQPANSKSKRSSTNK 47
PT PPP I+ P P + P +S ++ S N+
Sbjct: 107 PTPSPPPPPISKPPPPPRAQASPPHSNNRESHRNQ 141
>At1g73460
Length = 1157
Score = 29.3 bits (64), Expect = 2.0
Identities = 24/82 (29%), Positives = 32/82 (38%), Gaps = 21/82 (25%)
Query: 153 DEPIWT--------LFCNGKKMGYGVKRDPTDDDLYVIQMLHAVSVAVGV-LPSDMSD-- 201
DEP+W L G K G V R DD+YV H ++GV + SD +D
Sbjct: 539 DEPVWQGFVAQSNELLMLGDKKGINVHRKSHRDDVYVEDDQHDSVRSIGVGINSDAADFG 598
Query: 202 ----------PQDGELSYMRAH 213
+G+ Y R H
Sbjct: 599 SEVRDSLAGGSSEGDFEYSRDH 620
>At5g16780 putative protein
Length = 820
Score = 28.5 bits (62), Expect = 3.5
Identities = 18/56 (32%), Positives = 26/56 (46%), Gaps = 5/56 (8%)
Query: 171 KRDPTDDDLYVIQMLHAVSVAVGVLPSDMSDPQDGELSYMRAHFERVIGSKDSETY 226
K+D D + ML A +VA G+ D+ +DG M+ ER+ K S Y
Sbjct: 403 KKDKLD-----LSMLEAEAVASGLGAEDLGSRKDGRRQAMKEEKERIEYEKRSNAY 453
>At3g20520 Glycerophosphodiesterase-like (GPDL4)
Length = 729
Score = 28.5 bits (62), Expect = 3.5
Identities = 17/40 (42%), Positives = 21/40 (52%), Gaps = 5/40 (12%)
Query: 7 AQTMTLPTAQRPPPA-----ITSPKVPMQISLQPANSKSK 41
A M LP A+ P PA +T P +P S PA+S SK
Sbjct: 665 ANPMLLPPAEAPYPALLDSDVTEPPLPEARSQPPASSPSK 704
>At5g52290 unknown protein
Length = 1569
Score = 27.7 bits (60), Expect = 5.9
Identities = 23/97 (23%), Positives = 41/97 (41%), Gaps = 15/97 (15%)
Query: 65 SCKLPTMNGNHRTSETIIHG---GTRITGTLFGYRKARVNLAFQEDSKCHPFLLLELAIP 121
SC+ M+ N +T E I+H I + + K+ + L E L L+I
Sbjct: 538 SCRSGGMHPNPKTEEMILHSVRPSENIQALVGEFVKSYLTLVKDESENLSEDKLKLLSIS 597
Query: 122 TGKLLQDMGMGLNRIALECEKHSSNDKTKIVDEPIWT 158
GKL ++C + ++ KT++ D+ +T
Sbjct: 598 KGKL------------IDCIRKANVHKTQLADDKTFT 622
>At4g16290 FCA delta protein
Length = 533
Score = 27.7 bits (60), Expect = 5.9
Identities = 14/38 (36%), Positives = 21/38 (54%), Gaps = 4/38 (10%)
Query: 7 AQTMTLPTAQRPPPA----ITSPKVPMQISLQPANSKS 40
A + LPT+Q PP T+P+ P+ I+L+P S
Sbjct: 446 AYSSQLPTSQLPPQQNISRATAPQTPLNINLRPTTVSS 483
>At4g16280 FCA gamma protein
Length = 747
Score = 27.7 bits (60), Expect = 5.9
Identities = 14/38 (36%), Positives = 21/38 (54%), Gaps = 4/38 (10%)
Query: 7 AQTMTLPTAQRPPPA----ITSPKVPMQISLQPANSKS 40
A + LPT+Q PP T+P+ P+ I+L+P S
Sbjct: 446 AYSSQLPTSQLPPQQNISRATAPQTPLNINLRPTTVSS 483
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.135 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,006,459
Number of Sequences: 26719
Number of extensions: 262258
Number of successful extensions: 880
Number of sequences better than 10.0: 21
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 826
Number of HSP's gapped (non-prelim): 23
length of query: 247
length of database: 11,318,596
effective HSP length: 97
effective length of query: 150
effective length of database: 8,726,853
effective search space: 1309027950
effective search space used: 1309027950
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC148395.10


