

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148360.5 + phase: 0
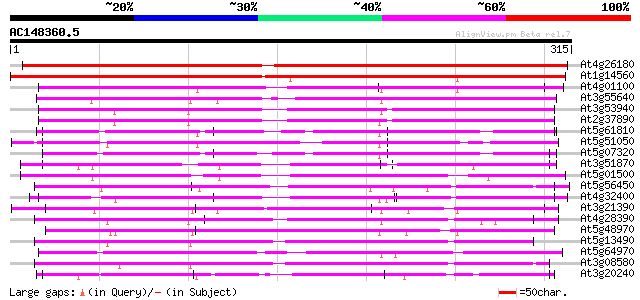
(315 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g26180 putative mitochondrial carrier protein 425 e-119
At1g14560 mitochondrial carrier like protein 359 1e-99
At4g01100 putative carrier protein 189 1e-48
At3g55640 Ca-dependent solute carrier - like protein 178 4e-45
At3g53940 unknown protein 169 2e-42
At2g37890 mitochondrial carrier like protein 168 4e-42
At5g61810 peroxisomal Ca-dependent solute carrier - like protein 164 8e-41
At5g51050 calcium-binding transporter-like protein 161 4e-40
At5g07320 peroxisomal Ca-dependent solute carrier-like protein 158 4e-39
At3g51870 carrier like protein 150 7e-37
At5g01500 unknown protein 147 7e-36
At5g56450 ADP/ATP translocase-like protein 145 4e-35
At4g32400 adenylate translocator (brittle-1) - like protein 143 1e-34
At3g21390 unknown protein 131 4e-31
At4g28390 ADP,ATP carrier-like protein 131 5e-31
At5g48970 mitochondrial carrier protein-like 130 1e-30
At5g13490 adenosine nucleotide translocator 128 4e-30
At5g64970 mitochondrial carrier protein-like 126 1e-29
At3g08580 adenylate translocator 126 2e-29
At3g20240 mitochondrial carrier protein, putative 120 1e-27
>At4g26180 putative mitochondrial carrier protein
Length = 325
Score = 425 bits (1092), Expect = e-119
Identities = 210/307 (68%), Positives = 251/307 (81%), Gaps = 7/307 (2%)
Query: 8 ILDHIPLFAKELLAGGLAGGFAKTVVAPLERLKILFQTRRTEFRSAGLSGSVRRIAKTEG 67
I+D IPLFAKEL+AGG+ GG AKT VAPLER+KILFQTRR EF+ GL GS+ +I KTEG
Sbjct: 10 IIDSIPLFAKELIAGGVTGGIAKTAVAPLERIKILFQTRRDEFKRIGLVGSINKIGKTEG 69
Query: 68 LLGFYRGNGASVARIIPYAGLHFMSYEEYRRLIMQAFPNVWKGPTLDLMAGSLSGGTAVL 127
L+GFYRGNGASVARI+PYA LH+M+YEEYRR I+ FP+ +GP LDL+AGS +GGTAVL
Sbjct: 70 LMGFYRGNGASVARIVPYAALHYMAYEEYRRWIIFGFPDTTRGPLLDLVAGSFAGGTAVL 129
Query: 128 FTYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIRGLYRGVAPT 187
FTYPLDL+RTKLAYQ V + + +YRGI DC S+TY+E G RGLYRGVAP+
Sbjct: 130 FTYPLDLVRTKLAYQ------TQVKAIPVEQIIYRGIVDCFSRTYRESGARGLYRGVAPS 183
Query: 188 LFGIFPYAGLKFYFYEEMKRRVPEDYKKSIMAKLTCGSVAGLLGQTFTYPLEVVRRQMQV 247
L+GIFPYAGLKFYFYEEMKR VP ++K+ I KL CGSVAGLLGQT TYPL+VVRRQMQV
Sbjct: 184 LYGIFPYAGLKFYFYEEMKRHVPPEHKQDISLKLVCGSVAGLLGQTLTYPLDVVRRQMQV 243
Query: 248 QNL-AASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSYL 306
+ L +A +E +GTM+++ IA+++GWK LFSGLSINY+KVVPS AIGFTVYD MK +L
Sbjct: 244 ERLYSAVKEETRRGTMQTLFKIAREEGWKQLFSGLSINYLKVVPSVAIGFTVYDIMKLHL 303
Query: 307 RVPSRDE 313
RVP R+E
Sbjct: 304 RVPPREE 310
>At1g14560 mitochondrial carrier like protein
Length = 331
Score = 359 bits (921), Expect = 1e-99
Identities = 180/319 (56%), Positives = 235/319 (73%), Gaps = 8/319 (2%)
Query: 1 MAASSESILDHIPLFAKELLAGGLAGGFAKTVVAPLERLKILFQTRRTEFRSAGLSGSVR 60
++A S++D +P+ AK L+AGG AG AKT VAPLER+KIL QTR +F++ G+S S++
Sbjct: 9 LSADVMSLVDTLPVLAKTLIAGGAAGAIAKTAVAPLERIKILLQTRTNDFKTLGVSQSLK 68
Query: 61 RIAKTEGLLGFYRGNGASVARIIPYAGLHFMSYEEYRRLIMQAFPNVWKGPTLDLMAGSL 120
++ + +G LGFY+GNGASV RIIPYA LH+M+YE YR I++ + GP +DL+AGS
Sbjct: 69 KVLQFDGPLGFYKGNGASVIRIIPYAALHYMTYEVYRDWILEKNLPLGSGPIVDLVAGSA 128
Query: 121 SGGTAVLFTYPLDLIRTKLAYQIVSPTKLNVSGMVN---NEQVYRGIRDCLSKTYKEGGI 177
+GGTAVL TYPLDL RTKLAYQ VS T+ ++ G N + Y GI++ L+ YKEGG
Sbjct: 129 AGGTAVLCTYPLDLARTKLAYQ-VSDTRQSLRGGANGFYRQPTYSGIKEVLAMAYKEGGP 187
Query: 178 RGLYRGVAPTLFGIFPYAGLKFYFYEEMKRRVPEDYKKSIMAKLTCGSVAGLLGQTFTYP 237
RGLYRG+ PTL GI PYAGLKFY YEE+KR VPE+++ S+ L CG++AGL GQT TYP
Sbjct: 188 RGLYRGIGPTLIGILPYAGLKFYIYEELKRHVPEEHQNSVRMHLPCGALAGLFGQTITYP 247
Query: 238 LEVVRRQMQVQNL----AASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAA 293
L+VVRRQMQV+NL + K T + I + QGWK LF+GLSINYIK+VPS A
Sbjct: 248 LDVVRRQMQVENLQPMTSEGNNKRYKNTFDGLNTIVRTQGWKQLFAGLSINYIKIVPSVA 307
Query: 294 IGFTVYDTMKSYLRVPSRD 312
IGFTVY++MKS++R+P R+
Sbjct: 308 IGFTVYESMKSWMRIPPRE 326
>At4g01100 putative carrier protein
Length = 352
Score = 189 bits (481), Expect = 1e-48
Identities = 114/321 (35%), Positives = 172/321 (53%), Gaps = 37/321 (11%)
Query: 17 KELLAGGLAGGFAKTVVAPLERLKILFQTRRTE-FRSAGLSGSVRRIAKTEGLLGFYRGN 75
K L AGG+AGG ++T VAPLER+KIL Q + + +G ++ I +TEGL G ++GN
Sbjct: 40 KSLFAGGVAGGVSRTAVAPLERMKILLQVQNPHNIKYSGTVQGLKHIWRTEGLRGLFKGN 99
Query: 76 GASVARIIPYAGLHFMSYEEYRRLIMQAF------PNVWKGPTLDLMAGSLSGGTAVLFT 129
G + ARI+P + + F SYE+ I+ + N P L L AG+ +G A+ T
Sbjct: 100 GTNCARIVPNSAVKFFSYEQASNGILYMYRQRTGNENAQLTPLLRLGAGATAGIIAMSAT 159
Query: 130 YPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIRGLYRGVAPTLF 189
YP+D++R +L Q N+ YRGI L+ +E G R LYRG P++
Sbjct: 160 YPMDMVRGRLTVQTA-----------NSPYQYRGIAHALATVLREEGPRALYRGWLPSVI 208
Query: 190 GIFPYAGLKFYFYEEMKR--------RVPEDYKKSIMAKLTCGSVAGLLGQTFTYPLEVV 241
G+ PY GL F YE +K + E+ + +++ +LTCG++AG +GQT YPL+V+
Sbjct: 209 GVVPYVGLNFSVYESLKDWLVKENPYGLVENNELTVVTRLTCGAIAGTVGQTIAYPLDVI 268
Query: 242 RRQMQVQNL-----------AASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVP 290
RR+MQ+ ++ E G + + + +G+ L+ GL N +KVVP
Sbjct: 269 RRRMQMVGWKDASAIVTGEGRSTASLEYTGMVDAFRKTVRHEGFGALYKGLVPNSVKVVP 328
Query: 291 SAAIGFTVYDTMKSYLRVPSR 311
S AI F Y+ +K L V R
Sbjct: 329 SIAIAFVTYEMVKDVLGVEFR 349
Score = 49.3 bits (116), Expect = 3e-06
Identities = 30/93 (32%), Positives = 49/93 (52%), Gaps = 3/93 (3%)
Query: 208 RVPEDYKKSIMAKLTCGSVAGLLGQTFTYPLEVVRRQMQVQNLAASEEAELKGTMRSMVL 267
+ P KSI L G VAG + +T PLE ++ +QVQN + GT++ +
Sbjct: 29 KAPSYAFKSICKSLFAGGVAGGVSRTAVAPLERMKILLQVQN---PHNIKYSGTVQGLKH 85
Query: 268 IAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYD 300
I + +G + LF G N ++VP++A+ F Y+
Sbjct: 86 IWRTEGLRGLFKGNGTNCARIVPNSAVKFFSYE 118
>At3g55640 Ca-dependent solute carrier - like protein
Length = 332
Score = 178 bits (451), Expect = 4e-45
Identities = 110/305 (36%), Positives = 162/305 (53%), Gaps = 26/305 (8%)
Query: 16 AKELLAGGLAGGFAKTVVAPLERLKILFQ-----TRRTEFRSAGLSGSVRRIAKTEGLLG 70
A +LLAGGLAG F+KT APL RL ILFQ T R + RI EGL
Sbjct: 35 ASQLLAGGLAGAFSKTCTAPLSRLTILFQVQGMHTNAAALRKPSILHEASRILNEEGLKA 94
Query: 71 FYRGNGASVARIIPYAGLHFMSYEEYRRLI--MQAFPNVWKGPTLDL----MAGSLSGGT 124
F++GN ++A +PY+ ++F +YE Y++ + + N +G + +L +AG L+G T
Sbjct: 95 FWKGNLVTIAHRLPYSSVNFYAYEHYKKFMYMVTGMENHKEGISSNLFVHFVAGGLAGIT 154
Query: 125 AVLFTYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIRGLYRGV 184
A TYPLDL+RT+LA Q TK+ Y GI L + GI GLY+G+
Sbjct: 155 AASATYPLDLVRTRLAAQ----TKVIY---------YSGIWHTLRSITTDEGILGLYKGL 201
Query: 185 APTLFGIFPYAGLKFYFYEEMKR--RVPEDYKKSIMAKLTCGSVAGLLGQTFTYPLEVVR 242
TL G+ P + F YE ++ R + IM L CGS++G+ T T+PL++VR
Sbjct: 202 GTTLVGVGPSIAISFSVYESLRSYWRSTRPHDSPIMVSLACGSLSGIASSTATFPLDLVR 261
Query: 243 RQMQVQNLAASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTM 302
R+ Q++ + G + ++ I Q +G + L+ G+ Y KVVP I F Y+T+
Sbjct: 262 RRKQLEGIGGRAVVYKTGLLGTLKRIVQTEGARGLYRGILPEYYKVVPGVGICFMTYETL 321
Query: 303 KSYLR 307
K Y +
Sbjct: 322 KLYFK 326
>At3g53940 unknown protein
Length = 365
Score = 169 bits (428), Expect = 2e-42
Identities = 104/305 (34%), Positives = 158/305 (51%), Gaps = 30/305 (9%)
Query: 17 KELLAGGLAGGFAKTVVAPLERLKILFQTRRTEFRSAGLSG-----SVRRIAKTEGLLGF 71
+ LLAGG+AG F+KT APL RL ILFQ + + +A LS RI K EG F
Sbjct: 71 ERLLAGGIAGAFSKTCTAPLARLTILFQIQGMQSEAAILSSPNIWHEASRIVKEEGFRAF 130
Query: 72 YRGNGASVARIIPYAGLHFMSYEEYRRL-----IMQAFP-NVWKGPTLDLMAGSLSGGTA 125
++GN +VA +PY ++F +YEEY+ ++Q++ N ++ ++G L+G TA
Sbjct: 131 WKGNLVTVAHRLPYGAVNFYAYEEYKTFLHSNPVLQSYKGNAGVDISVHFVSGGLAGLTA 190
Query: 126 VLFTYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIRGLYRGVA 185
TYPLDL+RT+L+ Q N Y+G+ +E GI GLY+G+
Sbjct: 191 ASATYPLDLVRTRLSAQ-------------RNSIYYQGVGHAFRTICREEGILGLYKGLG 237
Query: 186 PTLFGIFPYAGLKFYFYEEMKR----RVPEDYKKSIMAKLTCGSVAGLLGQTFTYPLEVV 241
TL G+ P + F YE K P D + + L CGS++G++ T T+PL++V
Sbjct: 238 ATLLGVGPSLAISFAAYETFKTFWLSHRPND--SNAVVSLGCGSLSGIVSSTATFPLDLV 295
Query: 242 RRQMQVQNLAASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDT 301
RR+MQ++ G + I + +G + L+ G+ Y KVVP I F ++
Sbjct: 296 RRRMQLEGAGGRARVYTTGLFGTFKHIFKTEGMRGLYRGIIPEYYKVVPGVGIAFMTFEE 355
Query: 302 MKSYL 306
+K L
Sbjct: 356 LKKLL 360
>At2g37890 mitochondrial carrier like protein
Length = 337
Score = 168 bits (425), Expect = 4e-42
Identities = 108/305 (35%), Positives = 152/305 (49%), Gaps = 30/305 (9%)
Query: 17 KELLAGGLAGGFAKTVVAPLERLKILFQTRRTEFRSAGLSG-----SVRRIAKTEGLLGF 71
+ LLAGG+AG +KT APL RL ILFQ + + A LS RI EG F
Sbjct: 43 QNLLAGGIAGAISKTCTAPLARLTILFQLQGMQSEGAVLSRPNLRREASRIINEEGYRAF 102
Query: 72 YRGNGASVARIIPYAGLHFMSYEEYRRL-----IMQAF-PNVWKGPTLDLMAGSLSGGTA 125
++GN +V IPY ++F +YE+Y ++Q+F N P + ++G L+G TA
Sbjct: 103 WKGNLVTVVHRIPYTAVNFYAYEKYNLFFNSNPVVQSFIGNTSGNPIVHFVSGGLAGITA 162
Query: 126 VLFTYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIRGLYRGVA 185
TYPLDL+RT+LA Q N Y+GI +E GI GLY+G+
Sbjct: 163 ATATYPLDLVRTRLAAQ-------------RNAIYYQGIEHTFRTICREEGILGLYKGLG 209
Query: 186 PTLFGIFPYAGLKFYFYEEMK----RRVPEDYKKSIMAKLTCGSVAGLLGQTFTYPLEVV 241
TL G+ P + F YE MK P D ++ L G +AG + T TYPL++V
Sbjct: 210 ATLLGVGPSLAINFAAYESMKLFWHSHRPND--SDLVVSLVSGGLAGAVSSTATYPLDLV 267
Query: 242 RRQMQVQNLAASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDT 301
RR+MQV+ G + I + +G+K ++ G+ Y KVVP I F YD
Sbjct: 268 RRRMQVEGAGGRARVYNTGLFGTFKHIFKSEGFKGIYRGILPEYYKVVPGVGIVFMTYDA 327
Query: 302 MKSYL 306
++ L
Sbjct: 328 LRRLL 332
Score = 34.7 bits (78), Expect = 0.069
Identities = 23/89 (25%), Positives = 39/89 (42%), Gaps = 3/89 (3%)
Query: 214 KKSIMAKLTCGSVAGLLGQTFTYPLEVVRRQMQVQNLAASEEAELK--GTMRSMVLIAQK 271
K L G +AG + +T T PL + Q+Q + SE A L R I +
Sbjct: 38 KLGTFQNLLAGGIAGAISKTCTAPLARLTILFQLQGM-QSEGAVLSRPNLRREASRIINE 96
Query: 272 QGWKTLFSGLSINYIKVVPSAAIGFTVYD 300
+G++ + G + + +P A+ F Y+
Sbjct: 97 EGYRAFWKGNLVTVVHRIPYTAVNFYAYE 125
>At5g61810 peroxisomal Ca-dependent solute carrier - like protein
Length = 478
Score = 164 bits (414), Expect = 8e-41
Identities = 99/296 (33%), Positives = 163/296 (54%), Gaps = 30/296 (10%)
Query: 16 AKELLAGGLAGGFAKTVVAPLERLKILFQTRRTEFRSAGLSGSVRRIAKTEGLLGFYRGN 75
+K LLAGG+AG ++T APL+RLK+ Q +RT G+ ++++I + + LLGF+RGN
Sbjct: 205 SKLLLAGGIAGAVSRTATAPLDRLKVALQVQRTNL---GVVPTIKKIWREDKLLGFFRGN 261
Query: 76 GASVARIIPYAGLHFMSYEEYRRLIMQAFPNVWKGPTLDLMAGSLSGGTAVLFTYPLDLI 135
G +VA++ P + + F +YE + +I A ++ G + L+AG L+G A YP+DL+
Sbjct: 262 GLNVAKVAPESAIKFAAYEMLKPIIGGADGDI--GTSGRLLAGGLAGAVAQTAIYPMDLV 319
Query: 136 RTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIRGLYRGVAPTLFGIFPYA 195
+T+L + V ++++ +D + + G R YRG+ P+L GI PYA
Sbjct: 320 KTRLQTFV---------SEVGTPKLWKLTKD----IWIQEGPRAFYRGLCPSLIGIIPYA 366
Query: 196 GLKFYFYEEMK-----RRVPEDYKKSIMAKLTCGSVAGLLGQTFTYPLEVVRRQMQVQNL 250
G+ YE +K + + + + +L CG +G LG + YPL+V+R +MQ +
Sbjct: 367 GIDLAAYETLKDLSRAHFLHDTAEPGPLIQLGCGMTSGALGASCVYPLQVIRTRMQADSS 426
Query: 251 AASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSYL 306
S E T+R +G K + G+ N+ KV+PSA+I + VY+ MK L
Sbjct: 427 KTSMGQEFLKTLRG-------EGLKGFYRGIFPNFFKVIPSASISYLVYEAMKKNL 475
Score = 94.0 bits (232), Expect = 1e-19
Identities = 64/196 (32%), Positives = 91/196 (45%), Gaps = 18/196 (9%)
Query: 19 LLAGGLAGGFAKTVVAPLERLKILFQTRRTEFRSAGLSGSVRRIAKTEGLLGFYRGNGAS 78
LLAGGLAG A+T + P++ +K QT +E + L + I EG FYRG S
Sbjct: 299 LLAGGLAGAVAQTAIYPMDLVKTRLQTFVSEVGTPKLWKLTKDIWIQEGPRAFYRGLCPS 358
Query: 79 VARIIPYAGLHFMSYEEYRRLIMQAF--PNVWKGPTLDLMAGSLSGGTAVLFTYPLDLIR 136
+ IIPYAG+ +YE + L F GP + L G SG YPL +IR
Sbjct: 359 LIGIIPYAGIDLAAYETLKDLSRAHFLHDTAEPGPLIQLGCGMTSGALGASCVYPLQVIR 418
Query: 137 TKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIRGLYRGVAPTLFGIFPYAG 196
T++ S T + + KT + G++G YRG+ P F + P A
Sbjct: 419 TRMQAD-SSKTSMGQEFL---------------KTLRGEGLKGFYRGIFPNFFKVIPSAS 462
Query: 197 LKFYFYEEMKRRVPED 212
+ + YE MK+ + D
Sbjct: 463 ISYLVYEAMKKNLALD 478
Score = 63.2 bits (152), Expect = 2e-10
Identities = 48/194 (24%), Positives = 87/194 (44%), Gaps = 21/194 (10%)
Query: 115 LMAGSLSGGTAVLFTYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKE 174
L+AG ++G + T PLD R K+A Q+ ++ G+ + K ++E
Sbjct: 208 LLAGGIAGAVSRTATAPLD--RLKVALQV--------------QRTNLGVVPTIKKIWRE 251
Query: 175 GGIRGLYRGVAPTLFGIFPYAGLKFYFYEEMKRRVP-EDYKKSIMAKLTCGSVAGLLGQT 233
+ G +RG + + P + +KF YE +K + D +L G +AG + QT
Sbjct: 252 DKLLGFFRGNGLNVAKVAPESAIKFAAYEMLKPIIGGADGDIGTSGRLLAGGLAGAVAQT 311
Query: 234 FTYPLEVVRRQMQVQNLAASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAA 293
YP+++V+ ++Q K T + ++G + + GL + I ++P A
Sbjct: 312 AIYPMDLVKTRLQTFVSEVGTPKLWKLTKDIWI----QEGPRAFYRGLCPSLIGIIPYAG 367
Query: 294 IGFTVYDTMKSYLR 307
I Y+T+K R
Sbjct: 368 IDLAAYETLKDLSR 381
>At5g51050 calcium-binding transporter-like protein
Length = 487
Score = 161 bits (408), Expect = 4e-40
Identities = 101/314 (32%), Positives = 167/314 (53%), Gaps = 30/314 (9%)
Query: 2 AASSESILDHIPLFAKELLAGGLAGGFAKTVVAPLERLKILFQTRRTEFRSAGLSGSVRR 61
A E I HI + +AGG+AG ++T APL+RLK+L Q ++T+ R + +++
Sbjct: 196 AVIPEGISKHIKR-SNYFIAGGIAGAASRTATAPLDRLKVLLQIQKTDAR---IREAIKL 251
Query: 62 IAKTEGLLGFYRGNGASVARIIPYAGLHFMSYEEYRRLIMQAF--PNVWKGPTLDLMAGS 119
I K G+ GF+RGNG ++ ++ P + + F +YE ++ I + G T+ L AG
Sbjct: 252 IWKQGGVRGFFRGNGLNIVKVAPESAIKFYAYELFKNAIGENMGEDKADIGTTVRLFAGG 311
Query: 120 LSGGTAVLFTYPLDLIRTKL-AYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIR 178
++G A YPLDL++T+L Y + + G + + + G R
Sbjct: 312 MAGAVAQASIYPLDLVKTRLQTYTSQAGVAVPRLGTLTKDILVHE------------GPR 359
Query: 179 GLYRGVAPTLFGIFPYAGLKFYFYEEMK----RRVPEDYKKSIMAKLTCGSVAGLLGQTF 234
Y+G+ P+L GI PYAG+ YE +K + +D + + +L CG+++G LG T
Sbjct: 360 AFYKGLFPSLLGIIPYAGIDLAAYETLKDLSRTYILQDAEPGPLVQLGCGTISGALGATC 419
Query: 235 TYPLEVVRRQMQVQNLAASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAI 294
YPL+VVR +MQ + S + G R + ++G++ L+ GL N +KVVP+A+I
Sbjct: 420 VYPLQVVRTRMQAERARTS----MSGVFRRTI---SEEGYRALYKGLLPNLLKVVPAASI 472
Query: 295 GFTVYDTMKSYLRV 308
+ VY+ MK L +
Sbjct: 473 TYMVYEAMKKSLEL 486
Score = 86.3 bits (212), Expect = 2e-17
Identities = 61/197 (30%), Positives = 95/197 (47%), Gaps = 19/197 (9%)
Query: 19 LLAGGLAGGFAKTVVAPLERLKILFQTRRTEFRSA--GLSGSVRRIAKTEGLLGFYRGNG 76
L AGG+AG A+ + PL+ +K QT ++ A L + I EG FY+G
Sbjct: 307 LFAGGMAGAVAQASIYPLDLVKTRLQTYTSQAGVAVPRLGTLTKDILVHEGPRAFYKGLF 366
Query: 77 ASVARIIPYAGLHFMSYEEYRRLIMQ-AFPNVWKGPTLDLMAGSLSGGTAVLFTYPLDLI 135
S+ IIPYAG+ +YE + L + GP + L G++SG YPL ++
Sbjct: 367 PSLLGIIPYAGIDLAAYETLKDLSRTYILQDAEPGPLVQLGCGTISGALGATCVYPLQVV 426
Query: 136 RTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIRGLYRGVAPTLFGIFPYA 195
RT++ + + ++SG V+R +T E G R LY+G+ P L + P A
Sbjct: 427 RTRMQAE---RARTSMSG------VFR-------RTISEEGYRALYKGLLPNLLKVVPAA 470
Query: 196 GLKFYFYEEMKRRVPED 212
+ + YE MK+ + D
Sbjct: 471 SITYMVYEAMKKSLELD 487
>At5g07320 peroxisomal Ca-dependent solute carrier-like protein
Length = 479
Score = 158 bits (399), Expect = 4e-39
Identities = 96/289 (33%), Positives = 162/289 (55%), Gaps = 28/289 (9%)
Query: 19 LLAGGLAGGFAKTVVAPLERLKILFQTRRTEFRSAGLSGSVRRIAKTEGLLGFYRGNGAS 78
LLAGGLAG ++T APL+RLK++ Q +R AG+ ++++I + + L+GF+RGNG +
Sbjct: 209 LLAGGLAGAVSRTATAPLDRLKVVLQVQRAH---AGVLPTIKKIWREDKLMGFFRGNGLN 265
Query: 79 VARIIPYAGLHFMSYEEYRRLIMQAFPNVWKGPTLDLMAGSLSGGTAVLFTYPLDLIRTK 138
V ++ P + + F +YE + +I ++ G + LMAG ++G A YP+DL++T+
Sbjct: 266 VMKVAPESAIKFCAYEMLKPMIGGEDGDI--GTSGRLMAGGMAGALAQTAIYPMDLVKTR 323
Query: 139 LAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIRGLYRGVAPTLFGIFPYAGLK 198
L + VS ++++ +D + G R Y+G+ P+L GI PYAG+
Sbjct: 324 L--------QTCVSEGGKAPKLWKLTKD----IWVREGPRAFYKGLFPSLLGIVPYAGID 371
Query: 199 FYFYEEMK----RRVPEDYKKSIMAKLTCGSVAGLLGQTFTYPLEVVRRQMQVQNLAASE 254
YE +K + +D + + +L+CG +G LG + YPL+VVR +MQ + +
Sbjct: 372 LAAYETLKDLSRTYILQDTEPGPLIQLSCGMTSGALGASCVYPLQVVRTRMQADSSKTTM 431
Query: 255 EAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMK 303
+ E TM+ +G + + GL N +KVVP+A+I + VY+ MK
Sbjct: 432 KQEFMNTMKG-------EGLRGFYRGLLPNLLKVVPAASITYIVYEAMK 473
Score = 87.0 bits (214), Expect = 1e-17
Identities = 59/196 (30%), Positives = 89/196 (45%), Gaps = 18/196 (9%)
Query: 19 LLAGGLAGGFAKTVVAPLERLKILFQTRRTEFRSAG-LSGSVRRIAKTEGLLGFYRGNGA 77
L+AGG+AG A+T + P++ +K QT +E A L + I EG FY+G
Sbjct: 300 LMAGGMAGALAQTAIYPMDLVKTRLQTCVSEGGKAPKLWKLTKDIWVREGPRAFYKGLFP 359
Query: 78 SVARIIPYAGLHFMSYEEYRRLIMQ-AFPNVWKGPTLDLMAGSLSGGTAVLFTYPLDLIR 136
S+ I+PYAG+ +YE + L + GP + L G SG YPL ++R
Sbjct: 360 SLLGIVPYAGIDLAAYETLKDLSRTYILQDTEPGPLIQLSCGMTSGALGASCVYPLQVVR 419
Query: 137 TKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIRGLYRGVAPTLFGIFPYAG 196
T++ T ++ T K G+RG YRG+ P L + P A
Sbjct: 420 TRMQADSSKTT----------------MKQEFMNTMKGEGLRGFYRGLLPNLLKVVPAAS 463
Query: 197 LKFYFYEEMKRRVPED 212
+ + YE MK+ + D
Sbjct: 464 ITYIVYEAMKKNMALD 479
Score = 73.6 bits (179), Expect = 1e-13
Identities = 51/194 (26%), Positives = 90/194 (46%), Gaps = 20/194 (10%)
Query: 115 LMAGSLSGGTAVLFTYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKE 174
L+AG L+G + T PLD R K+ Q+ ++ + G+ + K ++E
Sbjct: 209 LLAGGLAGAVSRTATAPLD--RLKVVLQV--------------QRAHAGVLPTIKKIWRE 252
Query: 175 GGIRGLYRGVAPTLFGIFPYAGLKFYFYEEMKRRVP-EDYKKSIMAKLTCGSVAGLLGQT 233
+ G +RG + + P + +KF YE +K + ED +L G +AG L QT
Sbjct: 253 DKLMGFFRGNGLNVMKVAPESAIKFCAYEMLKPMIGGEDGDIGTSGRLMAGGMAGALAQT 312
Query: 234 FTYPLEVVRRQMQVQNLAASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAA 293
YP+++V+ ++Q SE + + I ++G + + GL + + +VP A
Sbjct: 313 AIYPMDLVKTRLQT---CVSEGGKAPKLWKLTKDIWVREGPRAFYKGLFPSLLGIVPYAG 369
Query: 294 IGFTVYDTMKSYLR 307
I Y+T+K R
Sbjct: 370 IDLAAYETLKDLSR 383
>At3g51870 carrier like protein
Length = 381
Score = 150 bits (380), Expect = 7e-37
Identities = 95/309 (30%), Positives = 156/309 (49%), Gaps = 42/309 (13%)
Query: 7 SILDHIPLFAKELLAGGLAGGFAKTVVAPLERLKILFQT------RRTEFRSAGLSGSVR 60
+IL +P A AG LAG AKTV APL+R+K+L QT +++ ++ G ++
Sbjct: 79 AILALVPKDAAIFAAGALAGAAAKTVTAPLDRIKLLMQTHGIRLGQQSAKKAIGFIEAIT 138
Query: 61 RIAKTEGLLGFYRGNGASVARIIPYAGLHFMSYEEYRRLIMQAFPNVWKGPTLDLM---- 116
IAK EG+ G+++GN V R++PY+ + ++YE Y+ N++KG L
Sbjct: 139 LIAKEEGVKGYWKGNLPQVIRVLPYSAVQLLAYESYK--------NLFKGKDDQLSVIGR 190
Query: 117 --AGSLSGGTAVLFTYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKE 174
AG+ +G T+ L TYPLD++R +LA E YR + ++
Sbjct: 191 LAAGACAGMTSTLLTYPLDVLRLRLAV----------------EPRYRTMSQVALSMLRD 234
Query: 175 GGIRGLYRGVAPTLFGIFPYAGLKFYFYEEMKRRVPEDYKKSIMAKLTCGSVAGLLGQTF 234
GI Y G+ P+L GI PY + F ++ +K+ +PE+Y+K + L ++ +
Sbjct: 235 EGIASFYYGLGPSLVGIAPYIAVNFCIFDLVKKSLPEEYRKKAQSSLLTAVLSAGIATLT 294
Query: 235 TYPLEVVRRQMQVQNLAASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAI 294
YPL+ VRRQMQ++ K + I + G L+ G N +K +P+++I
Sbjct: 295 CYPLDTVRRQMQMRG------TPYKSIPEAFAGIIDRDGLIGLYRGFLPNALKTLPNSSI 348
Query: 295 GFTVYDTMK 303
T +D +K
Sbjct: 349 RLTTFDMVK 357
Score = 64.3 bits (155), Expect = 8e-11
Identities = 50/200 (25%), Positives = 85/200 (42%), Gaps = 27/200 (13%)
Query: 19 LLAGGLAGGFAKTVVAPLE--RLKILFQTRRTEFRSAGLSGSVRRIAKTEGLLGFYRGNG 76
L AG AG + + PL+ RL++ + R LS + + EG+ FY G G
Sbjct: 191 LAAGACAGMTSTLLTYPLDVLRLRLAVEPRYRTMSQVALS-----MLRDEGIASFYYGLG 245
Query: 77 ASVARIIPYAGLHFMSYEEYRRLIMQAFPNVW-KGPTLDLMAGSLSGGTAVLFTYPLDLI 135
S+ I PY ++F ++ L+ ++ P + K L+ LS G A L YPLD +
Sbjct: 246 PSLVGIAPYIAVNFCIFD----LVKKSLPEEYRKKAQSSLLTAVLSAGIATLTCYPLDTV 301
Query: 136 RTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIRGLYRGVAPTLFGIFPYA 195
R ++ + Y+ I + + G+ GLYRG P P +
Sbjct: 302 RRQMQMRGTP---------------YKSIPEAFAGIIDRDGLIGLYRGFLPNALKTLPNS 346
Query: 196 GLKFYFYEEMKRRVPEDYKK 215
++ ++ +KR + K+
Sbjct: 347 SIRLTTFDMVKRLIATSEKQ 366
Score = 44.7 bits (104), Expect = 7e-05
Identities = 27/101 (26%), Positives = 53/101 (51%), Gaps = 7/101 (6%)
Query: 209 VPEDYKKSIMAKLTCGSVAGLLGQTFTYPLEVVRRQMQVQNLAASEEAELK--GTMRSMV 266
VP+D A G++AG +T T PL+ ++ MQ + +++ K G + ++
Sbjct: 84 VPKD-----AAIFAAGALAGAAAKTVTAPLDRIKLLMQTHGIRLGQQSAKKAIGFIEAIT 138
Query: 267 LIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSYLR 307
LIA+++G K + G I+V+P +A+ Y++ K+ +
Sbjct: 139 LIAKEEGVKGYWKGNLPQVIRVLPYSAVQLLAYESYKNLFK 179
>At5g01500 unknown protein
Length = 415
Score = 147 bits (371), Expect = 7e-36
Identities = 95/321 (29%), Positives = 157/321 (48%), Gaps = 48/321 (14%)
Query: 7 SILDHIPLFAKELLAGGLAGGFAKTVVAPLERLKILFQT------RRTEFRSAGLSGSVR 60
++L +P A AG AG AK+V APL+R+K+L QT +++ ++ G ++
Sbjct: 107 ALLSIVPKDAALFFAGAFAGAAAKSVTAPLDRIKLLMQTHGVRAGQQSAKKAIGFIEAIT 166
Query: 61 RIAKTEGLLGFYRGNGASVARIIPYAGLHFMSYEEYRRLIMQAFPNVWKGPTLDLM---- 116
I K EG+ G+++GN V RI+PY+ + +YE Y++L K L ++
Sbjct: 167 LIGKEEGIKGYWKGNLPQVIRIVPYSAVQLFAYETYKKLFRG------KDGQLSVLGRLG 220
Query: 117 AGSLSGGTAVLFTYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGG 176
AG+ +G T+ L TYPLD++R +LA E YR + +E G
Sbjct: 221 AGACAGMTSTLITYPLDVLRLRLAV----------------EPGYRTMSQVALNMLREEG 264
Query: 177 IRGLYRGVAPTLFGIFPYAGLKFYFYEEMKRRVPEDYKKSIMAKLTCGSVAGLLGQTFTY 236
+ Y G+ P+L I PY + F ++ +K+ +PE Y++ + L VA + Y
Sbjct: 265 VASFYNGLGPSLLSIAPYIAINFCVFDLVKKSLPEKYQQKTQSSLLTAVVAAAIATGTCY 324
Query: 237 PLEVVRRQMQVQNLAASEEAELKGTMRSMVL-----IAQKQGWKTLFSGLSINYIKVVPS 291
PL+ +RRQMQ LKGT VL I ++G L+ G N +K +P+
Sbjct: 325 PLDTIRRQMQ-----------LKGTPYKSVLDAFSGIIAREGVVGLYRGFVPNALKSMPN 373
Query: 292 AAIGFTVYDTMKSYLRVPSRD 312
++I T +D +K + ++
Sbjct: 374 SSIKLTTFDIVKKLIAASEKE 394
>At5g56450 ADP/ATP translocase-like protein
Length = 330
Score = 145 bits (365), Expect = 4e-35
Identities = 96/309 (31%), Positives = 153/309 (49%), Gaps = 30/309 (9%)
Query: 15 FAKELLAGGLAGGFAKTVVAPLERLKILFQTRRTEF------------RSAGLSGSVRRI 62
F K+LLAG + GG T+VAP+ER K+L QT+ + R G+ + R
Sbjct: 30 FQKDLLAGAVMGGVVHTIVAPIERAKLLLQTQESNIAIVGDEGHAGKRRFKGMFDFIFRT 89
Query: 63 AKTEGLLGFYRGNGASVARIIPYAGLHFMSYEEYRRLIMQAFP---NVWKGPTLDLMAGS 119
+ EG+L +RGNG+SV R P L+F + YR ++ + +++ G + MAGS
Sbjct: 90 VREEGVLSLWRGNGSSVLRYYPSVALNFSLKDLYRSILRNSSSQENHIFSGALANFMAGS 149
Query: 120 LSGGTAVLFTYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIRG 179
+G TA++ YPLD+ T+LA I P + +RGI LS +K+ G+RG
Sbjct: 150 AAGCTALIVVYPLDIAHTRLAADIGKP----------EARQFRGIHHFLSTIHKKDGVRG 199
Query: 180 LYRGVAPTLFGIFPYAGLKFYFYEEMKRRVPEDYKKSIMAKLTCGSVAGLLGQT--FTYP 237
+YRG+ +L G+ + GL F ++ +K ED K + G + +YP
Sbjct: 200 IYRGLPASLHGVIIHRGLYFGGFDTVKEIFSEDTKPELALWKRWGLAQAVTTSAGLASYP 259
Query: 238 LEVVRRQMQVQNLAASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGFT 297
L+ VRR++ +Q + E + T+ I + +G + + G N + SAAI
Sbjct: 260 LDTVRRRIMMQ--SGMEHPMYRSTLDCWKKIYRSEGLASFYRGALSNMFRSTGSAAI-LV 316
Query: 298 VYDTMKSYL 306
YD +K +L
Sbjct: 317 FYDEVKRFL 325
Score = 59.7 bits (143), Expect = 2e-09
Identities = 46/218 (21%), Positives = 90/218 (41%), Gaps = 8/218 (3%)
Query: 103 AFPNVWKGPTLDLMAGSLSGGTAVLFTYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYR 162
+ P K DL+AG++ GG P++ + L Q + + G + ++
Sbjct: 22 SLPQTLKHFQKDLLAGAVMGGVVHTIVAPIERAKLLLQTQESNIAIVGDEGHAGKRR-FK 80
Query: 163 GIRDCLSKTYKEGGIRGLYRGVAPTLFGIFPYAGLKFYF---YEEMKRRVPEDYK---KS 216
G+ D + +T +E G+ L+RG ++ +P L F Y + R
Sbjct: 81 GMFDFIFRTVREEGVLSLWRGNGSSVLRYYPSVALNFSLKDLYRSILRNSSSQENHIFSG 140
Query: 217 IMAKLTCGSVAGLLGQTFTYPLEVVRRQMQVQNLAASEEAELKGTMRSMVLIAQKQGWKT 276
+A GS AG YPL++ ++ ++ E + +G + I +K G +
Sbjct: 141 ALANFMAGSAAGCTALIVVYPLDIAHTRLAA-DIGKPEARQFRGIHHFLSTIHKKDGVRG 199
Query: 277 LFSGLSINYIKVVPSAAIGFTVYDTMKSYLRVPSRDEV 314
++ GL + V+ + F +DT+K ++ E+
Sbjct: 200 IYRGLPASLHGVIIHRGLYFGGFDTVKEIFSEDTKPEL 237
>At4g32400 adenylate translocator (brittle-1) - like protein
Length = 392
Score = 143 bits (361), Expect = 1e-34
Identities = 97/296 (32%), Positives = 146/296 (48%), Gaps = 28/296 (9%)
Query: 17 KELLAGGLAGGFAKTVVAPLERLKILFQTRRTEFRSAGLSGS--VRRIAKTEGLLGFYRG 74
+ LL+G +AG ++TVVAPLE ++ S G S + I K EG G +RG
Sbjct: 112 RRLLSGAVAGAVSRTVVAPLETIRTHLMVG-----SGGNSSTEVFSDIMKHEGWTGLFRG 166
Query: 75 NGASVARIIPYAGLHFMSYEEYRRLIMQAFPNVWKGPT-LDLMAGSLSGGTAVLFTYPLD 133
N +V R+ P + +E + + K P L+AG+ +G + L TYPL+
Sbjct: 167 NLVNVIRVAPARAVELFVFETVNKKLSPPHGQESKIPIPASLLAGACAGVSQTLLTYPLE 226
Query: 134 LIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIRGLYRGVAPTLFGIFP 193
L++T+L Q VY+GI D K +E G LYRG+AP+L G+ P
Sbjct: 227 LVKTRLTIQ---------------RGVYKGIFDAFLKIIREEGPTELYRGLAPSLIGVVP 271
Query: 194 YAGLKFYFYEEMK---RRVPEDYKKSIMAKLTCGSVAGLLGQTFTYPLEVVRRQMQVQNL 250
YA ++ Y+ ++ R + K + L GS+AG L T T+PLEV R+ MQV
Sbjct: 272 YAATNYFAYDSLRKAYRSFSKQEKIGNIETLLIGSLAGALSSTATFPLEVARKHMQVG-- 329
Query: 251 AASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSYL 306
A S K + ++V I + +G + GL + +K+VP+A I F Y+ K L
Sbjct: 330 AVSGRVVYKNMLHALVTILEHEGILGWYKGLGPSCLKLVPAAGISFMCYEACKKIL 385
Score = 80.5 bits (197), Expect = 1e-15
Identities = 56/205 (27%), Positives = 96/205 (46%), Gaps = 14/205 (6%)
Query: 12 IPLFAKELLAGGLAGGFAKTVVAPLERLKILFQTRRTEFRSAGLSGSVRRIAKTEGLLGF 71
IP+ A LLAG AG + PLE +K +R ++ G+ + +I + EG
Sbjct: 202 IPIPAS-LLAGACAGVSQTLLTYPLELVKTRLTIQRGVYK--GIFDAFLKIIREEGPTEL 258
Query: 72 YRGNGASVARIIPYAGLHFMSYEEYRRLIMQAFPNVWKGPTLDLMAGSLSGGTAVLFTYP 131
YRG S+ ++PYA ++ +Y+ R+ G L+ GSL+G + T+P
Sbjct: 259 YRGLAPSLIGVVPYAATNYFAYDSLRKAYRSFSKQEKIGNIETLLIGSLAGALSSTATFP 318
Query: 132 LDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIRGLYRGVAPTLFGI 191
L++ R + G V+ VY+ + L + GI G Y+G+ P+ +
Sbjct: 319 LEVARKHMQV-----------GAVSGRVVYKNMLHALVTILEHEGILGWYKGLGPSCLKL 367
Query: 192 FPYAGLKFYFYEEMKRRVPEDYKKS 216
P AG+ F YE K+ + E+ +++
Sbjct: 368 VPAAGISFMCYEACKKILIENNQEA 392
Score = 70.1 bits (170), Expect = 1e-12
Identities = 53/203 (26%), Positives = 97/203 (47%), Gaps = 26/203 (12%)
Query: 115 LMAGSLSGGTAVLFTYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKE 174
L++G+++G + PL+ IRT L SG ++ +V+ I K
Sbjct: 114 LLSGAVAGAVSRTVVAPLETIRTHLMVG---------SGGNSSTEVFSDIM-------KH 157
Query: 175 GGIRGLYRGVAPTLFGIFPYAGLKFYFYEEMKRRVP----EDYKKSIMAKLTCGSVAGLL 230
G GL+RG + + P ++ + +E + +++ ++ K I A L G+ AG+
Sbjct: 158 EGWTGLFRGNLVNVIRVAPARAVELFVFETVNKKLSPPHGQESKIPIPASLLAGACAGVS 217
Query: 231 GQTFTYPLEVVRRQMQVQNLAASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVP 290
TYPLE+V+ ++ +Q KG + + I +++G L+ GL+ + I VVP
Sbjct: 218 QTLLTYPLELVKTRLTIQR------GVYKGIFDAFLKIIREEGPTELYRGLAPSLIGVVP 271
Query: 291 SAAIGFTVYDTMKSYLRVPSRDE 313
AA + YD+++ R S+ E
Sbjct: 272 YAATNYFAYDSLRKAYRSFSKQE 294
Score = 48.5 bits (114), Expect = 5e-06
Identities = 28/96 (29%), Positives = 45/96 (46%), Gaps = 7/96 (7%)
Query: 218 MAKLTCGSVAGLLGQTFTYPLEVVRRQMQVQNLAASEEAELKGTMRSMVLIAQKQGWKTL 277
+ +L G+VAG + +T PLE +R + V + S M+ +GW L
Sbjct: 111 LRRLLSGAVAGAVSRTVVAPLETIRTHLMVGSGGNSSTEVFSDIMKH-------EGWTGL 163
Query: 278 FSGLSINYIKVVPSAAIGFTVYDTMKSYLRVPSRDE 313
F G +N I+V P+ A+ V++T+ L P E
Sbjct: 164 FRGNLVNVIRVAPARAVELFVFETVNKKLSPPHGQE 199
>At3g21390 unknown protein
Length = 335
Score = 131 bits (330), Expect = 4e-31
Identities = 92/318 (28%), Positives = 148/318 (45%), Gaps = 52/318 (16%)
Query: 21 AGGLAGGFAKTVVAPLERLKILFQTR------------RTEFRSAGLSGSVRRIAKTEGL 68
AGG+AG ++ V +PL+ +KI FQ + + + + GL + + I + EGL
Sbjct: 21 AGGVAGAISRMVTSPLDVIKIRFQVQLEPTATWALKDSQLKPKYNGLFRTTKDIFREEGL 80
Query: 69 LGFYRGNGASVARIIPYAGLHFMSYEEYRRLIM---QAFPNVWKGPTLDLMAGSLSGGTA 125
GF+RGN ++ ++PY + F + + +A + P L ++G+L+G A
Sbjct: 81 SGFWRGNVPALLMVVPYTSIQFAVLHKVKSFAAGSSKAENHAQLSPYLSYISGALAGCAA 140
Query: 126 VLFTYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIRGLYRGVA 185
+ +YP DL+RT LA Q +VY +R + GI+GLY G++
Sbjct: 141 TVGSYPFDLLRTVLASQ-------------GEPKVYPNMRSAFLSIVQTRGIKGLYAGLS 187
Query: 186 PTLFGIFPYAGLKFYFYEEMKR----------------RVPEDYKKSIMAKLTCGSVAGL 229
PTL I PYAGL+F Y+ KR P D S L CG +G
Sbjct: 188 PTLIEIIPYAGLQFGTYDTFKRWSMVYNKRYRSSSSSSTNPSDSLSSFQLFL-CGLASGT 246
Query: 230 LGQTFTYPLEVVRRQMQVQNL-------AASEEAELKGTMRSMVLIAQKQGWKTLFSGLS 282
+ + +PL+VV+++ QV+ L A E K + I + +GW L+ G+
Sbjct: 247 VSKLVCHPLDVVKKRFQVEGLQRHPKYGARVELNAYKNMFDGLGQILRSEGWHGLYKGIV 306
Query: 283 INYIKVVPSAAIGFTVYD 300
+ IK P+ A+ F Y+
Sbjct: 307 PSTIKAAPAGAVTFVAYE 324
Score = 84.3 bits (207), Expect = 8e-17
Identities = 60/211 (28%), Positives = 99/211 (46%), Gaps = 13/211 (6%)
Query: 105 PNVWKGPTLDLMAGSLSGGTAVLFTYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGI 164
P K +D AG ++G + + T PLD+I+ + Q+ PT + Y G+
Sbjct: 9 PGKLKRAVIDASAGGVAGAISRMVTSPLDVIKIRFQVQL-EPTATWALKDSQLKPKYNGL 67
Query: 165 RDCLSKTYKEGGIRGLYRGVAPTLFGIFPYAGLKFYFYEEMKRRVPEDYKKSIMAKLT-- 222
++E G+ G +RG P L + PY ++F ++K K A+L+
Sbjct: 68 FRTTKDIFREEGLSGFWRGNVPALLMVVPYTSIQFAVLHKVKSFAAGSSKAENHAQLSPY 127
Query: 223 ----CGSVAGLLGQTFTYPLEVVRRQMQVQNLAASEEAELKGTMRSMVL-IAQKQGWKTL 277
G++AG +YP +++R LA+ E ++ MRS L I Q +G K L
Sbjct: 128 LSYISGALAGCAATVGSYPFDLLRTV-----LASQGEPKVYPNMRSAFLSIVQTRGIKGL 182
Query: 278 FSGLSINYIKVVPSAAIGFTVYDTMKSYLRV 308
++GLS I+++P A + F YDT K + V
Sbjct: 183 YAGLSPTLIEIIPYAGLQFGTYDTFKRWSMV 213
Score = 65.9 bits (159), Expect = 3e-11
Identities = 53/215 (24%), Positives = 93/215 (42%), Gaps = 15/215 (6%)
Query: 2 AASSESILDHIPLFAK-ELLAGGLAGGFAKTVVAPLERLKILFQTRRTEFRSAGLSGSVR 60
AA S +H L ++G LAG A P + L+ + ++ + +
Sbjct: 112 AAGSSKAENHAQLSPYLSYISGALAGCAATVGSYPFDLLRTVLASQGEPKVYPNMRSAFL 171
Query: 61 RIAKTEGLLGFYRGNGASVARIIPYAGLHFMSYEEYRRLIM------QAFPNVWKGPTLD 114
I +T G+ G Y G ++ IIPYAGL F +Y+ ++R M ++ + P+
Sbjct: 172 SIVQTRGIKGLYAGLSPTLIEIIPYAGLQFGTYDTFKRWSMVYNKRYRSSSSSSTNPSDS 231
Query: 115 L------MAGSLSGGTAVLFTYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCL 168
L + G SG + L +PLD+++ + +Q+ + G Y+ + D L
Sbjct: 232 LSSFQLFLCGLASGTVSKLVCHPLDVVKKR--FQVEGLQRHPKYGARVELNAYKNMFDGL 289
Query: 169 SKTYKEGGIRGLYRGVAPTLFGIFPYAGLKFYFYE 203
+ + G GLY+G+ P+ P + F YE
Sbjct: 290 GQILRSEGWHGLYKGIVPSTIKAAPAGAVTFVAYE 324
Score = 38.1 bits (87), Expect = 0.006
Identities = 30/107 (28%), Positives = 52/107 (48%), Gaps = 8/107 (7%)
Query: 215 KSIMAKLTCGSVAGLLGQTFTYPLEVVRRQMQVQ----NLAASEEAELK----GTMRSMV 266
K + + G VAG + + T PL+V++ + QVQ A ++++LK G R+
Sbjct: 13 KRAVIDASAGGVAGAISRMVTSPLDVIKIRFQVQLEPTATWALKDSQLKPKYNGLFRTTK 72
Query: 267 LIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSYLRVPSRDE 313
I +++G + G + VVP +I F V +KS+ S+ E
Sbjct: 73 DIFREEGLSGFWRGNVPALLMVVPYTSIQFAVLHKVKSFAAGSSKAE 119
>At4g28390 ADP,ATP carrier-like protein
Length = 379
Score = 131 bits (329), Expect = 5e-31
Identities = 87/290 (30%), Positives = 141/290 (48%), Gaps = 20/290 (6%)
Query: 15 FAKELLAGGLAGGFAKTVVAPLERLKILFQTRRTEFRSA-------GLSGSVRRIAKTEG 67
F + L GG++ +KT AP+ER+K+L Q + ++ G+S R K EG
Sbjct: 79 FLIDFLMGGVSAAVSKTAAAPIERVKLLIQNQDEMIKAGRLSEPYKGISDCFARTVKDEG 138
Query: 68 LLGFYRGNGASVARIIPYAGLHFMSYEEYRRL--IMQAFPNVWKGPTLDLMAGSLSGGTA 125
+L +RGN A+V R P L+F + ++RL + WK +L +G +G ++
Sbjct: 139 MLALWRGNTANVIRYFPTQALNFAFKDYFKRLFNFKKEKDGYWKWFAGNLASGGAAGASS 198
Query: 126 VLFTYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIRGLYRGVA 185
+LF Y LD RT+LA + K ++ + G+ D KT GI GLYRG
Sbjct: 199 LLFVYSLDYARTRLANDAKAAKK-------GGQRQFNGMVDVYKKTIASDGIVGLYRGFN 251
Query: 186 PTLFGIFPYAGLKFYFYEEMKRRVPED-YKKSIMAKLTCGSVAGLLGQTFTYPLEVVRRQ 244
+ GI Y GL F Y+ +K V D + S +A G + +YP++ VRR+
Sbjct: 252 ISCVGIVVYRGLYFGLYDSLKPVVLVDGLQDSFLASFLLGWGITIGAGLASYPIDTVRRR 311
Query: 245 MQVQNLAASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAI 294
M + + E + K ++++ I + +G K+LF G N ++ V A +
Sbjct: 312 MM---MTSGEAVKYKSSLQAFSQIVKNEGAKSLFKGAGANILRAVAGAGV 358
Score = 65.5 bits (158), Expect = 4e-11
Identities = 57/211 (27%), Positives = 95/211 (45%), Gaps = 23/211 (10%)
Query: 110 GPTLDLMAGSLSGGTAVLFTYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLS 169
G +D + G +S + P++ R KL Q + ++ +G ++ + Y+GI DC +
Sbjct: 78 GFLIDFLMGGVSAAVSKTAAAPIE--RVKLLIQ--NQDEMIKAGRLS--EPYKGISDCFA 131
Query: 170 KTYKEGGIRGLYRGVAPTLFGIFPYAGLKFYFYEEMKR-----RVPEDYKKSIMAKLTCG 224
+T K+ G+ L+RG + FP L F F + KR + + Y K L G
Sbjct: 132 RTVKDEGMLALWRGNTANVIRYFPTQALNFAFKDYFKRLFNFKKEKDGYWKWFAGNLASG 191
Query: 225 SVAGLLGQTFTYPLEVVRRQMQVQNLAASEEAELKGTMR---SMVLIAQK----QGWKTL 277
AG F Y L+ R + LA +A KG R MV + +K G L
Sbjct: 192 GAAGASSLLFVYSLDYARTR-----LANDAKAAKKGGQRQFNGMVDVYKKTIASDGIVGL 246
Query: 278 FSGLSINYIKVVPSAAIGFTVYDTMKSYLRV 308
+ G +I+ + +V + F +YD++K + V
Sbjct: 247 YRGFNISCVGIVVYRGLYFGLYDSLKPVVLV 277
>At5g48970 mitochondrial carrier protein-like
Length = 339
Score = 130 bits (326), Expect = 1e-30
Identities = 90/324 (27%), Positives = 151/324 (45%), Gaps = 51/324 (15%)
Query: 21 AGGLAGGFAKTVVAPLERLKILFQTRRTEFRSAGL-----SGS---------VRRIAKTE 66
AG ++GG +++V +PL+ +KI FQ + S GL SG+ + I + E
Sbjct: 24 AGAISGGVSRSVTSPLDVIKIRFQVQLEPTTSWGLVRGNLSGASKYTGMVQATKDIFREE 83
Query: 67 GLLGFYRGNGASVARIIPYAGLHFMSYEEYRRLIM---QAFPNVWKGPTLDLMAGSLSGG 123
G GF+RGN ++ ++PY + F + + + ++ P L ++G+L+G
Sbjct: 84 GFRGFWRGNVPALLMVMPYTSIQFTVLHKLKSFASGSTKTEDHIHLSPYLSFVSGALAGC 143
Query: 124 TAVLFTYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIRGLYRG 183
A L +YP DL+RT LA Q +VY +R + GIRGLY G
Sbjct: 144 AATLGSYPFDLLRTILASQ-------------GEPKVYPTMRSAFVDIIQSRGIRGLYNG 190
Query: 184 VAPTLFGIFPYAGLKFYFYEEMKRRVPEDYKKSIMAKL--------------TCGSVAGL 229
+ PTL I PYAGL+F Y+ KR + + + + +K+ CG AG
Sbjct: 191 LTPTLVEIVPYAGLQFGTYDMFKRWMMDWNRYKLSSKIPINVDTNLSSFQLFICGLGAGT 250
Query: 230 LGQTFTYPLEVVRRQMQVQNL-------AASEEAELKGTMRSMVLIAQKQGWKTLFSGLS 282
+ +PL+VV+++ Q++ L A E + + + I +GW L+ G+
Sbjct: 251 SAKLVCHPLDVVKKRFQIEGLQRHPRYGARVERRAYRNMLDGLRQIMISEGWHGLYKGIV 310
Query: 283 INYIKVVPSAAIGFTVYDTMKSYL 306
+ +K P+ A+ F Y+ +L
Sbjct: 311 PSTVKAAPAGAVTFVAYEFTSDWL 334
Score = 89.0 bits (219), Expect = 3e-18
Identities = 59/210 (28%), Positives = 100/210 (47%), Gaps = 13/210 (6%)
Query: 105 PNVWKGPTLDLMAGSLSGGTAVLFTYPLDLIRTKLAYQIVSPTKLN-VSGMVNNEQVYRG 163
P K +D AG++SGG + T PLD+I+ + Q+ T V G ++ Y G
Sbjct: 12 PGQIKRALIDASAGAISGGVSRSVTSPLDVIKIRFQVQLEPTTSWGLVRGNLSGASKYTG 71
Query: 164 IRDCLSKTYKEGGIRGLYRGVAPTLFGIFPYAGLKFYFYEEMK------RRVPEDYKKSI 217
+ ++E G RG +RG P L + PY ++F ++K + + S
Sbjct: 72 MVQATKDIFREEGFRGFWRGNVPALLMVMPYTSIQFTVLHKLKSFASGSTKTEDHIHLSP 131
Query: 218 MAKLTCGSVAGLLGQTFTYPLEVVRRQMQVQNLAASEEAELKGTMRS-MVLIAQKQGWKT 276
G++AG +YP +++R LA+ E ++ TMRS V I Q +G +
Sbjct: 132 YLSFVSGALAGCAATLGSYPFDLLR-----TILASQGEPKVYPTMRSAFVDIIQSRGIRG 186
Query: 277 LFSGLSINYIKVVPSAAIGFTVYDTMKSYL 306
L++GL+ +++VP A + F YD K ++
Sbjct: 187 LYNGLTPTLVEIVPYAGLQFGTYDMFKRWM 216
Score = 35.8 bits (81), Expect = 0.031
Identities = 25/111 (22%), Positives = 57/111 (50%), Gaps = 14/111 (12%)
Query: 215 KSIMAKLTCGSVAGLLGQTFTYPLEVVRRQMQVQ------------NLAASEEAELKGTM 262
K + + G+++G + ++ T PL+V++ + QVQ NL+ + ++ G +
Sbjct: 16 KRALIDASAGAISGGVSRSVTSPLDVIKIRFQVQLEPTTSWGLVRGNLSGA--SKYTGMV 73
Query: 263 RSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSYLRVPSRDE 313
++ I +++G++ + G + V+P +I FTV +KS+ ++ E
Sbjct: 74 QATKDIFREEGFRGFWRGNVPALLMVMPYTSIQFTVLHKLKSFASGSTKTE 124
>At5g13490 adenosine nucleotide translocator
Length = 385
Score = 128 bits (322), Expect = 4e-30
Identities = 87/290 (30%), Positives = 139/290 (47%), Gaps = 19/290 (6%)
Query: 15 FAKELLAGGLAGGFAKTVVAPLERLKILFQTRRTEFRSA-------GLSGSVRRIAKTEG 67
FA + + GG++ +KT AP+ER+K+L Q + ++ G+ R + EG
Sbjct: 84 FAIDFMMGGVSAAVSKTAAAPIERVKLLIQNQDEMLKAGRLTEPYKGIRDCFGRTIRDEG 143
Query: 68 LLGFYRGNGASVARIIPYAGLHFMSYEEYRRLI--MQAFPNVWKGPTLDLMAGSLSGGTA 125
+ +RGN A+V R P L+F + ++RL + WK +L +G +G ++
Sbjct: 144 IGSLWRGNTANVIRYFPTQALNFAFKDYFKRLFNFKKDKDGYWKWFAGNLASGGAAGASS 203
Query: 126 VLFTYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIRGLYRGVA 185
+LF Y LD RT+LA S K E+ + G+ D KT K GI GLYRG
Sbjct: 204 LLFVYSLDYARTRLANDSKSAKK------GGGERQFNGLVDVYKKTLKSDGIAGLYRGFN 257
Query: 186 PTLFGIFPYAGLKFYFYEEMKR-RVPEDYKKSIMAKLTCGSVAGLLGQTFTYPLEVVRRQ 244
+ GI Y GL F Y+ +K + D + S A G + +YP++ VRR+
Sbjct: 258 ISCAGIIVYRGLYFGLYDSVKPVLLTGDLQDSFFASFALGWLITNGAGLASYPIDTVRRR 317
Query: 245 MQVQNLAASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAI 294
M + + E + K + + I +K+G K+LF G N ++ V A +
Sbjct: 318 MM---MTSGEAVKYKSSFDAFSQIVKKEGAKSLFKGAGANILRAVAGAGV 364
>At5g64970 mitochondrial carrier protein-like
Length = 428
Score = 126 bits (317), Expect = 1e-29
Identities = 93/316 (29%), Positives = 153/316 (47%), Gaps = 44/316 (13%)
Query: 17 KELLAGGLAGGFAKTVVAPLERLKILFQTRRTEFRSAGLSGSVRRIAKTEGLLGFYRGNG 76
K L AG A ++T +APLER+K+ + R + L ++RIA EG+ GF++GN
Sbjct: 133 KHLWAGAFAAMVSRTCIAPLERMKLEYIVRGEQ---GNLLELIQRIATNEGIRGFWKGNL 189
Query: 77 ASVARIIPYAGLHFMSYEEYRRLIMQAFPNVWKGPTLDLMAGSLSGGTAVLFTYPLDLIR 136
++ R P+ ++F +Y+ YR +++ N +AG+ +G TA L PLD IR
Sbjct: 190 VNILRTAPFKSINFYAYDTYRGQLLKLSGNEETTNFERFVAGAAAGVTASLLCLPLDTIR 249
Query: 137 TKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIRGLYRGVAPTLFGIFPYAG 196
T + V+P + G+V + G LY+G+ P+L + P
Sbjct: 250 TVM----VAPGGEALGGVVG----------AFRHMIQTEGFFSLYKGLVPSLVSMAPSGA 295
Query: 197 LKFYFYEEMKR---RVPEDYKK-------------------SIMAKLTCGSVAGLLGQTF 234
+ + Y+ +K PE K+ M L G++AG +
Sbjct: 296 VFYGVYDILKSAYLHTPEGKKRLEHMKQEGEELNAFDQLELGPMRTLLYGAIAGACSEAA 355
Query: 235 TYPLEVVRRQMQVQNLAASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAI 294
TYP EVVRR++Q+Q+ A+ + + V I ++ G L++GL + ++V+PSAAI
Sbjct: 356 TYPFEVVRRRLQMQS-----HAKRLSAVATCVKIIEQGGVPALYAGLIPSLLQVLPSAAI 410
Query: 295 GFTVYDTMKSYLRVPS 310
+ VY+ MK L+V S
Sbjct: 411 SYFVYEFMKVVLKVES 426
>At3g08580 adenylate translocator
Length = 381
Score = 126 bits (316), Expect = 2e-29
Identities = 90/299 (30%), Positives = 143/299 (47%), Gaps = 20/299 (6%)
Query: 15 FAKELLAGGLAGGFAKTVVAPLERLKILFQTRRTEFRSAGLSGSVR-------RIAKTEG 67
FA + L GG++ +KT AP+ER+K+L Q + ++ LS + R K EG
Sbjct: 80 FALDFLMGGVSAAVSKTAAAPIERVKLLIQNQDEMIKAGRLSEPYKGIGDCFGRTIKDEG 139
Query: 68 LLGFYRGNGASVARIIPYAGLHFMSYEEYRRLI--MQAFPNVWKGPTLDLMAGSLSGGTA 125
+RGN A+V R P L+F + ++RL + WK +L +G +G ++
Sbjct: 140 FGSLWRGNTANVIRYFPTQALNFAFKDYFKRLFNFKKDRDGYWKWFAGNLASGGAAGASS 199
Query: 126 VLFTYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIRGLYRGVA 185
+LF Y LD RT+LA + K + + G+ D KT K GI GLYRG
Sbjct: 200 LLFVYSLDYARTRLANDAKAAKK------GGGGRQFDGLVDVYRKTLKTDGIAGLYRGFN 253
Query: 186 PTLFGIFPYAGLKFYFYEEMKR-RVPEDYKKSIMAKLTCGSVAGLLGQTFTYPLEVVRRQ 244
+ GI Y GL F Y+ +K + D + S A G V +YP++ VRR+
Sbjct: 254 ISCVGIIVYRGLYFGLYDSVKPVLLTGDLQDSFFASFALGWVITNGAGLASYPIDTVRRR 313
Query: 245 MQVQNLAASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMK 303
M + + E + K ++ + I + +G K+LF G N ++ V A + + YD ++
Sbjct: 314 MM---MTSGEAVKYKSSLDAFKQILKNEGAKSLFKGAGANILRAVAGAGV-LSGYDKLQ 368
>At3g20240 mitochondrial carrier protein, putative
Length = 348
Score = 120 bits (301), Expect = 1e-27
Identities = 85/315 (26%), Positives = 151/315 (46%), Gaps = 48/315 (15%)
Query: 16 AKELLAGGLAGGFAKTVVAPLERLKILFQTRR-TEFRSAGLSGSVRRIAKTEGLLGFYRG 74
A+E L+G LAG K V+APLE ++ TR S + GS + + +G G + G
Sbjct: 49 AREFLSGALAGAMTKAVLAPLETIR----TRMIVGVGSRSIPGSFLEVVQKQGWQGLWAG 104
Query: 75 NGASVARIIPYAGLHFMSYEEYRRLIMQA-------------------FPNV-WKGPTLD 114
N ++ RIIP + ++E +R + A P++ W P
Sbjct: 105 NEINMIRIIPTQAIELGTFEWVKRAMTSAQVKLKKIEDAKIEIGDFSFSPSISWISPVA- 163
Query: 115 LMAGSLSGGTAVLFTYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKE 174
+AG+ +G + L +PL++++ +L VSP ++Y + + + ++
Sbjct: 164 -VAGASAGIASTLVCHPLEVLKDRLT---VSP------------EIYPSLSLAIPRIFRA 207
Query: 175 GGIRGLYRGVAPTLFGIFPYAGLKFYFYEEMKRRVPEDYKKSIMAK---LTCGSVAGLLG 231
GIRG Y G+ PTL G+ PY+ ++ Y++MK + K +++ L G++AGL
Sbjct: 208 DGIRGFYAGLGPTLVGMLPYSTCYYFMYDKMKTSYCKSKNKKALSRPEMLVLGALAGLTA 267
Query: 232 QTFTYPLEVVRRQMQVQNLAASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPS 291
T ++PLEV R+++ V L + + +V +K+G L+ G + +KV+PS
Sbjct: 268 STISFPLEVARKRLMVGALKGECPPNMAAAIAEVV---KKEGVMGLYRGWGASCLKVMPS 324
Query: 292 AAIGFTVYDTMKSYL 306
+ I + Y+ K L
Sbjct: 325 SGITWVFYEAWKDIL 339
Score = 51.2 bits (121), Expect = 7e-07
Identities = 27/87 (31%), Positives = 52/87 (59%), Gaps = 3/87 (3%)
Query: 19 LLAGGLAGGFAKTVVAPLE--RLKILFQTRRTEFRSAGLSGSVRRIAKTEGLLGFYRGNG 76
L+ G LAG A T+ PLE R +++ + E ++ ++ + K EG++G YRG G
Sbjct: 257 LVLGALAGLTASTISFPLEVARKRLMVGALKGEC-PPNMAAAIAEVVKKEGVMGLYRGWG 315
Query: 77 ASVARIIPYAGLHFMSYEEYRRLIMQA 103
AS +++P +G+ ++ YE ++ +++ A
Sbjct: 316 ASCLKVMPSSGITWVFYEAWKDILLAA 342
Score = 45.8 bits (107), Expect = 3e-05
Identities = 28/94 (29%), Positives = 51/94 (53%), Gaps = 8/94 (8%)
Query: 211 EDYKKSIMAK-LTCGSVAGLLGQTFTYPLEVVRRQMQVQNLAASEEAELKGTMRSMVLIA 269
+D+ KS A+ G++AG + + PLE +R +M + + G S + +
Sbjct: 41 KDFFKSREAREFLSGALAGAMTKAVLAPLETIRTRM----IVGVGSRSIPG---SFLEVV 93
Query: 270 QKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMK 303
QKQGW+ L++G IN I+++P+ AI ++ +K
Sbjct: 94 QKQGWQGLWAGNEINMIRIIPTQAIELGTFEWVK 127
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.139 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,472,470
Number of Sequences: 26719
Number of extensions: 261472
Number of successful extensions: 1101
Number of sequences better than 10.0: 63
Number of HSP's better than 10.0 without gapping: 60
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 536
Number of HSP's gapped (non-prelim): 167
length of query: 315
length of database: 11,318,596
effective HSP length: 99
effective length of query: 216
effective length of database: 8,673,415
effective search space: 1873457640
effective search space used: 1873457640
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC148360.5


