

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148360.2 + phase: 0 /pseudo
(322 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
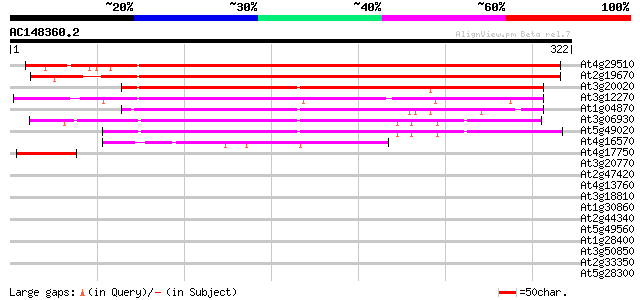
Score E
Sequences producing significant alignments: (bits) Value
At4g29510 arginine methyltransferase (pam1) 464 e-131
At2g19670 putative arginine N-methyltransferase 439 e-123
At3g20020 arginine methyltransferase like protein 213 9e-56
At3g12270 hypothetical protein 195 2e-50
At1g04870 unknown protein 155 3e-38
At3g06930 arginine methyltransferase like protein 151 4e-37
At5g49020 arginine methyltransferase-like protein 150 1e-36
At4g16570 unknown protein 58 8e-09
At4g17750 heat shock transcription factor HSF1 43 3e-04
At3g20770 ethylene-insensitive 3 37 0.011
At2g47420 putative dimethyladenosine transferase 37 0.019
At4g13760 putative polygalacturonase 35 0.042
At3g18810 protein kinase, putative 35 0.042
At1g30860 hypothetical protein 35 0.055
At2g44340 hypothetical protein 34 0.093
At5g49560 putative protein 34 0.12
At1g28400 unknown protein 34 0.12
At3g50850 unknown protein 33 0.16
At2g33350 hypothetical protein 33 0.16
At5g28300 GTL1 - like protein 33 0.21
>At4g29510 arginine methyltransferase (pam1)
Length = 390
Score = 464 bits (1195), Expect = e-131
Identities = 240/324 (74%), Positives = 271/324 (83%), Gaps = 20/324 (6%)
Query: 10 TNNTNMDNNN-----NNNNNHLRFEDADEVIEDVAETSNL-------DQSM--GGCDLDD 55
T N+N D N N N +RFEDADE ++VAE S + D+SM G D
Sbjct: 2 TKNSNHDENEFISFEPNQNTKIRFEDADE--DEVAEGSGVAGEETPQDESMFDAGESADT 59
Query: 56 S---NDKTSADYYFDSYSHFGNFHKEMLKDTVRTKTYQNVIYQNRFLFKNKVVLDVGAGT 112
+ +D TSADYYFDSYSHFG H+EMLKD VRTKTYQNVIYQN+FL K+K+VLDVGAGT
Sbjct: 60 AEVTDDTTSADYYFDSYSHFG-IHEEMLKDVVRTKTYQNVIYQNKFLIKDKIVLDVGAGT 118
Query: 113 GILSLFCAKAGAAHVYAVECSHMADRAKEIVETNGYSKVITVLKGKIEELELPVPKVDII 172
GILSLFCAKAGAAHVYAVECS MAD AKEIV+ NG+S VITVLKGKIEE+ELP PKVD+I
Sbjct: 119 GILSLFCAKAGAAHVYAVECSQMADMAKEIVKANGFSDVITVLKGKIEEIELPTPKVDVI 178
Query: 173 ISEWMGYFLLFENMLNSVLFARDKWLVDDGVILPDIASLYLTAIEDKDYKEDKIEFWNNV 232
ISEWMGYFLLFENML+SVL+ARDKWLV+ GV+LPD ASL+LTAIED +YKEDKIEFWN+V
Sbjct: 179 ISEWMGYFLLFENMLDSVLYARDKWLVEGGVVLPDKASLHLTAIEDSEYKEDKIEFWNSV 238
Query: 233 YGFDMSCIKKQALMEPLVDTVDQNQIATNCQLLKSMDISKMSSGDCSFTAPFKLVAARDD 292
YGFDMSCIKK+A+MEPLVDTVDQNQI T+ +LLK+MDISKMSSGD SFTAPFKLVA R+D
Sbjct: 239 YGFDMSCIKKKAMMEPLVDTVDQNQIVTDSRLLKTMDISKMSSGDASFTAPFKLVAQRND 298
Query: 293 FIHAFVAYFDVSFTKCHKLMGFST 316
+IHA VAYFDVSFT CHKL+GFST
Sbjct: 299 YIHALVAYFDVSFTMCHKLLGFST 322
>At2g19670 putative arginine N-methyltransferase
Length = 366
Score = 439 bits (1129), Expect = e-123
Identities = 218/308 (70%), Positives = 260/308 (83%), Gaps = 15/308 (4%)
Query: 13 TNMDNNNNNNNN----HLRFEDADEVIEDVAETSNLDQSMGGCDLDDSNDKTSADYYFDS 68
T+ +NNNN ++ L FEDADE + D + + D ++D TSADYYFDS
Sbjct: 2 TSTENNNNGSDETQTTKLHFEDADESMHDGDDNN----------ADVADDITSADYYFDS 51
Query: 69 YSHFGNFHKEMLKDTVRTKTYQNVIYQNRFLFKNKVVLDVGAGTGILSLFCAKAGAAHVY 128
YSHFG H+EMLKD VRTK+YQ+VIY+N+FL K+K+VLDVGAGTGILSLFCAKAGAAHVY
Sbjct: 52 YSHFG-IHEEMLKDVVRTKSYQDVIYKNKFLIKDKIVLDVGAGTGILSLFCAKAGAAHVY 110
Query: 129 AVECSHMADRAKEIVETNGYSKVITVLKGKIEELELPVPKVDIIISEWMGYFLLFENMLN 188
AVECS MAD AKEIV++NG+S VITVLKGKIEE+ELPVPKVD+IISEWMGYFLL+ENML+
Sbjct: 111 AVECSQMADTAKEIVKSNGFSDVITVLKGKIEEIELPVPKVDVIISEWMGYFLLYENMLD 170
Query: 189 SVLFARDKWLVDDGVILPDIASLYLTAIEDKDYKEDKIEFWNNVYGFDMSCIKKQALMEP 248
+VL+AR+KWLVD G++LPD ASLY+TAIED YK+DK+EFW++VYGFDMSCIK++A+ EP
Sbjct: 171 TVLYARNKWLVDGGIVLPDKASLYVTAIEDAHYKDDKVEFWDDVYGFDMSCIKRRAITEP 230
Query: 249 LVDTVDQNQIATNCQLLKSMDISKMSSGDCSFTAPFKLVAARDDFIHAFVAYFDVSFTKC 308
LVDTVD NQI T+ +LLK+MDISKM++GD SFTAPFKLVA R+D IHA VAYFDVSFT C
Sbjct: 231 LVDTVDGNQIVTDSKLLKTMDISKMAAGDASFTAPFKLVAQRNDHIHALVAYFDVSFTMC 290
Query: 309 HKLMGFST 316
HK MGFST
Sbjct: 291 HKKMGFST 298
>At3g20020 arginine methyltransferase like protein
Length = 435
Score = 213 bits (543), Expect = 9e-56
Identities = 113/247 (45%), Positives = 162/247 (64%), Gaps = 7/247 (2%)
Query: 65 YFDSYSHFGNFHKEMLKDTVRTKTYQNVIYQNRFLFKNKVVLDVGAGTGILSLFCAKAGA 124
YF SY+H G H+EM+KD RT+TY+ I Q++ L + KVV+DVG GTGILS+FCA+AGA
Sbjct: 83 YFHSYAHVG-IHEEMIKDRARTETYREAIMQHQSLIEGKVVVDVGCGTGILSIFCAQAGA 141
Query: 125 AHVYAVECSHMADRAKEIVETNGYSKVITVLKGKIEELELPVPKVDIIISEWMGYFLLFE 184
VYAV+ S +A +AKE+V+ NG S + VL G++E++E+ +VD+IISEWMGY LL+E
Sbjct: 142 KRVYAVDASDIAVQAKEVVKANGLSDKVIVLHGRVEDVEID-EEVDVIISEWMGYMLLYE 200
Query: 185 NMLNSVLFARDKWLVDDGVILPDIASLYLTAIEDKDYKEDKIEFWNNVYGFDMSCI---- 240
+ML SV+ ARD+WL G+ILP A+LY+ I D I+FW NVYG DMS +
Sbjct: 201 SMLGSVITARDRWLKPGGLILPSHATLYMAPISHPDRYSHSIDFWRNVYGIDMSAMMQLA 260
Query: 241 KKQALMEPLVDTVDQNQIATNCQLLKSMDISKMSSGDC-SFTAPFKLVAARDDFIHAFVA 299
K+ A EP V+++ + T +++K +D + + S TA +K + +H F
Sbjct: 261 KQCAFEEPSVESISGENVLTWPEVVKHIDCKTIKIQELDSVTARYKFNSMMRAPMHGFAF 320
Query: 300 YFDVSFT 306
+FDV F+
Sbjct: 321 WFDVEFS 327
>At3g12270 hypothetical protein
Length = 590
Score = 195 bits (496), Expect = 2e-50
Identities = 120/319 (37%), Positives = 176/319 (54%), Gaps = 24/319 (7%)
Query: 3 SRKNNNQTN-NTNMDNNNNNNNNHLRFEDADEVIEDVAETSNLDQSMGGCD--LDDSNDK 59
S N+++ N N + D + +N N L+ AD++I + D CD L + N +
Sbjct: 181 SMSNSDKCNINGSKDVTSLSNCNGLKQSSADDLI-----VNGKDAEPKVCDGRLVNRNIR 235
Query: 60 TSADYYFDSYSHFGNFHKEMLKDTVRTKTYQNVIYQNRFLFKNKVVLDVGAGTGILSLFC 119
+ YF SYS FG H+EML D VRT+ Y++ + +N L VV+DVG GTGILSLF
Sbjct: 236 KVNENYFGSYSSFG-IHREMLSDKVRTEAYRDALLKNPTLLNGSVVMDVGCGTGILSLFA 294
Query: 120 AKAGAAHVYAVECSHMADRAKEIVETNGYSKVITVLKGKIEELELPVP----KVDIIISE 175
AKAGA+ V AVE S + ++ N ++ V+ V +EEL+ + VD+++SE
Sbjct: 295 AKAGASRVVAVEASEKMAKDNKVFNDNEHNGVLEVAHSMVEELDKSIQIQPHSVDVLVSE 354
Query: 176 WMGYFLLFENMLNSVLFARDKWLVDDGVILPDIASLYLTAIEDKDYKEDKIEFWNNVYGF 235
WMGY LL+E+ML+SVL+ARD+WL G ILPD A++++ + FW +VYGF
Sbjct: 355 WMGYCLLYESMLSSVLYARDRWLKPGGAILPDTATMFVAGF---GKGATSLPFWEDVYGF 411
Query: 236 DMSCIKKQ----ALMEPLVDTVDQNQIATNCQLLKSMDISKMSSGDCSFTAPFKL----V 287
DMS I K+ P+VD + + + T LL++ D++ M + FTA L
Sbjct: 412 DMSSIGKEIHDDTTRLPIVDVIAERDLVTQPTLLQTFDLATMKPDEVDFTATATLEPTES 471
Query: 288 AARDDFIHAFVAYFDVSFT 306
A+ H V +FD FT
Sbjct: 472 EAKTRLCHGVVLWFDTGFT 490
>At1g04870 unknown protein
Length = 383
Score = 155 bits (392), Expect = 3e-38
Identities = 96/268 (35%), Positives = 148/268 (54%), Gaps = 31/268 (11%)
Query: 65 YFDSYSHFGNFHKEMLKDTVRTKTYQNVIYQNRFLFKNKVVLDVGAGTGILSLFCAKAGA 124
YF +YS F K+ML D VR Y N ++QN+ F+ K VLDVG G+GIL+++ A+AGA
Sbjct: 35 YFCTYS-FLYHQKDMLSDRVRMDAYFNAVFQNKHHFEGKTVLDVGTGSGILAIWSAQAGA 93
Query: 125 AHVYAVECSHMADRAKEIVETNGYSKVITVLKGKIEELELPVPKVDIIISEWMGYFLLFE 184
VYAVE + MAD A+ +V+ N ++ V++G +E++ LP KVD+IISEWMGYFLL E
Sbjct: 94 RKVYAVEATKMADHARALVKANNLDHIVEVIEGSVEDISLP-EKVDVIISEWMGYFLLRE 152
Query: 185 NMLNSVLFARDKWLVDDGVILPDIASLYLTAIEDKDYKEDKIEF------WNN------- 231
+M +SV+ ARD+WL GV+ P A ++L I+ + +F W+N
Sbjct: 153 SMFDSVISARDRWLKPTGVMYPSHARMWLAPIKSNIADRKRNDFDGAMADWHNFSDEIKS 212
Query: 232 VYGFDMSCI--------KKQALMEPLVDTVDQNQIATNCQLLKSMD-----ISKMSSGDC 278
YG DM + +K + + + ++ QI ++K MD +S++
Sbjct: 213 YYGVDMGVLTKPFAEEQEKYYIQTAMWNDLNPQQIIGTPTIVKEMDCLTASVSEIEEVRS 272
Query: 279 SFTAPFKLVAARDDFIHAFVAYFDVSFT 306
+ T+ + R + F +FDV F+
Sbjct: 273 NVTSVINMEHTR---LCGFGGWFDVQFS 297
>At3g06930 arginine methyltransferase like protein
Length = 534
Score = 151 bits (382), Expect = 4e-37
Identities = 103/307 (33%), Positives = 161/307 (51%), Gaps = 17/307 (5%)
Query: 12 NTNMDNNNNNNNNHLRFED--ADEVIEDVAETSNLDQSMGGCDLDDSNDKTSADYYFDSY 69
N N ++ + FE+ D V++ + N S D+ + +SA YF Y
Sbjct: 94 NIKFKNEKDSKDFCESFEEWRNDSVVQG-SSLQNGTVSANKSKFDNKIEASSAKMYFHYY 152
Query: 70 SHFGNFHKEMLKDTVRTKTYQNVIYQNRFLFKNKVVLDVGAGTGILSLFCAKAGAAHVYA 129
+ + ML+D VRT TY + +N F +VV+DVGAG+GILS+F A+AGA HVYA
Sbjct: 153 GQLLH-QQNMLQDYVRTGTYYAAVMENHSDFAGRVVVDVGAGSGILSMFAAQAGAKHVYA 211
Query: 130 VECSHMADRAKEIVETNG-YSKVITVLKGKIEELELPVPKVDIIISEWMGYFLLFENMLN 188
VE S MA+ A++++ N ++ ITV+KGK+E++ELP K DI+ISE MG L+ E ML
Sbjct: 212 VEASEMAEYARKLIAGNPLFADRITVIKGKVEDIELP-EKADILISEPMGTLLVNERMLE 270
Query: 189 SVLFARDKWLVDDGVILPDIASLYLTAIEDKDY---KEDKIEFW--NNVYGFDMSCIKKQ 243
S + ARD+++ G + P + +++ D+ +K FW N YG D++ +
Sbjct: 271 SYVIARDRFMTPKGKMFPTVGRIHMAPFSDEFLFIEMANKAMFWQQQNYYGVDLTPLYGS 330
Query: 244 A----LMEPLVDTVDQNQIATNCQLLKSMDISKMSSGD-CSFTAPFKLVAARDDFIHAFV 298
A +P+VD D + + + +D ++M D P K A+ +H
Sbjct: 331 AHQGYFSQPVVDAFDPRLLVAS-PMFHMIDFTQMKEEDFYEIDIPLKFTASMCTRMHGLA 389
Query: 299 AYFDVSF 305
+FDV F
Sbjct: 390 CWFDVLF 396
>At5g49020 arginine methyltransferase-like protein
Length = 528
Score = 150 bits (378), Expect = 1e-36
Identities = 100/275 (36%), Positives = 148/275 (53%), Gaps = 14/275 (5%)
Query: 54 DDSNDKTSADYYFDSYSHFGNFHKEMLKDTVRTKTYQNVIYQNRFLFKNKVVLDVGAGTG 113
DD + SA YF Y + + ML+D VRT TY + +NR F +VV+DVGAG+G
Sbjct: 140 DDKIEAASAKMYFHYYGQLLH-QQNMLQDYVRTGTYHAAVMENRSDFSGRVVVDVGAGSG 198
Query: 114 ILSLFCAKAGAAHVYAVECSHMADRAKEIVETNG-YSKVITVLKGKIEELELPVPKVDII 172
ILS+F A AGA HVYAVE S MA+ A++++ N ++ ITV+KGKIE++ELP K D++
Sbjct: 199 ILSMFAALAGAKHVYAVEASEMAEYARKLIAGNPLLAERITVIKGKIEDIELP-EKADVL 257
Query: 173 ISEWMGYFLLFENMLNSVLFARDKWLVDDGVILPDIASLYLTAIEDKDY---KEDKIEFW 229
ISE MG L+ E ML + + ARD++L +G + P + +++ D+ +K FW
Sbjct: 258 ISEPMGTLLVNERMLETYVIARDRFLSPNGKMFPTVGRIHMAPFADEFLFVEMANKALFW 317
Query: 230 --NNVYGFDMSCIKKQA----LMEPLVDTVDQNQIATNCQLLKSMDISKMSSGD-CSFTA 282
N YG D++ + A +P+VD D + + +D + M+
Sbjct: 318 QQQNYYGVDLTPLYVSAHQGYFSQPVVDAFDPRLLVAP-SMFHVIDFTMMTEEQFYEIDI 376
Query: 283 PFKLVAARDDFIHAFVAYFDVSFTKCHKLMGFSTA 317
P K A+ IH +FDV F F+TA
Sbjct: 377 PLKFTASVCTRIHGLACWFDVLFDGSTVQRWFTTA 411
>At4g16570 unknown protein
Length = 724
Score = 57.8 bits (138), Expect = 8e-09
Identities = 47/173 (27%), Positives = 84/173 (48%), Gaps = 16/173 (9%)
Query: 54 DDSNDKTSADYYFDSYSHFGNFHKEMLKDTVRTKTYQNVIYQNRFLFKNKVVLDVGAGTG 113
D+ TS D + S+ +ML D+ R Y+ I + + + VLD+GAGTG
Sbjct: 60 DNDQPGTSTDGLLATTSYL-----DMLNDSRRNIAYRLAI--EKTITEPCHVLDIGAGTG 112
Query: 114 ILSLFCAKA--GAAHVYAVECSH---MADRAKEIVETNGYSKVITVLKGKIEELELP--- 165
+LS+ +A G + C M ++++ NG +K I ++ + +EL++
Sbjct: 113 LLSMMAVRAMRGDSKGMVTACESYLPMVKLMRKVMHKNGMTKNINLINKRSDELKVGSED 172
Query: 166 -VPKVDIIISEWMGYFLLFENMLNSVLFARDKWLVDDGVILPDIASLYLTAIE 217
+ D+++SE + LL E ++ S+ A D LVD+ +P A+ Y +E
Sbjct: 173 IASRADVLVSEILDSELLGEGLIPSLQHAHDMLLVDNPKTVPYRATTYCQLVE 225
>At4g17750 heat shock transcription factor HSF1
Length = 495
Score = 42.7 bits (99), Expect = 3e-04
Identities = 17/34 (50%), Positives = 24/34 (70%)
Query: 5 KNNNQTNNTNMDNNNNNNNNHLRFEDADEVIEDV 38
+NNN NN N +NNNNNNNN+ D++IE++
Sbjct: 453 ENNNNNNNNNNNNNNNNNNNNTNGRHMDKLIEEL 486
>At3g20770 ethylene-insensitive 3
Length = 628
Score = 37.4 bits (85), Expect = 0.011
Identities = 17/42 (40%), Positives = 23/42 (54%)
Query: 6 NNNQTNNTNMDNNNNNNNNHLRFEDADEVIEDVAETSNLDQS 47
NNN +NN NNNNNN +F+ AD + A +N + S
Sbjct: 540 NNNSSNNQTFFQGNNNNNNVFKFDTADHNNFEAAHNNNNNSS 581
>At2g47420 putative dimethyladenosine transferase
Length = 353
Score = 36.6 bits (83), Expect = 0.019
Identities = 37/142 (26%), Positives = 61/142 (42%), Gaps = 7/142 (4%)
Query: 48 MGGCDLDDSNDKTSADYYFDSYSHFGNFHKEMLKDTVRTKTYQNVIYQNRFLFKNKVVLD 107
M G + K S + Y +FHK + ++ + I Q + V+L+
Sbjct: 1 MAGGKIRKEKPKASNRAPSNHYQGGISFHKSKGQHILKNPLLVDSIVQKAGIKSTDVILE 60
Query: 108 VGAGTGILSLFCAKAGAAHVYAVEC-SHMADRAKEIVETNGYSKVITVLKGKIEELELP- 165
+G GTG L+ +AG V AVE S M + + +S + V++G + + ELP
Sbjct: 61 IGPGTGNLTKKLLEAG-KEVIAVELDSRMVLELQRRFQGTPFSNRLKVIQGDVLKTELPR 119
Query: 166 ----VPKVDIIISEWMGYFLLF 183
V + IS + + LLF
Sbjct: 120 FDICVANIPYQISSPLTFKLLF 141
>At4g13760 putative polygalacturonase
Length = 375
Score = 35.4 bits (80), Expect = 0.042
Identities = 39/163 (23%), Positives = 65/163 (38%), Gaps = 30/163 (18%)
Query: 6 NNNQTNNTNMDNNNNNNNNHLRFEDADEVIEDVAETSNLDQSMGGC-------------- 51
N+ T+ M + +N + ++ D+ I ++ T+NLD S C
Sbjct: 168 NSPNTDGIKMGSCSNIHISNTNIGTGDDCIAILSGTTNLDISNVNCGPGHGISVGSLGKN 227
Query: 52 -DLDDSNDKTSADYYFDSYSHFGNFHKEMLKDTVRTKTYQNV---IYQNRFLFKNKVVLD 107
D D D T D F+ S D +R KT+++ I + FL++N ++D
Sbjct: 228 KDEKDVKDLTIRDVIFNGTS-----------DGIRIKTWESSASKILVSNFLYENIQMID 276
Query: 108 VGAGTGILSLFCAKAGAAHVYAVECSHMADRAKEIVETNGYSK 150
VG I +C H E SH+ + ++ G SK
Sbjct: 277 VGKPINIDQKYCPHPPCEHEQKGE-SHVQIQNLKLKNIYGTSK 318
>At3g18810 protein kinase, putative
Length = 700
Score = 35.4 bits (80), Expect = 0.042
Identities = 17/56 (30%), Positives = 29/56 (51%)
Query: 6 NNNQTNNTNMDNNNNNNNNHLRFEDADEVIEDVAETSNLDQSMGGCDLDDSNDKTS 61
NNN NN N NNNN+NNN ++ + + +N + + G + ++ND +
Sbjct: 80 NNNDNNNNNNGNNNNDNNNGNNKDNNNNGNNNNGNNNNGNDNNGNNNNGNNNDNNN 135
Score = 34.3 bits (77), Expect = 0.093
Identities = 13/24 (54%), Positives = 17/24 (70%)
Query: 2 GSRKNNNQTNNTNMDNNNNNNNNH 25
G+ NNN NN N + NNNN+NN+
Sbjct: 75 GNNGNNNNDNNNNNNGNNNNDNNN 98
Score = 33.1 bits (74), Expect = 0.21
Identities = 13/19 (68%), Positives = 15/19 (78%)
Query: 6 NNNQTNNTNMDNNNNNNNN 24
NNN NN N +N+NNNNNN
Sbjct: 71 NNNDGNNGNNNNDNNNNNN 89
Score = 32.7 bits (73), Expect = 0.27
Identities = 12/24 (50%), Positives = 15/24 (62%)
Query: 2 GSRKNNNQTNNTNMDNNNNNNNNH 25
G+ N+ N N DNNNNNN N+
Sbjct: 69 GNNNNDGNNGNNNNDNNNNNNGNN 92
Score = 32.3 bits (72), Expect = 0.36
Identities = 12/24 (50%), Positives = 16/24 (66%)
Query: 2 GSRKNNNQTNNTNMDNNNNNNNNH 25
G+ K+NN N N NNNN N+N+
Sbjct: 99 GNNKDNNNNGNNNNGNNNNGNDNN 122
Score = 32.0 bits (71), Expect = 0.46
Identities = 12/22 (54%), Positives = 15/22 (67%)
Query: 3 SRKNNNQTNNTNMDNNNNNNNN 24
+ NN+ NN N +NNN NNNN
Sbjct: 96 NNNGNNKDNNNNGNNNNGNNNN 117
Score = 31.6 bits (70), Expect = 0.61
Identities = 16/45 (35%), Positives = 22/45 (48%)
Query: 6 NNNQTNNTNMDNNNNNNNNHLRFEDADEVIEDVAETSNLDQSMGG 50
NNN N N +N NNNN N+ D + + N +Q+ GG
Sbjct: 96 NNNGNNKDNNNNGNNNNGNNNNGNDNNGNNNNGNNNDNNNQNNGG 140
Score = 31.2 bits (69), Expect = 0.79
Identities = 11/24 (45%), Positives = 16/24 (65%)
Query: 2 GSRKNNNQTNNTNMDNNNNNNNNH 25
G+ N+N NN +NN+NNN N+
Sbjct: 78 GNNNNDNNNNNNGNNNNDNNNGNN 101
Score = 30.4 bits (67), Expect = 1.3
Identities = 11/23 (47%), Positives = 14/23 (60%)
Query: 2 GSRKNNNQTNNTNMDNNNNNNNN 24
G+ N N N N + NNN+NNN
Sbjct: 113 GNNNNGNDNNGNNNNGNNNDNNN 135
Score = 30.0 bits (66), Expect = 1.8
Identities = 11/23 (47%), Positives = 14/23 (60%)
Query: 2 GSRKNNNQTNNTNMDNNNNNNNN 24
G+ N+N N +NNN NNNN
Sbjct: 90 GNNNNDNNNGNNKDNNNNGNNNN 112
Score = 29.6 bits (65), Expect = 2.3
Identities = 11/23 (47%), Positives = 15/23 (64%)
Query: 3 SRKNNNQTNNTNMDNNNNNNNNH 25
+ + N NN N +NNNNN NN+
Sbjct: 71 NNNDGNNGNNNNDNNNNNNGNNN 93
Score = 29.3 bits (64), Expect = 3.0
Identities = 11/23 (47%), Positives = 16/23 (68%)
Query: 3 SRKNNNQTNNTNMDNNNNNNNNH 25
S + NN + N +NNN+NNNN+
Sbjct: 66 SPQGNNNNDGNNGNNNNDNNNNN 88
Score = 28.9 bits (63), Expect = 3.9
Identities = 11/22 (50%), Positives = 12/22 (54%)
Query: 3 SRKNNNQTNNTNMDNNNNNNNN 24
+ NN N N NNNN NNN
Sbjct: 110 NNNGNNNNGNDNNGNNNNGNNN 131
Score = 28.9 bits (63), Expect = 3.9
Identities = 11/21 (52%), Positives = 12/21 (56%)
Query: 2 GSRKNNNQTNNTNMDNNNNNN 22
G+ N N N N DNNN NN
Sbjct: 118 GNDNNGNNNNGNNNDNNNQNN 138
Score = 28.5 bits (62), Expect = 5.1
Identities = 13/27 (48%), Positives = 17/27 (62%), Gaps = 4/27 (14%)
Query: 3 SRKNNNQTNNTNMDNNN----NNNNNH 25
++ NNN NN N +NNN N NNN+
Sbjct: 101 NKDNNNNGNNNNGNNNNGNDNNGNNNN 127
Score = 28.1 bits (61), Expect = 6.7
Identities = 10/23 (43%), Positives = 14/23 (60%)
Query: 3 SRKNNNQTNNTNMDNNNNNNNNH 25
+ NN NN N + NN +NNN+
Sbjct: 85 NNNNNGNNNNDNNNGNNKDNNNN 107
Score = 28.1 bits (61), Expect = 6.7
Identities = 10/23 (43%), Positives = 15/23 (64%)
Query: 3 SRKNNNQTNNTNMDNNNNNNNNH 25
+ NNN NN + +N NN +NN+
Sbjct: 84 NNNNNNGNNNNDNNNGNNKDNNN 106
>At1g30860 hypothetical protein
Length = 739
Score = 35.0 bits (79), Expect = 0.055
Identities = 18/90 (20%), Positives = 42/90 (46%)
Query: 10 TNNTNMDNNNNNNNNHLRFEDADEVIEDVAETSNLDQSMGGCDLDDSNDKTSADYYFDSY 69
+ + + N + NN ++ E V E++ +DQ + + N + Y D
Sbjct: 421 SRESRLRQNQDENNVGKYMQETRETEGLVHESNEMDQCLDQQETSYLNRWGEQEEYADEQ 480
Query: 70 SHFGNFHKEMLKDTVRTKTYQNVIYQNRFL 99
S++G ++ + L + R ++Y + ++R+L
Sbjct: 481 SYYGEYNDDWLSEIARPRSYWEELRKSRYL 510
>At2g44340 hypothetical protein
Length = 188
Score = 34.3 bits (77), Expect = 0.093
Identities = 13/19 (68%), Positives = 14/19 (73%)
Query: 6 NNNQTNNTNMDNNNNNNNN 24
NNN NN N +NNNNNN N
Sbjct: 161 NNNNNNNNNNNNNNNNNTN 179
Score = 32.7 bits (73), Expect = 0.27
Identities = 12/19 (63%), Positives = 15/19 (78%)
Query: 11 NNTNMDNNNNNNNNHLRFE 29
NN N +NNNNNNNN+ F+
Sbjct: 163 NNNNNNNNNNNNNNNTNFD 181
Score = 32.3 bits (72), Expect = 0.36
Identities = 12/19 (63%), Positives = 14/19 (73%)
Query: 7 NNQTNNTNMDNNNNNNNNH 25
NN NN N +NNNNNNN +
Sbjct: 161 NNNNNNNNNNNNNNNNNTN 179
Score = 27.7 bits (60), Expect = 8.7
Identities = 10/19 (52%), Positives = 12/19 (62%)
Query: 3 SRKNNNQTNNTNMDNNNNN 21
+ NNN NN N +NNN N
Sbjct: 161 NNNNNNNNNNNNNNNNNTN 179
>At5g49560 putative protein
Length = 274
Score = 33.9 bits (76), Expect = 0.12
Identities = 25/95 (26%), Positives = 51/95 (53%), Gaps = 9/95 (9%)
Query: 105 VLDVGAGTGILSLFCAKAGAAHVYAVECSHMADRAK-------EIVETNGYSKVITVLK- 156
+L++G+GTG++ + A +A+V + H+ D EIVE G + L+
Sbjct: 109 ILELGSGTGLVGIAAAITLSANVTVTDLPHVLDNLNFNAEANAEIVERFGGKVNVAPLRW 168
Query: 157 GKIEELELPVPKVDIIISEWMGYF-LLFENMLNSV 190
G+ +++E+ VD+I++ + Y L+E +L ++
Sbjct: 169 GEADDVEVLGQNVDLILASDVVYHDHLYEPLLKTL 203
>At1g28400 unknown protein
Length = 335
Score = 33.9 bits (76), Expect = 0.12
Identities = 24/96 (25%), Positives = 44/96 (45%), Gaps = 6/96 (6%)
Query: 6 NNNQ--TNNTNMDNNNNNNNNHLRFEDADEVIEDVAETSNLDQSMGGCDLDDSNDKTSAD 63
NNN+ N + NNNNNNN+ ++ + V E+ +N D + + + +
Sbjct: 168 NNNKYDENYAKEEFNNNNNNNNYNYKYDENVKEESFPENNEDNKKNVYNSNAYGTELERE 227
Query: 64 YYFDSYSHFGNFHKEMLKDT--VRTKTYQNVIYQNR 97
+ YSH N ++ + DT + +Y +Y +R
Sbjct: 228 TPYKGYSH--NLERQGMSDTRFMEKGSYYYDLYNDR 261
>At3g50850 unknown protein
Length = 251
Score = 33.5 bits (75), Expect = 0.16
Identities = 23/97 (23%), Positives = 51/97 (51%), Gaps = 9/97 (9%)
Query: 105 VLDVGAGTGILSLFCAKAGAAHVYAVECSHMADRAKEIVETN--------GYSKVITVLK 156
++++G+GTGI+ + A A+V + ++ + K + N G V ++
Sbjct: 92 IVELGSGTGIVGIAAAATLGANVTVTDLPNVIENLKFNADANAQVVAKFGGKVHVASLRW 151
Query: 157 GKIEELELPVPKVDIIISEWMGYFL-LFENMLNSVLF 192
G+I+++E VD+I++ + Y + L+E +L ++ F
Sbjct: 152 GEIDDVESLGQNVDLILASDVVYHVHLYEPLLKTLRF 188
>At2g33350 hypothetical protein
Length = 440
Score = 33.5 bits (75), Expect = 0.16
Identities = 21/69 (30%), Positives = 32/69 (45%), Gaps = 6/69 (8%)
Query: 2 GSRKNNNQTNNTNMDNNNNNNNNHLRFEDADEVIEDVAETSNLDQSMGGCDLDDSNDKTS 61
GS +N T T ++NNNNNN + +D D + +N D S+ +D + +
Sbjct: 87 GSFHSNTNTTTTTENSNNNNNNKNTNLQD------DEDDNNNTDLSIIFDSQEDFENDIT 140
Query: 62 ADYYFDSYS 70
A F S S
Sbjct: 141 ASIDFSSSS 149
>At5g28300 GTL1 - like protein
Length = 619
Score = 33.1 bits (74), Expect = 0.21
Identities = 18/60 (30%), Positives = 29/60 (48%), Gaps = 8/60 (13%)
Query: 6 NNNQTNNTN----MDNNNNNNNNHLRFEDADEVIEDVAETSNLDQSMGGCDLDDSNDKTS 61
NNN NNTN + N NN NN+ F + +E + ++ +G D+ N +T+
Sbjct: 161 NNNNNNNTNDHQHIGNYNNKGNNYRIFSEVEEFYHHGHDNEHVSSEVG----DNQNKRTN 216
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.135 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,715,826
Number of Sequences: 26719
Number of extensions: 345646
Number of successful extensions: 2269
Number of sequences better than 10.0: 76
Number of HSP's better than 10.0 without gapping: 52
Number of HSP's successfully gapped in prelim test: 25
Number of HSP's that attempted gapping in prelim test: 1789
Number of HSP's gapped (non-prelim): 227
length of query: 322
length of database: 11,318,596
effective HSP length: 99
effective length of query: 223
effective length of database: 8,673,415
effective search space: 1934171545
effective search space used: 1934171545
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC148360.2


