

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148360.13 + phase: 0
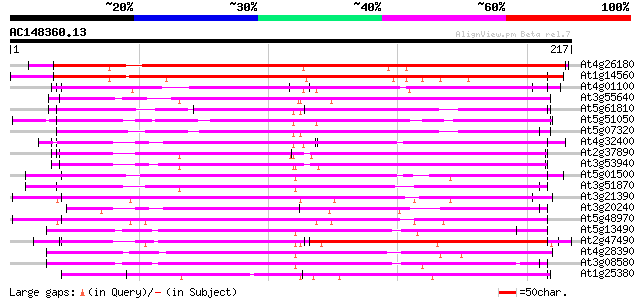
(217 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g26180 putative mitochondrial carrier protein 234 3e-62
At1g14560 mitochondrial carrier like protein 211 2e-55
At4g01100 putative carrier protein 103 5e-23
At3g55640 Ca-dependent solute carrier - like protein 100 5e-22
At5g61810 peroxisomal Ca-dependent solute carrier - like protein 96 1e-20
At5g51050 calcium-binding transporter-like protein 96 1e-20
At5g07320 peroxisomal Ca-dependent solute carrier-like protein 95 2e-20
At4g32400 adenylate translocator (brittle-1) - like protein 95 3e-20
At2g37890 mitochondrial carrier like protein 94 4e-20
At3g53940 unknown protein 91 5e-19
At5g01500 unknown protein 87 9e-18
At3g51870 carrier like protein 85 3e-17
At3g21390 unknown protein 84 7e-17
At3g20240 mitochondrial carrier protein, putative 82 2e-16
At5g48970 mitochondrial carrier protein-like 81 5e-16
At5g13490 adenosine nucleotide translocator 80 8e-16
At2g47490 mitochondrial carrier like protein 79 2e-15
At4g28390 ADP,ATP carrier-like protein 77 7e-15
At3g08580 adenylate translocator 75 3e-14
At1g25380 unknown protein 75 3e-14
>At4g26180 putative mitochondrial carrier protein
Length = 325
Score = 234 bits (596), Expect = 3e-62
Identities = 120/201 (59%), Positives = 150/201 (73%), Gaps = 9/201 (4%)
Query: 18 ELLAGGLAGGFAKTVVAPLE--RLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYK 75
+L+AG AGG A PL+ R K+ +Q V + + +YRGI DC S+TY+
Sbjct: 116 DLVAGSFAGGTAVLFTYPLDLVRTKLAYQT------QVKAIPVEQIIYRGIVDCFSRTYR 169
Query: 76 EGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYKKSIMAKLTCGSVAGLLGQT 135
E G +G+YRGVAP+L+GIFPYAGLKFYFYEEMKRHVP ++K+ I KL CGSVAGLLGQT
Sbjct: 170 ESGARGLYRGVAPSLYGIFPYAGLKFYFYEEMKRHVPPEHKQDISLKLVCGSVAGLLGQT 229
Query: 136 FTYFLEVVRRQMQVQNL-PASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSA 194
TY L+VVRRQMQV+ L A +E +GTM+++ IA+++GWK LFSGLSINY+KVVPS
Sbjct: 230 LTYPLDVVRRQMQVERLYSAVKEETRRGTMQTLFKIAREEGWKQLFSGLSINYLKVVPSV 289
Query: 195 AIGFTVYDTMKSYLRVPSRDE 215
AIGFTVYD MK +LRVP R+E
Sbjct: 290 AIGFTVYDIMKLHLRVPPREE 310
Score = 109 bits (273), Expect = 1e-24
Identities = 74/216 (34%), Positives = 112/216 (51%), Gaps = 19/216 (8%)
Query: 8 ILDHIPLFAKELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIR 67
I+D IPLFAKEL+AGG+ GG AKT VAPLER+KILFQ +E G+
Sbjct: 10 IIDSIPLFAKELIAGGVTGGIAKTAVAPLERIKILFQT-----------RRDEFKRIGLV 58
Query: 68 DCLSKTYKEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVP---EDYKKSIMAKLT 124
++K K G+ G YRG ++ I PYA L + YEE +R + D + + L
Sbjct: 59 GSINKIGKTEGLMGFYRGNGASVARIVPYAALHYMAYEEYRRWIIFGFPDTTRGPLLDLV 118
Query: 125 CGSVAGLLGQTFTYFLEVVRR----QMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLF 180
GS AG FTY L++VR Q QV+ +P E+ +G + ++ G + L+
Sbjct: 119 AGSFAGGTAVLFTYPLDLVRTKLAYQTQVKAIPV-EQIIYRGIVDCFSRTYRESGARGLY 177
Query: 181 SGLSINYIKVVPSAAIGFTVYDTMKSYLRVPSRDEV 216
G++ + + P A + F Y+ MK ++ + ++
Sbjct: 178 RGVAPSLYGIFPYAGLKFYFYEEMKRHVPPEHKQDI 213
Score = 36.6 bits (83), Expect = 0.010
Identities = 30/114 (26%), Positives = 50/114 (43%), Gaps = 17/114 (14%)
Query: 6 ESILDHIPLFAKE-----LLAGGLAGGFAKTVVAPLERLKILFQV---VSPTKLNVSGMV 57
E + H+P K+ L+ G +AG +T+ PL+ ++ QV S K
Sbjct: 199 EEMKRHVPPEHKQDISLKLVCGSVAGLLGQTLTYPLDVVRRQMQVERLYSAVK------- 251
Query: 58 NNEQVYRGIRDCLSKTYKEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHV 111
E+ RG L K +E G K ++ G++ + P + F Y+ MK H+
Sbjct: 252 --EETRRGTMQTLFKIAREEGWKQLFSGLSINYLKVVPSVAIGFTVYDIMKLHL 303
>At1g14560 mitochondrial carrier like protein
Length = 331
Score = 211 bits (538), Expect = 2e-55
Identities = 110/206 (53%), Positives = 145/206 (69%), Gaps = 10/206 (4%)
Query: 18 ELLAGGLAGGFAKTVVAPLE--RLKILFQVVSPTKLNVSGMVNN---EQVYRGIRDCLSK 72
+L+AG AGG A PL+ R K+ +QV S T+ ++ G N + Y GI++ L+
Sbjct: 122 DLVAGSAAGGTAVLCTYPLDLARTKLAYQV-SDTRQSLRGGANGFYRQPTYSGIKEVLAM 180
Query: 73 TYKEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYKKSIMAKLTCGSVAGLL 132
YKEGG +G+YRG+ PTL GI PYAGLKFY YEE+KRHVPE+++ S+ L CG++AGL
Sbjct: 181 AYKEGGPRGLYRGIGPTLIGILPYAGLKFYIYEELKRHVPEEHQNSVRMHLPCGALAGLF 240
Query: 133 GQTFTYFLEVVRRQMQVQNL-PASEEA---ELKGTMRSMVLIAQKQGWKTLFSGLSINYI 188
GQT TY L+VVRRQMQV+NL P + E K T + I + QGWK LF+GLSINYI
Sbjct: 241 GQTITYPLDVVRRQMQVENLQPMTSEGNNKRYKNTFDGLNTIVRTQGWKQLFAGLSINYI 300
Query: 189 KVVPSAAIGFTVYDTMKSYLRVPSRD 214
K+VPS AIGFTVY++MKS++R+P R+
Sbjct: 301 KIVPSVAIGFTVYESMKSWMRIPPRE 326
Score = 87.8 bits (216), Expect = 4e-18
Identities = 66/222 (29%), Positives = 105/222 (46%), Gaps = 25/222 (11%)
Query: 1 MAASSESILDHIPLFAKELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNE 60
++A S++D +P+ AK L+AGG AG AKT VAPLER+KIL Q + N+
Sbjct: 9 LSADVMSLVDTLPVLAKTLIAGGAAGAIAKTAVAPLERIKILLQTRT-----------ND 57
Query: 61 QVYRGIRDCLSKTYKEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPE---DYKK 117
G+ L K + G G Y+G ++ I PYA L + YE + + E
Sbjct: 58 FKTLGVSQSLKKVLQFDGPLGFYKGNGASVIRIIPYAALHYMTYEVYRDWILEKNLPLGS 117
Query: 118 SIMAKLTCGSVAGLLGQTFTYFLEVVRRQM--QVQNLPASEEAELKGTMR--------SM 167
+ L GS AG TY L++ R ++ QV + S G R +
Sbjct: 118 GPIVDLVAGSAAGGTAVLCTYPLDLARTKLAYQVSDTRQSLRGGANGFYRQPTYSGIKEV 177
Query: 168 VLIAQKQGW-KTLFSGLSINYIKVVPSAAIGFTVYDTMKSYL 208
+ +A K+G + L+ G+ I ++P A + F +Y+ +K ++
Sbjct: 178 LAMAYKEGGPRGLYRGIGPTLIGILPYAGLKFYIYEELKRHV 219
>At4g01100 putative carrier protein
Length = 352
Score = 103 bits (258), Expect = 5e-23
Identities = 59/201 (29%), Positives = 109/201 (53%), Gaps = 21/201 (10%)
Query: 17 KELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKE 76
K L AGG+AGG ++T VAPLER+KIL QV +P + SG V L ++
Sbjct: 40 KSLFAGGVAGGVSRTAVAPLERMKILLQVQNPHNIKYSGTVQG----------LKHIWRT 89
Query: 77 GGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYKK---------SIMAKLTCGS 127
G++G+++G I P + +KF+ YE+ + Y++ + + +L G+
Sbjct: 90 EGLRGLFKGNGTNCARIVPNSAVKFFSYEQASNGILYMYRQRTGNENAQLTPLLRLGAGA 149
Query: 128 VAGLLGQTFTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINY 187
AG++ + TY +++VR ++ VQ A+ + +G ++ + +++G + L+ G +
Sbjct: 150 TAGIIAMSATYPMDMVRGRLTVQT--ANSPYQYRGIAHALATVLREEGPRALYRGWLPSV 207
Query: 188 IKVVPSAAIGFTVYDTMKSYL 208
I VVP + F+VY+++K +L
Sbjct: 208 IGVVPYVGLNFSVYESLKDWL 228
Score = 103 bits (258), Expect = 5e-23
Identities = 65/212 (30%), Positives = 105/212 (48%), Gaps = 28/212 (13%)
Query: 21 AGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIK 80
AG AG A + P++ ++ V + N+ YRGI L+ +E G +
Sbjct: 147 AGATAGIIAMSATYPMDMVRGRLTVQT---------ANSPYQYRGIAHALATVLREEGPR 197
Query: 81 GIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVP--------EDYKKSIMAKLTCGSVAGLL 132
+YRG P++ G+ PY GL F YE +K + E+ + +++ +LTCG++AG +
Sbjct: 198 ALYRGWLPSVIGVVPYVGLNFSVYESLKDWLVKENPYGLVENNELTVVTRLTCGAIAGTV 257
Query: 133 GQTFTYFLEVVRRQMQVQNLP-----------ASEEAELKGTMRSMVLIAQKQGWKTLFS 181
GQT Y L+V+RR+MQ+ ++ E G + + + +G+ L+
Sbjct: 258 GQTIAYPLDVIRRRMQMVGWKDASAIVTGEGRSTASLEYTGMVDAFRKTVRHEGFGALYK 317
Query: 182 GLSINYIKVVPSAAIGFTVYDTMKSYLRVPSR 213
GL N +KVVPS AI F Y+ +K L V R
Sbjct: 318 GLVPNSVKVVPSIAIAFVTYEMVKDVLGVEFR 349
Score = 44.7 bits (104), Expect = 4e-05
Identities = 28/86 (32%), Positives = 46/86 (52%), Gaps = 3/86 (3%)
Query: 117 KSIMAKLTCGSVAGLLGQTFTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGW 176
KSI L G VAG + +T LE ++ +QVQN + GT++ + I + +G
Sbjct: 36 KSICKSLFAGGVAGGVSRTAVAPLERMKILLQVQN---PHNIKYSGTVQGLKHIWRTEGL 92
Query: 177 KTLFSGLSINYIKVVPSAAIGFTVYD 202
+ LF G N ++VP++A+ F Y+
Sbjct: 93 RGLFKGNGTNCARIVPNSAVKFFSYE 118
Score = 41.2 bits (95), Expect = 4e-04
Identities = 24/94 (25%), Positives = 41/94 (43%), Gaps = 4/94 (4%)
Query: 19 LLAGGLAGGFAKTVVAPLERLKILFQVV----SPTKLNVSGMVNNEQVYRGIRDCLSKTY 74
L G +AG +T+ PL+ ++ Q+V + + G Y G+ D KT
Sbjct: 248 LTCGAIAGTVGQTIAYPLDVIRRRMQMVGWKDASAIVTGEGRSTASLEYTGMVDAFRKTV 307
Query: 75 KEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMK 108
+ G +Y+G+ P + P + F YE +K
Sbjct: 308 RHEGFGALYKGLVPNSVKVVPSIAIAFVTYEMVK 341
>At3g55640 Ca-dependent solute carrier - like protein
Length = 332
Score = 100 bits (250), Expect = 5e-22
Identities = 70/205 (34%), Positives = 104/205 (50%), Gaps = 23/205 (11%)
Query: 16 AKELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYR--GIRDCLSKT 73
A +LLAGGLAG F+KT APL RL ILFQ V GM N R I S+
Sbjct: 35 ASQLLAGGLAGAFSKTCTAPLSRLTILFQ--------VQGMHTNAAALRKPSILHEASRI 86
Query: 74 YKEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHV-----PEDYKKSIMAKL----T 124
E G+K ++G T+ PY+ + FY YE K+ + E++K+ I + L
Sbjct: 87 LNEEGLKAFWKGNLVTIAHRLPYSSVNFYAYEHYKKFMYMVTGMENHKEGISSNLFVHFV 146
Query: 125 CGSVAGLLGQTFTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLS 184
G +AG+ + TY L++VR ++ Q ++ G ++ I +G L+ GL
Sbjct: 147 AGGLAGITAASATYPLDLVRTRLAAQ----TKVIYYSGIWHTLRSITTDEGILGLYKGLG 202
Query: 185 INYIKVVPSAAIGFTVYDTMKSYLR 209
+ V PS AI F+VY++++SY R
Sbjct: 203 TTLVGVGPSIAISFSVYESLRSYWR 227
Score = 94.7 bits (234), Expect = 3e-20
Identities = 58/192 (30%), Positives = 97/192 (50%), Gaps = 13/192 (6%)
Query: 20 LAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGI 79
+AGGLAG A + PL+ ++ ++ + TK+ Y GI L + GI
Sbjct: 146 VAGGLAGITAASATYPLDLVRT--RLAAQTKVIY---------YSGIWHTLRSITTDEGI 194
Query: 80 KGIYRGVAPTLFGIFPYAGLKFYFYEEMKRH--VPEDYKKSIMAKLTCGSVAGLLGQTFT 137
G+Y+G+ TL G+ P + F YE ++ + + IM L CGS++G+ T T
Sbjct: 195 LGLYKGLGTTLVGVGPSIAISFSVYESLRSYWRSTRPHDSPIMVSLACGSLSGIASSTAT 254
Query: 138 YFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIG 197
+ L++VRR+ Q++ + G + ++ I Q +G + L+ G+ Y KVVP I
Sbjct: 255 FPLDLVRRRKQLEGIGGRAVVYKTGLLGTLKRIVQTEGARGLYRGILPEYYKVVPGVGIC 314
Query: 198 FTVYDTMKSYLR 209
F Y+T+K Y +
Sbjct: 315 FMTYETLKLYFK 326
>At5g61810 peroxisomal Ca-dependent solute carrier - like protein
Length = 478
Score = 95.9 bits (237), Expect = 1e-20
Identities = 61/195 (31%), Positives = 98/195 (49%), Gaps = 23/195 (11%)
Query: 19 LLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGG 78
LLAGGLAG A+T + P++ +K Q V ++++ +D + + G
Sbjct: 299 LLAGGLAGAVAQTAIYPMDLVKTRLQTFVSE-------VGTPKLWKLTKDI----WIQEG 347
Query: 79 IKGIYRGVAPTLFGIFPYAGLKFYFYEEMK-----RHVPEDYKKSIMAKLTCGSVAGLLG 133
+ YRG+ P+L GI PYAG+ YE +K + + + + +L CG +G LG
Sbjct: 348 PRAFYRGLCPSLIGIIPYAGIDLAAYETLKDLSRAHFLHDTAEPGPLIQLGCGMTSGALG 407
Query: 134 QTFTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPS 193
+ Y L+V+R +MQ + S E T+R +G K + G+ N+ KV+PS
Sbjct: 408 ASCVYPLQVIRTRMQADSSKTSMGQEFLKTLRG-------EGLKGFYRGIFPNFFKVIPS 460
Query: 194 AAIGFTVYDTMKSYL 208
A+I + VY+ MK L
Sbjct: 461 ASISYLVYEAMKKNL 475
Score = 74.3 bits (181), Expect = 5e-14
Identities = 52/195 (26%), Positives = 91/195 (46%), Gaps = 19/195 (9%)
Query: 16 AKELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYK 75
+K LLAGG+AG ++T APL+RLK+ QV ++ G+ + K ++
Sbjct: 205 SKLLLAGGIAGAVSRTATAPLDRLKVALQV--------------QRTNLGVVPTIKKIWR 250
Query: 76 EGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHV-PEDYKKSIMAKLTCGSVAGLLGQ 134
E + G +RG + + P + +KF YE +K + D +L G +AG + Q
Sbjct: 251 EDKLLGFFRGNGLNVAKVAPESAIKFAAYEMLKPIIGGADGDIGTSGRLLAGGLAGAVAQ 310
Query: 135 TFTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSA 194
T Y +++V+ ++Q K T + ++G + + GL + I ++P A
Sbjct: 311 TAIYPMDLVKTRLQTFVSEVGTPKLWKLTKDIWI----QEGPRAFYRGLCPSLIGIIPYA 366
Query: 195 AIGFTVYDTMKSYLR 209
I Y+T+K R
Sbjct: 367 GIDLAAYETLKDLSR 381
Score = 42.4 bits (98), Expect = 2e-04
Identities = 17/43 (39%), Positives = 26/43 (59%)
Query: 72 KTYKEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPED 114
KT + G+KG YRG+ P F + P A + + YE MK+++ D
Sbjct: 436 KTLRGEGLKGFYRGIFPNFFKVIPSASISYLVYEAMKKNLALD 478
>At5g51050 calcium-binding transporter-like protein
Length = 487
Score = 95.9 bits (237), Expect = 1e-20
Identities = 64/196 (32%), Positives = 105/196 (52%), Gaps = 20/196 (10%)
Query: 19 LLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGG 78
L AGG+AG A+ + PL+ +K Q T + +G V ++ +D L G
Sbjct: 307 LFAGGMAGAVAQASIYPLDLVKTRLQ----TYTSQAG-VAVPRLGTLTKDILVHE----G 357
Query: 79 IKGIYRGVAPTLFGIFPYAGLKFYFYEEMK----RHVPEDYKKSIMAKLTCGSVAGLLGQ 134
+ Y+G+ P+L GI PYAG+ YE +K ++ +D + + +L CG+++G LG
Sbjct: 358 PRAFYKGLFPSLLGIIPYAGIDLAAYETLKDLSRTYILQDAEPGPLVQLGCGTISGALGA 417
Query: 135 TFTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSA 194
T Y L+VVR +MQ + S + G R + ++G++ L+ GL N +KVVP+A
Sbjct: 418 TCVYPLQVVRTRMQAERARTS----MSGVFRRTI---SEEGYRALYKGLLPNLLKVVPAA 470
Query: 195 AIGFTVYDTMKSYLRV 210
+I + VY+ MK L +
Sbjct: 471 SITYMVYEAMKKSLEL 486
Score = 92.4 bits (228), Expect = 2e-19
Identities = 58/213 (27%), Positives = 102/213 (47%), Gaps = 22/213 (10%)
Query: 2 AASSESILDHIPLFAKELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQ 61
A E I HI + +AGG+AG ++T APL+RLK+L Q+ ++
Sbjct: 196 AVIPEGISKHIKR-SNYFIAGGIAGAASRTATAPLDRLKVLLQI--------------QK 240
Query: 62 VYRGIRDCLSKTYKEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYKK---- 117
IR+ + +K+GG++G +RG + + P + +KFY YE K + E+ +
Sbjct: 241 TDARIREAIKLIWKQGGVRGFFRGNGLNIVKVAPESAIKFYAYELFKNAIGENMGEDKAD 300
Query: 118 -SIMAKLTCGSVAGLLGQTFTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGW 176
+L G +AG + Q Y L++V+ ++Q A GT+ +L+ +G
Sbjct: 301 IGTTVRLFAGGMAGAVAQASIYPLDLVKTRLQTYTSQAGVAVPRLGTLTKDILV--HEGP 358
Query: 177 KTLFSGLSINYIKVVPSAAIGFTVYDTMKSYLR 209
+ + GL + + ++P A I Y+T+K R
Sbjct: 359 RAFYKGLFPSLLGIIPYAGIDLAAYETLKDLSR 391
Score = 39.7 bits (91), Expect = 0.001
Identities = 26/97 (26%), Positives = 46/97 (46%), Gaps = 14/97 (14%)
Query: 18 ELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEG 77
+L G ++G T V PL+ ++ Q + ++SG V+R +T E
Sbjct: 405 QLGCGTISGALGATCVYPLQVVRTRMQA-ERARTSMSG------VFR-------RTISEE 450
Query: 78 GIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPED 114
G + +Y+G+ P L + P A + + YE MK+ + D
Sbjct: 451 GYRALYKGLLPNLLKVVPAASITYMVYEAMKKSLELD 487
>At5g07320 peroxisomal Ca-dependent solute carrier-like protein
Length = 479
Score = 95.1 bits (235), Expect = 2e-20
Identities = 59/191 (30%), Positives = 101/191 (51%), Gaps = 21/191 (10%)
Query: 19 LLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGG 78
L+AGG+AG A+T + P++ +K Q VS ++++ +D + G
Sbjct: 300 LMAGGMAGALAQTAIYPMDLVKTRLQTC------VSEGGKAPKLWKLTKDI----WVREG 349
Query: 79 IKGIYRGVAPTLFGIFPYAGLKFYFYEEMK----RHVPEDYKKSIMAKLTCGSVAGLLGQ 134
+ Y+G+ P+L GI PYAG+ YE +K ++ +D + + +L+CG +G LG
Sbjct: 350 PRAFYKGLFPSLLGIVPYAGIDLAAYETLKDLSRTYILQDTEPGPLIQLSCGMTSGALGA 409
Query: 135 TFTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSA 194
+ Y L+VVR +MQ + + + E TM+ +G + + GL N +KVVP+A
Sbjct: 410 SCVYPLQVVRTRMQADSSKTTMKQEFMNTMKG-------EGLRGFYRGLLPNLLKVVPAA 462
Query: 195 AIGFTVYDTMK 205
+I + VY+ MK
Sbjct: 463 SITYIVYEAMK 473
Score = 84.3 bits (207), Expect = 4e-17
Identities = 55/192 (28%), Positives = 94/192 (48%), Gaps = 18/192 (9%)
Query: 19 LLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGG 78
LLAGGLAG ++T APL+RLK++ QV ++ + G+ + K ++E
Sbjct: 209 LLAGGLAGAVSRTATAPLDRLKVVLQV--------------QRAHAGVLPTIKKIWREDK 254
Query: 79 IKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHV-PEDYKKSIMAKLTCGSVAGLLGQTFT 137
+ G +RG + + P + +KF YE +K + ED +L G +AG L QT
Sbjct: 255 LMGFFRGNGLNVMKVAPESAIKFCAYEMLKPMIGGEDGDIGTSGRLMAGGMAGALAQTAI 314
Query: 138 YFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIG 197
Y +++V+ ++Q SE + + I ++G + + GL + + +VP A I
Sbjct: 315 YPMDLVKTRLQT---CVSEGGKAPKLWKLTKDIWVREGPRAFYKGLFPSLLGIVPYAGID 371
Query: 198 FTVYDTMKSYLR 209
Y+T+K R
Sbjct: 372 LAAYETLKDLSR 383
Score = 39.3 bits (90), Expect = 0.002
Identities = 25/97 (25%), Positives = 41/97 (41%), Gaps = 14/97 (14%)
Query: 18 ELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEG 77
+L G +G + V PL+ ++ Q S +N T K
Sbjct: 397 QLSCGMTSGALGASCVYPLQVVRTRMQADSSKTTMKQEFMN--------------TMKGE 442
Query: 78 GIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPED 114
G++G YRG+ P L + P A + + YE MK+++ D
Sbjct: 443 GLRGFYRGLLPNLLKVVPAASITYIVYEAMKKNMALD 479
>At4g32400 adenylate translocator (brittle-1) - like protein
Length = 392
Score = 94.7 bits (234), Expect = 3e-20
Identities = 67/200 (33%), Positives = 99/200 (49%), Gaps = 19/200 (9%)
Query: 12 IPLFAKELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLS 71
IP+ A LLAG AG + PLE +K T+L + VY+GI D
Sbjct: 202 IPIPAS-LLAGACAGVSQTLLTYPLELVK--------TRLTIQ-----RGVYKGIFDAFL 247
Query: 72 KTYKEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMK---RHVPEDYKKSIMAKLTCGSV 128
K +E G +YRG+AP+L G+ PYA ++ Y+ ++ R + K + L GS+
Sbjct: 248 KIIREEGPTELYRGLAPSLIGVVPYAATNYFAYDSLRKAYRSFSKQEKIGNIETLLIGSL 307
Query: 129 AGLLGQTFTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYI 188
AG L T T+ LEV R+ MQV S K + ++V I + +G + GL + +
Sbjct: 308 AGALSSTATFPLEVARKHMQVG--AVSGRVVYKNMLHALVTILEHEGILGWYKGLGPSCL 365
Query: 189 KVVPSAAIGFTVYDTMKSYL 208
K+VP+A I F Y+ K L
Sbjct: 366 KLVPAAGISFMCYEACKKIL 385
Score = 73.2 bits (178), Expect = 1e-13
Identities = 55/203 (27%), Positives = 99/203 (48%), Gaps = 24/203 (11%)
Query: 17 KELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKE 76
+ LL+G +AG ++TVVAPLE ++ V S G ++ +V+ I K
Sbjct: 112 RRLLSGAVAGAVSRTVVAPLETIRTHLMVGS-------GGNSSTEVFSDIM-------KH 157
Query: 77 GGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVP----EDYKKSIMAKLTCGSVAGLL 132
G G++RG + + P ++ + +E + + + ++ K I A L G+ AG+
Sbjct: 158 EGWTGLFRGNLVNVIRVAPARAVELFVFETVNKKLSPPHGQESKIPIPASLLAGACAGVS 217
Query: 133 GQTFTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVP 192
TY LE+V+ ++ +Q KG + + I +++G L+ GL+ + I VVP
Sbjct: 218 QTLLTYPLELVKTRLTIQR------GVYKGIFDAFLKIIREEGPTELYRGLAPSLIGVVP 271
Query: 193 SAAIGFTVYDTMKSYLRVPSRDE 215
AA + YD+++ R S+ E
Sbjct: 272 YAATNYFAYDSLRKAYRSFSKQE 294
Score = 49.7 bits (117), Expect = 1e-06
Identities = 31/100 (31%), Positives = 48/100 (48%), Gaps = 9/100 (9%)
Query: 19 LLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGG 78
LL G LAG + T PLE + QV G V+ VY+ + L + G
Sbjct: 302 LLIGSLAGALSSTATFPLEVARKHMQV---------GAVSGRVVYKNMLHALVTILEHEG 352
Query: 79 IKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYKKS 118
I G Y+G+ P+ + P AG+ F YE K+ + E+ +++
Sbjct: 353 ILGWYKGLGPSCLKLVPAAGISFMCYEACKKILIENNQEA 392
Score = 43.5 bits (101), Expect = 9e-05
Identities = 27/96 (28%), Positives = 44/96 (45%), Gaps = 7/96 (7%)
Query: 120 MAKLTCGSVAGLLGQTFTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTL 179
+ +L G+VAG + +T LE +R + V + S M+ +GW L
Sbjct: 111 LRRLLSGAVAGAVSRTVVAPLETIRTHLMVGSGGNSSTEVFSDIMKH-------EGWTGL 163
Query: 180 FSGLSINYIKVVPSAAIGFTVYDTMKSYLRVPSRDE 215
F G +N I+V P+ A+ V++T+ L P E
Sbjct: 164 FRGNLVNVIRVAPARAVELFVFETVNKKLSPPHGQE 199
>At2g37890 mitochondrial carrier like protein
Length = 337
Score = 94.4 bits (233), Expect = 4e-20
Identities = 62/193 (32%), Positives = 92/193 (47%), Gaps = 17/193 (8%)
Query: 20 LAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGI 79
++GGLAG A T PL+ ++ T+L N Y+GI +E GI
Sbjct: 153 VSGGLAGITAATATYPLDLVR--------TRLAAQ---RNAIYYQGIEHTFRTICREEGI 201
Query: 80 KGIYRGVAPTLFGIFPYAGLKFYFYEEMK----RHVPEDYKKSIMAKLTCGSVAGLLGQT 135
G+Y+G+ TL G+ P + F YE MK H P D ++ L G +AG + T
Sbjct: 202 LGLYKGLGATLLGVGPSLAINFAAYESMKLFWHSHRPND--SDLVVSLVSGGLAGAVSST 259
Query: 136 FTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAA 195
TY L++VRR+MQV+ G + I + +G+K ++ G+ Y KVVP
Sbjct: 260 ATYPLDLVRRRMQVEGAGGRARVYNTGLFGTFKHIFKSEGFKGIYRGILPEYYKVVPGVG 319
Query: 196 IGFTVYDTMKSYL 208
I F YD ++ L
Sbjct: 320 IVFMTYDALRRLL 332
Score = 77.8 bits (190), Expect = 4e-15
Identities = 59/202 (29%), Positives = 92/202 (45%), Gaps = 23/202 (11%)
Query: 17 KELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYR--GIRDCLSKTY 74
+ LLAGG+AG +KT APL RL ILFQ + GM + V +R S+
Sbjct: 43 QNLLAGGIAGAISKTCTAPLARLTILFQ--------LQGMQSEGAVLSRPNLRREASRII 94
Query: 75 KEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEM-----KRHVPEDY----KKSIMAKLTC 125
E G + ++G T+ PY + FY YE+ V + + + +
Sbjct: 95 NEEGYRAFWKGNLVTVVHRIPYTAVNFYAYEKYNLFFNSNPVVQSFIGNTSGNPIVHFVS 154
Query: 126 GSVAGLLGQTFTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSI 185
G +AG+ T TY L++VR ++ Q +G + I +++G L+ GL
Sbjct: 155 GGLAGITAATATYPLDLVRTRLAAQR----NAIYYQGIEHTFRTICREEGILGLYKGLGA 210
Query: 186 NYIKVVPSAAIGFTVYDTMKSY 207
+ V PS AI F Y++MK +
Sbjct: 211 TLLGVGPSLAINFAAYESMKLF 232
Score = 52.4 bits (124), Expect = 2e-07
Identities = 28/92 (30%), Positives = 46/92 (49%), Gaps = 9/92 (9%)
Query: 19 LLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYR-GIRDCLSKTYKEG 77
L++GGLAG + T PL+ ++ ++ V G +VY G+ +K
Sbjct: 247 LVSGGLAGAVSSTATYPLDLVR--------RRMQVEGAGGRARVYNTGLFGTFKHIFKSE 298
Query: 78 GIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKR 109
G KGIYRG+ P + + P G+ F Y+ ++R
Sbjct: 299 GFKGIYRGILPEYYKVVPGVGIVFMTYDALRR 330
>At3g53940 unknown protein
Length = 365
Score = 90.9 bits (224), Expect = 5e-19
Identities = 57/193 (29%), Positives = 94/193 (48%), Gaps = 17/193 (8%)
Query: 20 LAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGI 79
++GGLAG A + PL+ ++ T+L+ N Y+G+ +E GI
Sbjct: 181 VSGGLAGLTAASATYPLDLVR--------TRLSAQ---RNSIYYQGVGHAFRTICREEGI 229
Query: 80 KGIYRGVAPTLFGIFPYAGLKFYFYEEMKR----HVPEDYKKSIMAKLTCGSVAGLLGQT 135
G+Y+G+ TL G+ P + F YE K H P D + + L CGS++G++ T
Sbjct: 230 LGLYKGLGATLLGVGPSLAISFAAYETFKTFWLSHRPND--SNAVVSLGCGSLSGIVSST 287
Query: 136 FTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAA 195
T+ L++VRR+MQ++ G + I + +G + L+ G+ Y KVVP
Sbjct: 288 ATFPLDLVRRRMQLEGAGGRARVYTTGLFGTFKHIFKTEGMRGLYRGIIPEYYKVVPGVG 347
Query: 196 IGFTVYDTMKSYL 208
I F ++ +K L
Sbjct: 348 IAFMTFEELKKLL 360
Score = 90.1 bits (222), Expect = 8e-19
Identities = 65/202 (32%), Positives = 95/202 (46%), Gaps = 23/202 (11%)
Query: 17 KELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYR--GIRDCLSKTY 74
+ LLAGG+AG F+KT APL RL ILFQ + GM + + I S+
Sbjct: 71 ERLLAGGIAGAFSKTCTAPLARLTILFQ--------IQGMQSEAAILSSPNIWHEASRIV 122
Query: 75 KEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMK-----RHVPEDYKKS----IMAKLTC 125
KE G + ++G T+ PY + FY YEE K V + YK + I
Sbjct: 123 KEEGFRAFWKGNLVTVAHRLPYGAVNFYAYEEYKTFLHSNPVLQSYKGNAGVDISVHFVS 182
Query: 126 GSVAGLLGQTFTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSI 185
G +AGL + TY L++VR ++ Q +G + I +++G L+ GL
Sbjct: 183 GGLAGLTAASATYPLDLVRTRLSAQR----NSIYYQGVGHAFRTICREEGILGLYKGLGA 238
Query: 186 NYIKVVPSAAIGFTVYDTMKSY 207
+ V PS AI F Y+T K++
Sbjct: 239 TLLGVGPSLAISFAAYETFKTF 260
>At5g01500 unknown protein
Length = 415
Score = 86.7 bits (213), Expect = 9e-18
Identities = 60/204 (29%), Positives = 105/204 (51%), Gaps = 15/204 (7%)
Query: 7 SILDHIPLFAKELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGI 66
++L +P A AG AG AK+V APL+R+K+L Q V + + G
Sbjct: 107 ALLSIVPKDAALFFAGAFAGAAAKSVTAPLDRIKLLMQT-----HGVRAGQQSAKKAIGF 161
Query: 67 RDCLSKTYKEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKR-HVPEDYKKSIMAKLTC 125
+ ++ KE GIKG ++G P + I PY+ ++ + YE K+ +D + S++ +L
Sbjct: 162 IEAITLIGKEEGIKGYWKGNLPQVIRIVPYSAVQLFAYETYKKLFRGKDGQLSVLGRLGA 221
Query: 126 GSVAGLLGQTFTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVL-IAQKQGWKTLFSGLS 184
G+ AG+ TY L+V+R ++ V+ P TM + L + +++G + ++GL
Sbjct: 222 GACAGMTSTLITYPLDVLRLRLAVE--PGYR------TMSQVALNMLREEGVASFYNGLG 273
Query: 185 INYIKVVPSAAIGFTVYDTMKSYL 208
+ + + P AI F V+D +K L
Sbjct: 274 PSLLSIAPYIAINFCVFDLVKKSL 297
Score = 77.0 bits (188), Expect = 7e-15
Identities = 48/194 (24%), Positives = 87/194 (44%), Gaps = 20/194 (10%)
Query: 21 AGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIK 80
AG AG + + PL+ L++ V E YR + +E G+
Sbjct: 221 AGACAGMTSTLITYPLDVLRLRLAV--------------EPGYRTMSQVALNMLREEGVA 266
Query: 81 GIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYKKSIMAKLTCGSVAGLLGQTFTYFL 140
Y G+ P+L I PY + F ++ +K+ +PE Y++ + L VA + Y L
Sbjct: 267 SFYNGLGPSLLSIAPYIAINFCVFDLVKKSLPEKYQQKTQSSLLTAVVAAAIATGTCYPL 326
Query: 141 EVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGFTV 200
+ +RRQMQ++ P K + + I ++G L+ G N +K +P+++I T
Sbjct: 327 DTIRRQMQLKGTP------YKSVLDAFSGIIAREGVVGLYRGFVPNALKSMPNSSIKLTT 380
Query: 201 YDTMKSYLRVPSRD 214
+D +K + ++
Sbjct: 381 FDIVKKLIAASEKE 394
Score = 40.0 bits (92), Expect = 0.001
Identities = 27/101 (26%), Positives = 50/101 (48%), Gaps = 7/101 (6%)
Query: 111 VPEDYKKSIMAKLTCGSVAGLLGQTFTYFLEVVRRQMQVQNLPASEEAELK--GTMRSMV 168
VP+D A G+ AG ++ T L+ ++ MQ + A +++ K G + ++
Sbjct: 112 VPKD-----AALFFAGAFAGAAAKSVTAPLDRIKLLMQTHGVRAGQQSAKKAIGFIEAIT 166
Query: 169 LIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSYLR 209
LI +++G K + G I++VP +A+ Y+T K R
Sbjct: 167 LIGKEEGIKGYWKGNLPQVIRIVPYSAVQLFAYETYKKLFR 207
>At3g51870 carrier like protein
Length = 381
Score = 85.1 bits (209), Expect = 3e-17
Identities = 57/206 (27%), Positives = 102/206 (48%), Gaps = 19/206 (9%)
Query: 7 SILDHIPLFAKELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYR-- 64
+IL +P A AG LAG AKTV APL+R+K+L Q G+ +Q +
Sbjct: 79 AILALVPKDAAIFAAGALAGAAAKTVTAPLDRIKLLMQ--------THGIRLGQQSAKKA 130
Query: 65 -GIRDCLSKTYKEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKR-HVPEDYKKSIMAK 122
G + ++ KE G+KG ++G P + + PY+ ++ YE K +D + S++ +
Sbjct: 131 IGFIEAITLIAKEEGVKGYWKGNLPQVIRVLPYSAVQLLAYESYKNLFKGKDDQLSVIGR 190
Query: 123 LTCGSVAGLLGQTFTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSG 182
L G+ AG+ TY L+V+R ++ V E + + + + + +G + + G
Sbjct: 191 LAAGACAGMTSTLLTYPLDVLRLRLAV-------EPRYRTMSQVALSMLRDEGIASFYYG 243
Query: 183 LSINYIKVVPSAAIGFTVYDTMKSYL 208
L + + + P A+ F ++D +K L
Sbjct: 244 LGPSLVGIAPYIAVNFCIFDLVKKSL 269
Score = 77.0 bits (188), Expect = 7e-15
Identities = 50/187 (26%), Positives = 85/187 (44%), Gaps = 20/187 (10%)
Query: 19 LLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGG 78
L AG AG + + PL+ L++ V E YR + ++ G
Sbjct: 191 LAAGACAGMTSTLLTYPLDVLRLRLAV--------------EPRYRTMSQVALSMLRDEG 236
Query: 79 IKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYKKSIMAKLTCGSVAGLLGQTFTY 138
I Y G+ P+L GI PY + F ++ +K+ +PE+Y+K + L ++ + Y
Sbjct: 237 IASFYYGLGPSLVGIAPYIAVNFCIFDLVKKSLPEEYRKKAQSSLLTAVLSAGIATLTCY 296
Query: 139 FLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGF 198
L+ VRRQMQ++ P K + I + G L+ G N +K +P+++I
Sbjct: 297 PLDTVRRQMQMRGTP------YKSIPEAFAGIIDRDGLIGLYRGFLPNALKTLPNSSIRL 350
Query: 199 TVYDTMK 205
T +D +K
Sbjct: 351 TTFDMVK 357
Score = 40.0 bits (92), Expect = 0.001
Identities = 26/101 (25%), Positives = 52/101 (50%), Gaps = 7/101 (6%)
Query: 111 VPEDYKKSIMAKLTCGSVAGLLGQTFTYFLEVVRRQMQVQNLPASEEAELK--GTMRSMV 168
VP+D A G++AG +T T L+ ++ MQ + +++ K G + ++
Sbjct: 84 VPKD-----AAIFAAGALAGAAAKTVTAPLDRIKLLMQTHGIRLGQQSAKKAIGFIEAIT 138
Query: 169 LIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSYLR 209
LIA+++G K + G I+V+P +A+ Y++ K+ +
Sbjct: 139 LIAKEEGVKGYWKGNLPQVIRVLPYSAVQLLAYESYKNLFK 179
Score = 29.3 bits (64), Expect = 1.7
Identities = 20/99 (20%), Positives = 38/99 (38%), Gaps = 13/99 (13%)
Query: 19 LLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGG 78
LL L+ G A PL+ ++ Q+ Y+ I + + G
Sbjct: 281 LLTAVLSAGIATLTCYPLDTVRRQMQMRGTP-------------YKSIPEAFAGIIDRDG 327
Query: 79 IKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYKK 117
+ G+YRG P P + ++ ++ +KR + K+
Sbjct: 328 LIGLYRGFLPNALKTLPNSSIRLTTFDMVKRLIATSEKQ 366
>At3g21390 unknown protein
Length = 335
Score = 83.6 bits (205), Expect = 7e-17
Identities = 59/198 (29%), Positives = 96/198 (47%), Gaps = 13/198 (6%)
Query: 21 AGGLAGGFAKTVVAPLERLKILFQV-VSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGI 79
AGG+AG ++ V +PL+ +KI FQV + PT + Y G+ ++E G+
Sbjct: 21 AGGVAGAISRMVTSPLDVIKIRFQVQLEPTATWALKDSQLKPKYNGLFRTTKDIFREEGL 80
Query: 80 KGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYKKSIMAKLT------CGSVAGLLG 133
G +RG P L + PY ++F ++K K A+L+ G++AG
Sbjct: 81 SGFWRGNVPALLMVVPYTSIQFAVLHKVKSFAAGSSKAENHAQLSPYLSYISGALAGCAA 140
Query: 134 QTFTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVL-IAQKQGWKTLFSGLSINYIKVVP 192
+Y +++R + Q P ++ MRS L I Q +G K L++GLS I+++P
Sbjct: 141 TVGSYPFDLLRTVLASQGEP-----KVYPNMRSAFLSIVQTRGIKGLYAGLSPTLIEIIP 195
Query: 193 SAAIGFTVYDTMKSYLRV 210
A + F YDT K + V
Sbjct: 196 YAGLQFGTYDTFKRWSMV 213
Score = 73.6 bits (179), Expect = 8e-14
Identities = 61/225 (27%), Positives = 92/225 (40%), Gaps = 36/225 (16%)
Query: 2 AASSESILDHIPLFAK-ELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNE 60
AA S +H L ++G LAG A P + L+ V
Sbjct: 112 AAGSSKAENHAQLSPYLSYISGALAGCAATVGSYPFDLLR-----------TVLASQGEP 160
Query: 61 QVYRGIRDCLSKTYKEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHV--------- 111
+VY +R + GIKG+Y G++PTL I PYAGL+F Y+ KR
Sbjct: 161 KVYPNMRSAFLSIVQTRGIKGLYAGLSPTLIEIIPYAGLQFGTYDTFKRWSMVYNKRYRS 220
Query: 112 -------PEDYKKSIMAKLTCGSVAGLLGQTFTYFLEVVRRQMQVQNLPAS-------EE 157
P D S L CG +G + + + L+VV+++ QV+ L E
Sbjct: 221 SSSSSTNPSDSLSSFQLFL-CGLASGTVSKLVCHPLDVVKKRFQVEGLQRHPKYGARVEL 279
Query: 158 AELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYD 202
K + I + +GW L+ G+ + IK P+ A+ F Y+
Sbjct: 280 NAYKNMFDGLGQILRSEGWHGLYKGIVPSTIKAAPAGAVTFVAYE 324
Score = 36.6 bits (83), Expect = 0.010
Identities = 30/107 (28%), Positives = 52/107 (48%), Gaps = 8/107 (7%)
Query: 117 KSIMAKLTCGSVAGLLGQTFTYFLEVVRRQMQVQNLP----ASEEAELK----GTMRSMV 168
K + + G VAG + + T L+V++ + QVQ P A ++++LK G R+
Sbjct: 13 KRAVIDASAGGVAGAISRMVTSPLDVIKIRFQVQLEPTATWALKDSQLKPKYNGLFRTTK 72
Query: 169 LIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSYLRVPSRDE 215
I +++G + G + VVP +I F V +KS+ S+ E
Sbjct: 73 DIFREEGLSGFWRGNVPALLMVVPYTSIQFAVLHKVKSFAAGSSKAE 119
>At3g20240 mitochondrial carrier protein, putative
Length = 348
Score = 82.0 bits (201), Expect = 2e-16
Identities = 54/190 (28%), Positives = 95/190 (49%), Gaps = 20/190 (10%)
Query: 23 GLAGGFAKTVVA-PLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIKG 81
G + G A T+V PLE LK +L VS ++Y + + + ++ GI+G
Sbjct: 166 GASAGIASTLVCHPLEVLK--------DRLTVS-----PEIYPSLSLAIPRIFRADGIRG 212
Query: 82 IYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYKKSIMAK---LTCGSVAGLLGQTFTY 138
Y G+ PTL G+ PY+ ++ Y++MK + K +++ L G++AGL T ++
Sbjct: 213 FYAGLGPTLVGMLPYSTCYYFMYDKMKTSYCKSKNKKALSRPEMLVLGALAGLTASTISF 272
Query: 139 FLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGF 198
LEV R+++ V L + + +V +K+G L+ G + +KV+PS+ I +
Sbjct: 273 PLEVARKRLMVGALKGECPPNMAAAIAEVV---KKEGVMGLYRGWGASCLKVMPSSGITW 329
Query: 199 TVYDTMKSYL 208
Y+ K L
Sbjct: 330 VFYEAWKDIL 339
Score = 41.6 bits (96), Expect = 3e-04
Identities = 28/98 (28%), Positives = 52/98 (52%), Gaps = 16/98 (16%)
Query: 113 EDYKKSIMAK-LTCGSVAGLLGQTFTYFLEVVRRQMQV----QNLPASEEAELKGTMRSM 167
+D+ KS A+ G++AG + + LE +R +M V +++P S
Sbjct: 41 KDFFKSREAREFLSGALAGAMTKAVLAPLETIRTRMIVGVGSRSIPGS-----------F 89
Query: 168 VLIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMK 205
+ + QKQGW+ L++G IN I+++P+ AI ++ +K
Sbjct: 90 LEVVQKQGWQGLWAGNEINMIRIIPTQAIELGTFEWVK 127
Score = 38.1 bits (87), Expect = 0.004
Identities = 47/215 (21%), Positives = 89/215 (40%), Gaps = 47/215 (21%)
Query: 16 AKELLAGGLAGGFAKTVVAPLE--RLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKT 73
A+E L+G LAG K V+APLE R +++ V S R I +
Sbjct: 49 AREFLSGALAGAMTKAVLAPLETIRTRMIVGVGS----------------RSIPGSFLEV 92
Query: 74 YKEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPE---DYKKSIMAKL------- 123
++ G +G++ G + I P ++ +E +KR + KK AK+
Sbjct: 93 VQKQGWQGLWAGNEINMIRIIPTQAIELGTFEWVKRAMTSAQVKLKKIEDAKIEIGDFSF 152
Query: 124 -----------TCGSVAGLLGQTFTYFLEVVRRQMQVQNLPASEEAELKGTMR-SMVLIA 171
G+ AG+ + LEV++ ++ V E+ ++ ++ I
Sbjct: 153 SPSISWISPVAVAGASAGIASTLVCHPLEVLKDRLTV-------SPEIYPSLSLAIPRIF 205
Query: 172 QKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMKS 206
+ G + ++GL + ++P + + +YD MK+
Sbjct: 206 RADGIRGFYAGLGPTLVGMLPYSTCYYFMYDKMKT 240
Score = 37.7 bits (86), Expect = 0.005
Identities = 25/90 (27%), Positives = 43/90 (47%), Gaps = 10/90 (11%)
Query: 19 LLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGG 78
L+ G LAG A T+ PLE V+ +L V + + + +++ K+ G
Sbjct: 257 LVLGALAGLTASTISFPLE--------VARKRLMVGAL--KGECPPNMAAAIAEVVKKEG 306
Query: 79 IKGIYRGVAPTLFGIFPYAGLKFYFYEEMK 108
+ G+YRG + + P +G+ + FYE K
Sbjct: 307 VMGLYRGWGASCLKVMPSSGITWVFYEAWK 336
>At5g48970 mitochondrial carrier protein-like
Length = 339
Score = 80.9 bits (198), Expect = 5e-16
Identities = 56/198 (28%), Positives = 97/198 (48%), Gaps = 15/198 (7%)
Query: 21 AGGLAGGFAKTVVAPLERLKILFQV-VSPTKL--NVSGMVNNEQVYRGIRDCLSKTYKEG 77
AG ++GG +++V +PL+ +KI FQV + PT V G ++ Y G+ ++E
Sbjct: 24 AGAISGGVSRSVTSPLDVIKIRFQVQLEPTTSWGLVRGNLSGASKYTGMVQATKDIFREE 83
Query: 78 GIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYKK------SIMAKLTCGSVAGL 131
G +G +RG P L + PY ++F ++K K S G++AG
Sbjct: 84 GFRGFWRGNVPALLMVMPYTSIQFTVLHKLKSFASGSTKTEDHIHLSPYLSFVSGALAGC 143
Query: 132 LGQTFTYFLEVVRRQMQVQNLPASEEAELKGTMRS-MVLIAQKQGWKTLFSGLSINYIKV 190
+Y +++R + Q E ++ TMRS V I Q +G + L++GL+ +++
Sbjct: 144 AATLGSYPFDLLRTILASQG-----EPKVYPTMRSAFVDIIQSRGIRGLYNGLTPTLVEI 198
Query: 191 VPSAAIGFTVYDTMKSYL 208
VP A + F YD K ++
Sbjct: 199 VPYAGLQFGTYDMFKRWM 216
Score = 73.2 bits (178), Expect = 1e-13
Identities = 57/229 (24%), Positives = 96/229 (41%), Gaps = 33/229 (14%)
Query: 2 AASSESILDHIPLFAK-ELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNE 60
A+ S DHI L ++G LAG A P + L+ + K
Sbjct: 117 ASGSTKTEDHIHLSPYLSFVSGALAGCAATLGSYPFDLLRTILASQGEPK---------- 166
Query: 61 QVYRGIRDCLSKTYKEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYKKSIM 120
VY +R + GI+G+Y G+ PTL I PYAGL+F Y+ KR + + + +
Sbjct: 167 -VYPTMRSAFVDIIQSRGIRGLYNGLTPTLVEIVPYAGLQFGTYDMFKRWMMDWNRYKLS 225
Query: 121 AKL--------------TCGSVAGLLGQTFTYFLEVVRRQMQVQNLPAS-------EEAE 159
+K+ CG AG + + L+VV+++ Q++ L E
Sbjct: 226 SKIPINVDTNLSSFQLFICGLGAGTSAKLVCHPLDVVKKRFQIEGLQRHPRYGARVERRA 285
Query: 160 LKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSYL 208
+ + + I +GW L+ G+ + +K P+ A+ F Y+ +L
Sbjct: 286 YRNMLDGLRQIMISEGWHGLYKGIVPSTVKAAPAGAVTFVAYEFTSDWL 334
Score = 33.5 bits (75), Expect = 0.088
Identities = 24/109 (22%), Positives = 54/109 (49%), Gaps = 10/109 (9%)
Query: 117 KSIMAKLTCGSVAGLLGQTFTYFLEVVRRQMQVQNLPA----------SEEAELKGTMRS 166
K + + G+++G + ++ T L+V++ + QVQ P S ++ G +++
Sbjct: 16 KRALIDASAGAISGGVSRSVTSPLDVIKIRFQVQLEPTTSWGLVRGNLSGASKYTGMVQA 75
Query: 167 MVLIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSYLRVPSRDE 215
I +++G++ + G + V+P +I FTV +KS+ ++ E
Sbjct: 76 TKDIFREEGFRGFWRGNVPALLMVMPYTSIQFTVLHKLKSFASGSTKTE 124
>At5g13490 adenosine nucleotide translocator
Length = 385
Score = 80.1 bits (196), Expect = 8e-16
Identities = 54/202 (26%), Positives = 94/202 (45%), Gaps = 12/202 (5%)
Query: 15 FAKELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTY 74
FA + + GG++ +KT AP+ER+K+L Q + ++ +G + + Y+GIRDC +T
Sbjct: 84 FAIDFMMGGVSAAVSKTAAAPIERVKLLIQ--NQDEMLKAGRLT--EPYKGIRDCFGRTI 139
Query: 75 KEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKR-----HVPEDYKKSIMAKLTCGSVA 129
++ GI ++RG + FP L F F + KR + Y K L G A
Sbjct: 140 RDEGIGSLWRGNTANVIRYFPTQALNFAFKDYFKRLFNFKKDKDGYWKWFAGNLASGGAA 199
Query: 130 GLLGQTFTYFLEVVRRQMQVQNLPASE---EAELKGTMRSMVLIAQKQGWKTLFSGLSIN 186
G F Y L+ R ++ + A + E + G + + G L+ G +I+
Sbjct: 200 GASSLLFVYSLDYARTRLANDSKSAKKGGGERQFNGLVDVYKKTLKSDGIAGLYRGFNIS 259
Query: 187 YIKVVPSAAIGFTVYDTMKSYL 208
++ + F +YD++K L
Sbjct: 260 CAGIIVYRGLYFGLYDSVKPVL 281
Score = 69.3 bits (168), Expect = 1e-12
Identities = 52/183 (28%), Positives = 85/183 (46%), Gaps = 8/183 (4%)
Query: 15 FAKELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTY 74
FA L +GG AG + V L+ + ++ + +K G E+ + G+ D KT
Sbjct: 189 FAGNLASGGAAGASSLLFVYSLDYART--RLANDSKSAKKG--GGERQFNGLVDVYKKTL 244
Query: 75 KEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKR-HVPEDYKKSIMAKLTCGSVAGLLG 133
K GI G+YRG + GI Y GL F Y+ +K + D + S A G +
Sbjct: 245 KSDGIAGLYRGFNISCAGIIVYRGLYFGLYDSVKPVLLTGDLQDSFFASFALGWLITNGA 304
Query: 134 QTFTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPS 193
+Y ++ VRR+M + + E + K + + I +K+G K+LF G N ++ V
Sbjct: 305 GLASYPIDTVRRRMM---MTSGEAVKYKSSFDAFSQIVKKEGAKSLFKGAGANILRAVAG 361
Query: 194 AAI 196
A +
Sbjct: 362 AGV 364
>At2g47490 mitochondrial carrier like protein
Length = 312
Score = 79.0 bits (193), Expect = 2e-15
Identities = 61/201 (30%), Positives = 97/201 (47%), Gaps = 14/201 (6%)
Query: 21 AGGLAGGFAKTVVAPLERLKILFQVVSPTKL---NVSGMVNNEQVYRGIRDCLSKTYKEG 77
AG AG A T V PL+ +K FQV KL N+ G + I L + +K
Sbjct: 19 AGAAAGVVAATFVCPLDVIKTRFQVHGLPKLGDANIKGSL--------IVGSLEQIFKRE 70
Query: 78 GIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHV-PEDYKKSIMAKLTCGSVAGLLGQTF 136
G++G+YRG++PT+ + + F Y+++K + D+K S+ A + S AG
Sbjct: 71 GMRGLYRGLSPTVMALLSNWAIYFTMYDQLKSFLCSNDHKLSVGANVLAASGAGAATTIA 130
Query: 137 TYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAI 196
T L VV+ ++Q Q + K T ++ IA ++G + L+SGL + + + AI
Sbjct: 131 TNPLWVVKTRLQTQGMRVG-IVPYKSTFSALRRIAYEEGIRGLYSGL-VPALAGISHVAI 188
Query: 197 GFTVYDTMKSYLRVPSRDEVD 217
F Y+ +K YL VD
Sbjct: 189 QFPTYEMIKVYLAKKGDKSVD 209
Score = 68.2 bits (165), Expect = 3e-12
Identities = 56/210 (26%), Positives = 90/210 (42%), Gaps = 18/210 (8%)
Query: 10 DHIPLFAKELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDC 69
DH +LA AG PL +K T+L GM Y+
Sbjct: 108 DHKLSVGANVLAASGAGAATTIATNPLWVVK--------TRLQTQGMRVGIVPYKSTFSA 159
Query: 70 LSKTYKEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYKKSI-----MAKLT 124
L + E GI+G+Y G+ P L GI + ++F YE +K ++ + KS+
Sbjct: 160 LRRIAYEEGIRGLYSGLVPALAGI-SHVAIQFPTYEMIKVYLAKKGDKSVDNLNARDVAV 218
Query: 125 CGSVAGLLGQTFTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLS 184
S+A + T TY EVVR ++Q Q E G + + +K G+ + G +
Sbjct: 219 ASSIAKIFASTLTYPHEVVRARLQEQG--HHSEKRYSGVRDCIKKVFEKDGFPGFYRGCA 276
Query: 185 INYIKVVPSAAIGFTVYDTMKSYL--RVPS 212
N ++ P+A I FT ++ + +L +PS
Sbjct: 277 TNLLRTTPAAVITFTSFEMVHRFLVTHIPS 306
Score = 60.8 bits (146), Expect = 5e-10
Identities = 31/94 (32%), Positives = 58/94 (60%), Gaps = 2/94 (2%)
Query: 117 KSIMAKLTCGSVAGLLGQTFTYFLEVVRRQMQVQNLPASEEAELKGTM--RSMVLIAQKQ 174
K+++ G+ AG++ TF L+V++ + QV LP +A +KG++ S+ I +++
Sbjct: 11 KNVLCNAAAGAAAGVVAATFVCPLDVIKTRFQVHGLPKLGDANIKGSLIVGSLEQIFKRE 70
Query: 175 GWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSYL 208
G + L+ GLS + ++ + AI FT+YD +KS+L
Sbjct: 71 GMRGLYRGLSPTVMALLSNWAIYFTMYDQLKSFL 104
Score = 47.4 bits (111), Expect = 6e-06
Identities = 30/99 (30%), Positives = 49/99 (49%), Gaps = 13/99 (13%)
Query: 20 LAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGI 79
+A +A FA T+ P E ++ +L G ++E+ Y G+RDC+ K +++ G
Sbjct: 218 VASSIAKIFASTLTYPHEVVR--------ARLQEQGH-HSEKRYSGVRDCIKKVFEKDGF 268
Query: 80 KGIYRGVAPTLFGIFPYAGLKFYFYEEMKR----HVPED 114
G YRG A L P A + F +E + R H+P +
Sbjct: 269 PGFYRGCATNLLRTTPAAVITFTSFEMVHRFLVTHIPSE 307
>At4g28390 ADP,ATP carrier-like protein
Length = 379
Score = 77.0 bits (188), Expect = 7e-15
Identities = 59/208 (28%), Positives = 98/208 (46%), Gaps = 21/208 (10%)
Query: 15 FAKELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTY 74
F + L GG++ +KT AP+ER+K+L Q + ++ +G ++ + Y+GI DC ++T
Sbjct: 79 FLIDFLMGGVSAAVSKTAAAPIERVKLLIQ--NQDEMIKAGRLS--EPYKGISDCFARTV 134
Query: 75 KEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKR-----HVPEDYKKSIMAKLTCGSVA 129
K+ G+ ++RG + FP L F F + KR + Y K L G A
Sbjct: 135 KDEGMLALWRGNTANVIRYFPTQALNFAFKDYFKRLFNFKKEKDGYWKWFAGNLASGGAA 194
Query: 130 GLLGQTFTYFLEVVRRQMQVQNLPASEEAELKGTMR---SMVLIAQK----QGWKTLFSG 182
G F Y L+ R + L +A KG R MV + +K G L+ G
Sbjct: 195 GASSLLFVYSLDYARTR-----LANDAKAAKKGGQRQFNGMVDVYKKTIASDGIVGLYRG 249
Query: 183 LSINYIKVVPSAAIGFTVYDTMKSYLRV 210
+I+ + +V + F +YD++K + V
Sbjct: 250 FNISCVGIVVYRGLYFGLYDSLKPVVLV 277
>At3g08580 adenylate translocator
Length = 381
Score = 75.1 bits (183), Expect = 3e-14
Identities = 53/202 (26%), Positives = 92/202 (45%), Gaps = 12/202 (5%)
Query: 15 FAKELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTY 74
FA + L GG++ +KT AP+ER+K+L Q + ++ +G ++ + Y+GI DC +T
Sbjct: 80 FALDFLMGGVSAAVSKTAAAPIERVKLLIQ--NQDEMIKAGRLS--EPYKGIGDCFGRTI 135
Query: 75 KEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKR-----HVPEDYKKSIMAKLTCGSVA 129
K+ G ++RG + FP L F F + KR + Y K L G A
Sbjct: 136 KDEGFGSLWRGNTANVIRYFPTQALNFAFKDYFKRLFNFKKDRDGYWKWFAGNLASGGAA 195
Query: 130 GLLGQTFTYFLEVVRRQMQVQNLPASEEA---ELKGTMRSMVLIAQKQGWKTLFSGLSIN 186
G F Y L+ R ++ A + + G + + G L+ G +I+
Sbjct: 196 GASSLLFVYSLDYARTRLANDAKAAKKGGGGRQFDGLVDVYRKTLKTDGIAGLYRGFNIS 255
Query: 187 YIKVVPSAAIGFTVYDTMKSYL 208
+ ++ + F +YD++K L
Sbjct: 256 CVGIIVYRGLYFGLYDSVKPVL 277
Score = 65.5 bits (158), Expect = 2e-11
Identities = 53/192 (27%), Positives = 88/192 (45%), Gaps = 9/192 (4%)
Query: 15 FAKELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTY 74
FA L +GG AG + V L+ + ++ + K G + + G+ D KT
Sbjct: 185 FAGNLASGGAAGASSLLFVYSLDYART--RLANDAKAAKKG--GGGRQFDGLVDVYRKTL 240
Query: 75 KEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKR-HVPEDYKKSIMAKLTCGSVAGLLG 133
K GI G+YRG + GI Y GL F Y+ +K + D + S A G V
Sbjct: 241 KTDGIAGLYRGFNISCVGIIVYRGLYFGLYDSVKPVLLTGDLQDSFFASFALGWVITNGA 300
Query: 134 QTFTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPS 193
+Y ++ VRR+M + + E + K ++ + I + +G K+LF G N ++ V
Sbjct: 301 GLASYPIDTVRRRMM---MTSGEAVKYKSSLDAFKQILKNEGAKSLFKGAGANILRAVAG 357
Query: 194 AAIGFTVYDTMK 205
A + + YD ++
Sbjct: 358 AGV-LSGYDKLQ 368
>At1g25380 unknown protein
Length = 363
Score = 75.1 bits (183), Expect = 3e-14
Identities = 45/170 (26%), Positives = 86/170 (50%), Gaps = 8/170 (4%)
Query: 46 VSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIKGIYRGVAPTLFGIFPYAGLKFYFYE 105
V T+L G+ Y+ + S+ E G++G+Y G+ P+L G+ + ++F YE
Sbjct: 140 VVKTRLMTQGIRPGVVPYKSVMSAFSRICHEEGVRGLYSGILPSLAGV-SHVAIQFPAYE 198
Query: 106 EMKRHVPEDYKKSIMAKLTCG------SVAGLLGQTFTYFLEVVRRQMQVQNLPASEEAE 159
++K+++ + S+ L+ G S+A ++ TY EV+R ++Q Q + E +
Sbjct: 199 KIKQYMAKMDNTSV-ENLSPGNVAIASSIAKVIASILTYPHEVIRAKLQEQGQIRNAETK 257
Query: 160 LKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSYLR 209
G + + + + +G L+ G + N ++ PSA I FT Y+ M + R
Sbjct: 258 YSGVIDCITKVFRSEGIPGLYRGCATNLLRTTPSAVITFTTYEMMLRFFR 307
Score = 68.6 bits (166), Expect = 2e-12
Identities = 56/191 (29%), Positives = 88/191 (45%), Gaps = 12/191 (6%)
Query: 21 AGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRG--IRDCLSKTYKEGG 78
AG AG A T V PL+ +K QV+ + SG RG I L KE G
Sbjct: 23 AGATAGAIAATFVCPLDVIKTRLQVLGLPEAPASGQ-------RGGVIITSLKNIIKEEG 75
Query: 79 IKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHV-PEDYKKSIMAKLTCGSVAGLLGQTFT 137
+G+YRG++PT+ + P + F Y ++K + D K SI + + + AG T
Sbjct: 76 YRGMYRGLSPTIIALLPNWAVYFSVYGKLKDVLQSSDGKLSIGSNMIAAAGAGAATSIAT 135
Query: 138 YFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIG 197
L VV+ ++ Q + K M + I ++G + L+SG+ + + V AI
Sbjct: 136 NPLWVVKTRLMTQGIRPG-VVPYKSVMSAFSRICHEEGVRGLYSGI-LPSLAGVSHVAIQ 193
Query: 198 FTVYDTMKSYL 208
F Y+ +K Y+
Sbjct: 194 FPAYEKIKQYM 204
Score = 52.4 bits (124), Expect = 2e-07
Identities = 29/100 (29%), Positives = 58/100 (58%), Gaps = 4/100 (4%)
Query: 114 DYK--KSIMAKLTCGSVAGLLGQTFTYFLEVVRRQMQVQNLPASEEAELKG--TMRSMVL 169
DY+ + + A G+ AG + TF L+V++ ++QV LP + + +G + S+
Sbjct: 10 DYRSIREVAANAGAGATAGAIAATFVCPLDVIKTRLQVLGLPEAPASGQRGGVIITSLKN 69
Query: 170 IAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSYLR 209
I +++G++ ++ GLS I ++P+ A+ F+VY +K L+
Sbjct: 70 IIKEEGYRGMYRGLSPTIIALLPNWAVYFSVYGKLKDVLQ 109
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.138 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,503,014
Number of Sequences: 26719
Number of extensions: 179880
Number of successful extensions: 660
Number of sequences better than 10.0: 64
Number of HSP's better than 10.0 without gapping: 56
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 359
Number of HSP's gapped (non-prelim): 159
length of query: 217
length of database: 11,318,596
effective HSP length: 95
effective length of query: 122
effective length of database: 8,780,291
effective search space: 1071195502
effective search space used: 1071195502
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Medicago: description of AC148360.13


