

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148349.9 - phase: 0
(149 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
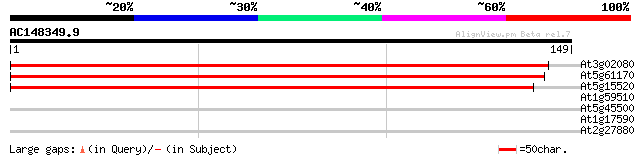
Score E
Sequences producing significant alignments: (bits) Value
At3g02080 putative 40S ribosomal protein S19 253 3e-68
At5g61170 40S ribsomal protein S19 - like 251 1e-67
At5g15520 40S RIBOSOMAL PROTEIN S19 - like 246 3e-66
At1g59510 unknown protein 27 4.1
At5g45500 unknown protein 27 5.3
At1g17590 transcription factor, putative 27 5.3
At2g27880 Argonaute (AGO1)-like protein 26 9.0
>At3g02080 putative 40S ribosomal protein S19
Length = 143
Score = 253 bits (646), Expect = 3e-68
Identities = 116/143 (81%), Positives = 133/143 (92%)
Query: 1 MASSKTVKDVSPREFVNAYSAHLKRSGKLELPEWTDLVKTAKFKELAPYDPDWYYVRAAS 60
MA+ KTVKDVSP +FV AY++HLKRSGK+ELP WTD+VKT K KELAPYDPDWYY+RAAS
Sbjct: 1 MATGKTVKDVSPHDFVKAYASHLKRSGKIELPTWTDIVKTGKLKELAPYDPDWYYIRAAS 60
Query: 61 MARKIYVRGGLGVGSFQRIYGGSQRNGSRPPHFCKSSGAIARHILQQLENMNLIEMDTKG 120
MARK+Y+RGGLGVG+F+RIYGGS+RNGSRPPHFCKSSG IARHILQQLE MN++E+DTKG
Sbjct: 61 MARKVYLRGGLGVGAFRRIYGGSKRNGSRPPHFCKSSGGIARHILQQLETMNIVELDTKG 120
Query: 121 GRKITSSGRRDLDQVAGRILVAP 143
GR+ITSSG+RDLDQVAGRI V P
Sbjct: 121 GRRITSSGQRDLDQVAGRIAVEP 143
>At5g61170 40S ribsomal protein S19 - like
Length = 143
Score = 251 bits (640), Expect = 1e-67
Identities = 115/142 (80%), Positives = 132/142 (91%)
Query: 1 MASSKTVKDVSPREFVNAYSAHLKRSGKLELPEWTDLVKTAKFKELAPYDPDWYYVRAAS 60
MA+ KTVKDVSP EFV AY+AHLKRSGK+ELP WTD+VKT K KELAPYDPDWYY+RAAS
Sbjct: 1 MATGKTVKDVSPHEFVKAYAAHLKRSGKIELPLWTDIVKTGKLKELAPYDPDWYYIRAAS 60
Query: 61 MARKIYVRGGLGVGSFQRIYGGSQRNGSRPPHFCKSSGAIARHILQQLENMNLIEMDTKG 120
MARK+Y+RGGLGVG+F+RIYGGS+RNGSRPPHFCKSSG +ARHILQQL+ MN++++DTKG
Sbjct: 61 MARKVYLRGGLGVGAFRRIYGGSKRNGSRPPHFCKSSGGVARHILQQLQTMNIVDLDTKG 120
Query: 121 GRKITSSGRRDLDQVAGRILVA 142
GRKITSSG+RDLDQVAGRI A
Sbjct: 121 GRKITSSGQRDLDQVAGRIAAA 142
>At5g15520 40S RIBOSOMAL PROTEIN S19 - like
Length = 143
Score = 246 bits (628), Expect = 3e-66
Identities = 113/139 (81%), Positives = 131/139 (93%)
Query: 1 MASSKTVKDVSPREFVNAYSAHLKRSGKLELPEWTDLVKTAKFKELAPYDPDWYYVRAAS 60
MA+ KTVKDVSP +FV AY++HLKRSGK+ELP WTD+VKT + KELAPYDPDWYY+RAAS
Sbjct: 1 MATGKTVKDVSPHDFVKAYASHLKRSGKIELPLWTDIVKTGRLKELAPYDPDWYYIRAAS 60
Query: 61 MARKIYVRGGLGVGSFQRIYGGSQRNGSRPPHFCKSSGAIARHILQQLENMNLIEMDTKG 120
MARKIY+RGGLGVG+F+RIYGGS+RNGSRPPHFCKSSG IARHILQQLE M+++E+DTKG
Sbjct: 61 MARKIYLRGGLGVGAFRRIYGGSKRNGSRPPHFCKSSGGIARHILQQLETMSIVELDTKG 120
Query: 121 GRKITSSGRRDLDQVAGRI 139
GR+ITSSG+RDLDQVAGRI
Sbjct: 121 GRRITSSGQRDLDQVAGRI 139
>At1g59510 unknown protein
Length = 381
Score = 26.9 bits (58), Expect = 4.1
Identities = 29/121 (23%), Positives = 49/121 (39%), Gaps = 17/121 (14%)
Query: 26 SGKLEL-------PEWTDLVKTAKFKELAPYDPDWYYVRAASMARKIYVRGGLGVGSFQR 78
SG++E+ P W +L+ K +EL + R S A +Y+ +GV ++ +
Sbjct: 168 SGEIEIGSDDSLKPRWMNLLSDDKCRELLADCIE----RFTSSAVSVYIDKTVGVNTYDQ 223
Query: 79 IYGGSQRNGSRPPHFCKSSGAIARHILQQLENMNLIEMD--TKGGRKITSSGRRDLDQVA 136
I+ G + P H + + LE D T G K SS R+ ++
Sbjct: 224 IFAGL----TNPKHRDSARDVLVSVCNGALETFMRTSHDVFTSSGEKTDSSLRKSENREN 279
Query: 137 G 137
G
Sbjct: 280 G 280
>At5g45500 unknown protein
Length = 489
Score = 26.6 bits (57), Expect = 5.3
Identities = 19/78 (24%), Positives = 33/78 (41%), Gaps = 4/78 (5%)
Query: 3 SSKTVKDVSPREFVNAYSAHLKRSGKLELPEWTDLVKTAKFKELAPYD----PDWYYVRA 58
S K ++V P+E + K + P +L KT K +L + P+W
Sbjct: 343 SEKGDENVKPKENIGILERAATMFRKEKDPTAPELPKTLKKLDLQCFPGEHLPEWLEPDN 402
Query: 59 ASMARKIYVRGGLGVGSF 76
K+Y++GG+ + F
Sbjct: 403 LLNVEKLYIKGGIKLTGF 420
>At1g17590 transcription factor, putative
Length = 328
Score = 26.6 bits (57), Expect = 5.3
Identities = 29/116 (25%), Positives = 49/116 (42%), Gaps = 12/116 (10%)
Query: 15 FVNAYSAHL---KRSGKLELPEWTDLVKTAKFKELAPYDPDWYYVRAASMARKIYVRGGL 71
FVNA H +R + +L L+K K PY + +V A R GG
Sbjct: 175 FVNAKQFHAIMRRRQQRAKLEAQNKLIKARK-----PYLHESRHVHALKRPRG---SGGR 226
Query: 72 GVGSFQRIYGGSQRNGSRPPHFCKSSGAIARHILQQLENMNLIEMDTKGGRKITSS 127
+ + +++ + P ++G ++R +L QL+N N + T ITS+
Sbjct: 227 FLNT-KKLQESTDPKQDMPIQQQHATGNMSRFVLYQLQNSNDCDCSTTSRSDITSA 281
>At2g27880 Argonaute (AGO1)-like protein
Length = 997
Score = 25.8 bits (55), Expect = 9.0
Identities = 24/75 (32%), Positives = 31/75 (41%), Gaps = 3/75 (4%)
Query: 68 RGGLGVGSFQRIYGGSQRNGSRPPHFCKSSGAIARHILQQLENMNLIEMDTKGGRKITSS 127
RGG G G R GG +R+ R +SSG +A LQQ + GR
Sbjct: 4 RGGGGHGGASRGRGGGRRSDQRQD---QSSGQVAWPGLQQSYGGRGGSVSAGRGRGNVGR 60
Query: 128 GRRDLDQVAGRILVA 142
G D A ++ VA
Sbjct: 61 GENTGDLTATQVPVA 75
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.137 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,184,997
Number of Sequences: 26719
Number of extensions: 125391
Number of successful extensions: 289
Number of sequences better than 10.0: 7
Number of HSP's better than 10.0 without gapping: 3
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 286
Number of HSP's gapped (non-prelim): 7
length of query: 149
length of database: 11,318,596
effective HSP length: 90
effective length of query: 59
effective length of database: 8,913,886
effective search space: 525919274
effective search space used: 525919274
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 55 (25.8 bits)
Medicago: description of AC148349.9


