

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148349.4 - phase: 0
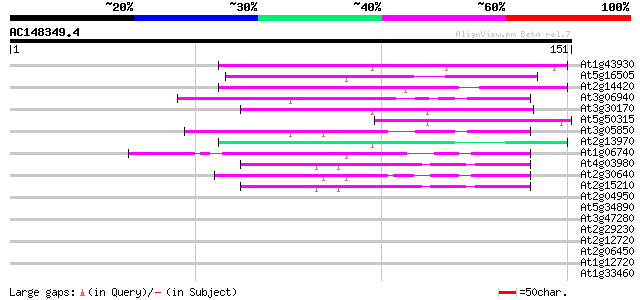
(151 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g43930 hypothetical protein 52 2e-07
At5g16505 mutator-like transposase-like protein (MQK4.25) 51 3e-07
At2g14420 Mutator-like transposase 47 3e-06
At3g06940 putative mudrA protein 45 1e-05
At3g30170 unknown protein 44 4e-05
At5g50315 unknown protein 41 3e-04
At3g05850 unknown protein 41 3e-04
At2g13970 Mutator-like transposase 40 4e-04
At1g06740 mudrA-like protein 40 4e-04
At4g03980 putative transposon protein 39 8e-04
At2g30640 Mutator-like transposase 39 8e-04
At2g15210 putaive transposase of transposon FARE2.7 (cds1) 39 8e-04
At2g04950 putative transposase of transposon FARE2.9 (cds1) 39 0.001
At5g34890 putative protein 39 0.001
At3g47280 putative protein 38 0.002
At2g29230 Mutator-like transposase 38 0.002
At2g12720 putative protein 37 0.004
At2g06450 putative transposase of FARE2.11 37 0.004
At1g12720 hypothetical protein 37 0.004
At1g33460 mutator transposase MUDRA, putative 37 0.005
>At1g43930 hypothetical protein
Length = 946
Score = 51.6 bits (122), Expect = 2e-07
Identities = 33/103 (32%), Positives = 46/103 (44%), Gaps = 9/103 (8%)
Query: 57 WLTLGSYRATYNYFIQPVNSQIYWEPTPYEKSAPPKVRRA-AGRPKKNRRKDGNKEPIGR 115
W T ++ TYN I PV ++ W PP RR GRPK + R+ G E
Sbjct: 727 WYTTSMWKQTYNDGIGPVQGKLLWPTVNKVGVLPPPWRRGNPGRPKNHARRKGVFESSTA 786
Query: 116 S-----QMKRTYNDTQCGRCGLIGHNLRGCNKQGV---ARRPK 150
S ++ R + C C GHN +GC + V A+RP+
Sbjct: 787 SSSSTTELSRLHRVMTCSNCQGEGHNKQGCKNETVAPPAKRPR 829
>At5g16505 mutator-like transposase-like protein (MQK4.25)
Length = 597
Score = 50.8 bits (120), Expect = 3e-07
Identities = 30/92 (32%), Positives = 42/92 (45%), Gaps = 16/92 (17%)
Query: 59 TLGSYRATYNYFIQPVNSQIYWEPTPYEKSA--------PPKVRRAAGRPKKNRRKDGNK 110
T+ SY+ TY+ I + + W+ E PPK RR GRPKK + N
Sbjct: 512 TVSSYQQTYSQMIGEIPDRSLWKDEGEEAGGGVESLRIRPPKTRRPPGRPKKKVVRVEN- 570
Query: 111 EPIGRSQMKRTYNDTQCGRCGLIGHNLRGCNK 142
+KR QCGRC L+GH+ + C +
Sbjct: 571 -------LKRPKRIVQCGRCHLLGHSQKKCTQ 595
>At2g14420 Mutator-like transposase
Length = 241
Score = 47.4 bits (111), Expect = 3e-06
Identities = 29/101 (28%), Positives = 42/101 (40%), Gaps = 12/101 (11%)
Query: 57 WLTLGSYRATYNYFIQPVNSQIYWEPTPYEKSAPPKVRRAAGRPKKNRR-------KDGN 109
W ++ ++ Y+ PVN W+ PP R GRPK N R +
Sbjct: 117 WYSIEKWKLCYSSLFFPVNGMELWKTHIDVVVMPPPDRIMPGRPKNNDRIRDPTEDRPPQ 176
Query: 110 KEPIGRSQMKRTYNDTQCGRCGLIGHNLRGCNKQGVARRPK 150
K P R +++ T C C IGHN R C ++ V + K
Sbjct: 177 KVPSTREKLQMT-----CSNCQQIGHNKRSCKRKAVPKPSK 212
>At3g06940 putative mudrA protein
Length = 749
Score = 45.1 bits (105), Expect = 1e-05
Identities = 32/99 (32%), Positives = 45/99 (45%), Gaps = 14/99 (14%)
Query: 46 MQRYQIEHCHAWLTLGSYRATYNYFIQPV----NSQIYWEPTPYEKSAPPKVRRAAGRPK 101
+++ E+C +LT+ S+R Y IQPV + P PP RR GRPK
Sbjct: 658 IEKSPYEYCSTYLTVESHRLMYAESIQPVPNMDRMMLDDPPEGLVCVTPPPTRRTPGRPK 717
Query: 102 KNRRKDGNKEPIGRSQMKRTYNDTQCGRCGLIGHNLRGC 140
+ EP+ MKR QC +C +GHN + C
Sbjct: 718 IKK-----VEPL--DMMKR---QLQCSKCKGLGHNKKTC 746
>At3g30170 unknown protein
Length = 995
Score = 43.5 bits (101), Expect = 4e-05
Identities = 29/83 (34%), Positives = 35/83 (41%), Gaps = 4/83 (4%)
Query: 63 YRATYNYFIQPVNSQIYWEPTPYEKSAPPKVRRA-AGRPKKNRRKDGNKE---PIGRSQM 118
+R TY I+PV W T PP RR AGRP R+ G E P ++M
Sbjct: 704 WRETYLKGIRPVQGMPLWCRTNRLPVLPPPWRRGNAGRPSNYARRKGRNEAAAPSNPNKM 763
Query: 119 KRTYNDTQCGRCGLIGHNLRGCN 141
R C C GHN + CN
Sbjct: 764 SREKRIMTCSNCHQEGHNKQRCN 786
>At5g50315 unknown protein
Length = 327
Score = 40.8 bits (94), Expect = 3e-04
Identities = 23/56 (41%), Positives = 32/56 (57%), Gaps = 3/56 (5%)
Query: 99 RPKKNRRKDGNKE--PIGRSQMKRTYNDTQCGRCGLIGHNLRGCNKQGVAR-RPKD 151
R K +RK +E P+ +++ R +CGRCG+IGHN R C GV RPK+
Sbjct: 246 RSKNPKRKKEVQESPPVKVTKVTREGRIVRCGRCGVIGHNTRKCGNVGVEYIRPKN 301
>At3g05850 unknown protein
Length = 777
Score = 40.8 bits (94), Expect = 3e-04
Identities = 31/98 (31%), Positives = 41/98 (41%), Gaps = 15/98 (15%)
Query: 48 RYQIEHCHAWLTLGSYRATYNYFIQPV---NSQIYWEPT--PYEKSAPPKVRRAAGRPKK 102
R ++C + T+ YR+TY I PV ++ E + PP RR GRP K
Sbjct: 683 RNPYDYCSKYFTVAYYRSTYAQSINPVPLLEGEMCRESSGGSAVTVTPPPTRRPPGRPPK 742
Query: 103 NRRKDGNKEPIGRSQMKRTYNDTQCGRCGLIGHNLRGC 140
K+ MKR QC RC +GHN C
Sbjct: 743 -------KKTPAEEVMKR---QLQCSRCKGLGHNKSTC 770
>At2g13970 Mutator-like transposase
Length = 597
Score = 40.4 bits (93), Expect = 4e-04
Identities = 29/95 (30%), Positives = 37/95 (38%), Gaps = 14/95 (14%)
Query: 57 WLTLGSYRATYNYFIQPVNSQIYWEPTPYEKSAPPKVRRA-AGRPKKNRRKDGNKEPIGR 115
W T ++ TY I PV ++ W PP RR GRPK + R+ G E
Sbjct: 379 WYTTYMWKQTYYDGIMPVQGKMLWPIVNRVGVLPPPWRRGNPGRPKNHDRRKGIYESATA 438
Query: 116 SQMKRTYNDTQCGRCGLIGHNLRGCNKQGVARRPK 150
S + GHN GC + VA PK
Sbjct: 439 STSRE-------------GHNKAGCKNETVAPPPK 460
>At1g06740 mudrA-like protein
Length = 726
Score = 40.4 bits (93), Expect = 4e-04
Identities = 30/113 (26%), Positives = 50/113 (43%), Gaps = 19/113 (16%)
Query: 33 SHNWTPLWSGDTPMQRYQIEHCHAWLTLGSYRATYNYFIQPVNSQIYWEPTPYEKSA--- 89
SH L S + + RY E C T+ +YR Y ++P++ ++ W+ E+ +
Sbjct: 624 SHAVGALLSCEEDVYRYT-ESC---FTVENYRRAYAETLEPISDKVQWKENDSERDSENV 679
Query: 90 --PPKVRRAAGRPKKNRRKDGNKEPIGRSQMKRTYNDTQCGRCGLIGHNLRGC 140
PK + A R ++ R +D R +++R CGRC GH C
Sbjct: 680 IKTPKAMKGAPRKRRVRAED-------RDRVRRV---VHCGRCNQTGHFRTTC 722
>At4g03980 putative transposon protein
Length = 580
Score = 39.3 bits (90), Expect = 8e-04
Identities = 29/81 (35%), Positives = 36/81 (43%), Gaps = 7/81 (8%)
Query: 63 YRATYNYFIQPVNSQIYWE-PTPYEK--SAPPKVRRAAGRPKKNRRKDGNKEPIGRSQMK 119
+R Y+ I P YWE P + PP R +GR KK R + GRSQ K
Sbjct: 500 WRQAYSESIHPNGDMEYWEIPDTVSEVICLPPSTRVPSGRRKKKRIPSVWEH--GRSQPK 557
Query: 120 RTYNDTQCGRCGLIGHNLRGC 140
+ +C RCG GHN C
Sbjct: 558 PKLH--KCSRCGQSGHNKSTC 576
>At2g30640 Mutator-like transposase
Length = 754
Score = 39.3 bits (90), Expect = 8e-04
Identities = 31/95 (32%), Positives = 41/95 (42%), Gaps = 18/95 (18%)
Query: 56 AWLTLGSYRATYNYFIQPVNSQIYWEPT-PYEKSA---------PPKVRRAAGRPKKNRR 105
++ T+ +YR TY I PV + W+ T P +S PP+ R RPKK RR
Sbjct: 664 SYFTVANYRRTYAETIHPVPDKTEWKTTEPAGESGEDGEEIVIRPPRDLRQPPRPKK-RR 722
Query: 106 KDGNKEPIGRSQMKRTYNDTQCGRCGLIGHNLRGC 140
G R + KR +C RC GH C
Sbjct: 723 SQGE----DRGRQKRV---VRCSRCNQAGHFRTTC 750
>At2g15210 putaive transposase of transposon FARE2.7 (cds1)
Length = 580
Score = 39.3 bits (90), Expect = 8e-04
Identities = 29/81 (35%), Positives = 36/81 (43%), Gaps = 7/81 (8%)
Query: 63 YRATYNYFIQPVNSQIYWE-PTPYEK--SAPPKVRRAAGRPKKNRRKDGNKEPIGRSQMK 119
+R Y+ I P YWE P + PP R +GR KK R + GRSQ K
Sbjct: 500 WRQAYSESIHPNGDMEYWEIPETISEVICLPPSTRVPSGRRKKKRIPSVWEH--GRSQPK 557
Query: 120 RTYNDTQCGRCGLIGHNLRGC 140
+ +C RCG GHN C
Sbjct: 558 PKLH--KCSRCGQSGHNKSTC 576
>At2g04950 putative transposase of transposon FARE2.9 (cds1)
Length = 616
Score = 38.9 bits (89), Expect = 0.001
Identities = 29/81 (35%), Positives = 36/81 (43%), Gaps = 7/81 (8%)
Query: 63 YRATYNYFIQPVNSQIYWE-PTPYEK--SAPPKVRRAAGRPKKNRRKDGNKEPIGRSQMK 119
+R Y+ I P YWE P + PP R +GR KK R + GRSQ K
Sbjct: 536 WRQAYSKSIHPNGDMEYWEIPETISEVICLPPSTRVPSGRRKKKRIPSVWEH--GRSQPK 593
Query: 120 RTYNDTQCGRCGLIGHNLRGC 140
+ +C RCG GHN C
Sbjct: 594 PKLH--KCSRCGQSGHNNSTC 612
>At5g34890 putative protein
Length = 681
Score = 38.5 bits (88), Expect = 0.001
Identities = 27/81 (33%), Positives = 38/81 (46%), Gaps = 7/81 (8%)
Query: 63 YRATYNYFIQPVNSQIYWE-PTPYEK--SAPPKVRRAAGRPKKNRRKDGNKEPIGRSQMK 119
+R Y+ I P YWE P + PP R +GR +N+++ + GRSQ K
Sbjct: 601 WRQAYSESIHPNGDMKYWEIPETISEVICLPPSTRVPSGR--RNKKRIPSVWEHGRSQPK 658
Query: 120 RTYNDTQCGRCGLIGHNLRGC 140
+ +C RCG GHN C
Sbjct: 659 PKLH--KCSRCGQSGHNKSTC 677
>At3g47280 putative protein
Length = 739
Score = 37.7 bits (86), Expect = 0.002
Identities = 28/81 (34%), Positives = 35/81 (42%), Gaps = 7/81 (8%)
Query: 63 YRATYNYFIQPVNSQIYWE-PTPYEK--SAPPKVRRAAGRPKKNRRKDGNKEPIGRSQMK 119
+R Y+ I P YWE P + PP R G+ KK R + GRSQ K
Sbjct: 659 WRQAYSESIHPNGDMEYWEIPETISEVICLPPSTRVPTGKRKKKRIPSVWEH--GRSQPK 716
Query: 120 RTYNDTQCGRCGLIGHNLRGC 140
+ +C RCG GHN C
Sbjct: 717 PKLH--KCSRCGQSGHNKSTC 735
>At2g29230 Mutator-like transposase
Length = 915
Score = 37.7 bits (86), Expect = 0.002
Identities = 27/83 (32%), Positives = 38/83 (45%), Gaps = 10/83 (12%)
Query: 62 SYRATYNYFIQPVNS--QIYWEPTPYE--KSAPPKVRRAAGRPKKNRRKDGNKEPIGRSQ 117
S+R +Y I+PV ++ P P + PP RR +GRPKK R + + Q
Sbjct: 835 SWRMSYLGTIKPVPEVGDVFALPEPIASLRLFPPATRRPSGRPKKKRITSRGEFTCPQRQ 894
Query: 118 MKRTYNDTQCGRCGLIGHNLRGC 140
+ T+C RC GHN C
Sbjct: 895 V------TRCSRCTGAGHNRATC 911
>At2g12720 putative protein
Length = 819
Score = 37.0 bits (84), Expect = 0.004
Identities = 19/51 (37%), Positives = 25/51 (48%), Gaps = 3/51 (5%)
Query: 90 PPKVRRAAGRPKKNRRKDGNKEPIGRSQMKRTYNDTQCGRCGLIGHNLRGC 140
PP V GRPKK R K + + + KR + C RC GHN++ C
Sbjct: 770 PPIVANQPGRPKKLRMKSALEVAV---ETKRPRKEHACSRCKETGHNVKTC 817
>At2g06450 putative transposase of FARE2.11
Length = 580
Score = 37.0 bits (84), Expect = 0.004
Identities = 27/81 (33%), Positives = 34/81 (41%), Gaps = 7/81 (8%)
Query: 63 YRATYNYFIQPVNSQIYWEPTPYEKSA---PPKVRRAAGRPKKNRRKDGNKEPIGRSQMK 119
+R Y+ I P YWE PP R +GR +K R + GRSQ K
Sbjct: 500 WRQAYSESIHPNGDMEYWEILETISEVICLPPSTRVPSGRREKKRIPSVWEH--GRSQPK 557
Query: 120 RTYNDTQCGRCGLIGHNLRGC 140
+ +C RCG GHN C
Sbjct: 558 PKLH--KCSRCGRSGHNKSTC 576
>At1g12720 hypothetical protein
Length = 585
Score = 37.0 bits (84), Expect = 0.004
Identities = 27/91 (29%), Positives = 32/91 (34%), Gaps = 4/91 (4%)
Query: 63 YRATYNYFIQPVNSQIYWEPTPYEKSAPPKVRRA-AGRPKKNRRKDGNKEPI---GRSQM 118
+R TY I+PV W PP RR GR RK G E +++M
Sbjct: 390 WRETYRDGIRPVQGMPLWPRMSRLPVLPPPWRRGNPGRQSNYARKKGRYETASSSNKNKM 449
Query: 119 KRTYNDTQCGRCGLIGHNLRGCNKQGVARRP 149
R C C GHN C V P
Sbjct: 450 SRANRIMTCSNCKQEGHNKSSCKNATVLLPP 480
>At1g33460 mutator transposase MUDRA, putative
Length = 826
Score = 36.6 bits (83), Expect = 0.005
Identities = 26/92 (28%), Positives = 37/92 (39%), Gaps = 13/92 (14%)
Query: 62 SYRATYNYFIQPVNSQIYWEPT-------PYEKSAPPKVRRAAGRPKKNRRKDGNKEPIG 114
++R Y+ PV Q+YW PT P + PP R+ + +R K N+ P
Sbjct: 669 AWREQYDTGPDPVRGQMYW-PTGLGLITAPLQDPVPPG-RKKGEKKNYHRIKGPNESPKK 726
Query: 115 RSQMKRTYNDT----QCGRCGLIGHNLRGCNK 142
+ + C CG GHN GC K
Sbjct: 727 KGPQRLKVGKKGVVMHCKSCGEAGHNAAGCKK 758
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.134 0.442
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,820,497
Number of Sequences: 26719
Number of extensions: 162569
Number of successful extensions: 547
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 28
Number of HSP's successfully gapped in prelim test: 27
Number of HSP's that attempted gapping in prelim test: 488
Number of HSP's gapped (non-prelim): 65
length of query: 151
length of database: 11,318,596
effective HSP length: 90
effective length of query: 61
effective length of database: 8,913,886
effective search space: 543747046
effective search space used: 543747046
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 55 (25.8 bits)
Medicago: description of AC148349.4


