

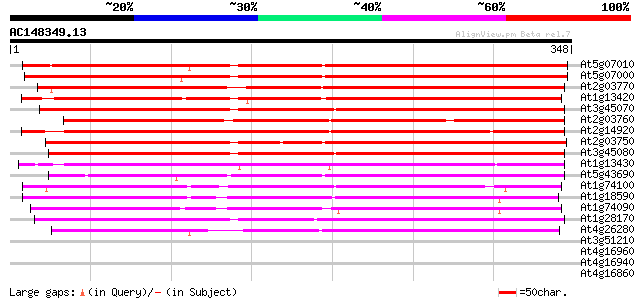
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148349.13 - phase: 0
(348 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g07010 steroid sulfotransferase-like protein 345 3e-95
At5g07000 steroid sulfotransferase-like protein 335 2e-92
At2g03770 putative steroid sulfotransferase 286 1e-77
At1g13420 unknown protein 285 3e-77
At3g45070 sulfotransferase-like protein 282 2e-76
At2g03760 putative steroid sulfotransferase 272 2e-73
At2g14920 putative steroid sulfotransferase 270 9e-73
At2g03750 steroid sulfotransferase like protein 270 9e-73
At3g45080 sulfotransferase-like protein 269 1e-72
At1g13430 unknown protein 264 6e-71
At5g43690 steroid sulfotransferase-like 261 3e-70
At1g74100 putative flavonol sulfotransferase 257 8e-69
At1g18590 unknown protein 254 4e-68
At1g74090 flavonol sulfotransferase like protein 252 2e-67
At1g28170 steroid sulfotransferase, putative 252 2e-67
At4g26280 steroid sulfotransferase - like protein 244 5e-65
At3g51210 putative protein 37 0.012
At4g16960 disease resistance RPP5 like protein 30 1.5
At4g16940 disease resistance RPP5 like protein 30 1.5
At4g16860 disease resistance RPP5 like protein 30 2.0
>At5g07010 steroid sulfotransferase-like protein
Length = 359
Score = 345 bits (884), Expect = 3e-95
Identities = 157/340 (46%), Positives = 242/340 (71%), Gaps = 8/340 (2%)
Query: 9 MKEDQSRDGQETTSKVNKVSHDYNNLILSLPRENGCETQYMYFFHGFWCPSTLIQSVNSF 68
+KE ++RD + +S ++ ++ SLP+E G T+Y+Y F GFWC + IQ++ SF
Sbjct: 25 LKEGKTRDVPKAEEDEG-LSCEFQEMLDSLPKERGWRTRYLYLFQGFWCQAKEIQAIMSF 83
Query: 69 QNNFHAKDSDIVVASIPKSGTTWLKGLAYAIVNRQHFTSLEN--NHPLLLFNPHELVPQF 126
Q +F + ++D+V+A+IPKSGTTWLK L + I+NR F + + NHPL NPH+LVP F
Sbjct: 84 QKHFQSLENDVVLATIPKSGTTWLKALTFTILNRHRFDPVASSTNHPLFTSNPHDLVPFF 143
Query: 127 EVNLYGDKDGPLPQIDVSNMTEPRLFGTHMPFPSLPKSVKESNCKIIYICRNPFDTFVSY 186
E LY + D P D+S + PR F TH+PF SL +++++ K++Y+CRNPFDTF+S
Sbjct: 144 EYKLYANGDVP----DLSGLASPRTFATHLPFGSLKETIEKPGVKVVYLCRNPFDTFISS 199
Query: 187 WIFINKIRLRKSLTELTLEESFERYCKGICLFGPFWDNMLGYLKESIERPDRVLFLKYED 246
W + N I+ +S++ + L+++F+ YC+G+ FGPFW++MLGY +ES++RP++V FL+YED
Sbjct: 200 WHYTNNIK-SESVSPVLLDQAFDLYCRGVIGFGPFWEHMLGYWRESLKRPEKVFFLRYED 258
Query: 247 LKEDVNFHTKRIAEFVGIPFTQEEENNGVIENIIKLCSFESMKEIEGNQSGTISGDIEKE 306
LK+D+ + KR+A F+ +PFT+EEE GV++ I +LCSFE++K++E N+S + E
Sbjct: 259 LKDDIETNLKRLATFLELPFTEEEERKGVVKAIAELCSFENLKKLEVNKSNKSIKNFENR 318
Query: 307 FYFRKGEIGDWANYLSSSMVEKLSKVMEEKLNGSSLSFKV 346
F FRKGE+ DW NYLS S VE+LS ++++KL GS L+F++
Sbjct: 319 FLFRKGEVSDWVNYLSPSQVERLSALVDDKLGGSGLTFRL 358
>At5g07000 steroid sulfotransferase-like protein
Length = 347
Score = 335 bits (860), Expect = 2e-92
Identities = 157/341 (46%), Positives = 240/341 (70%), Gaps = 9/341 (2%)
Query: 10 KEDQSRDGQETTSKVNKVSHDYNNLILSLPRENGCETQYMYFFHGFWCPSTLIQSVNSFQ 69
K + ++G+ + +S+++ ++ SLP+E G +Y+Y F GF C + IQ++ SFQ
Sbjct: 11 KPELLKEGKSEGQEEEGLSYEFQEMLDSLPKERGRRNRYLYLFQGFRCQAKEIQAITSFQ 70
Query: 70 NNFHAKDSDIVVASIPKSGTTWLKGLAYAIVNRQHF---TSLENNHPLLLFNPHELVPQF 126
+F + D+V+A+IPKSGTTWLK L + I+ R F +S ++HPLL NPH+LVP F
Sbjct: 71 KHFQSLPDDVVLATIPKSGTTWLKALTFTILTRHRFDPVSSSSSDHPLLTSNPHDLVPFF 130
Query: 127 EVNLYGDKDGPLPQIDVSNMTEPRLFGTHMPFPSLPKSVKESNCKIIYICRNPFDTFVSY 186
E LY + + P D+S + PR F TH+PF +L SV+ + K++Y+CRNPFDTF+S
Sbjct: 131 EYKLYANGNVP----DLSGLASPRTFATHVPFGALKDSVENPSVKVVYLCRNPFDTFISM 186
Query: 187 WIFINKIRLRKSLTELTLEESFERYCKGICL-FGPFWDNMLGYLKESIERPDRVLFLKYE 245
W +IN I +S++ + L+E+F+ YC+G+ + FGPFW++MLGY +ES++RP++VLFLKYE
Sbjct: 187 WHYINNIT-SESVSAVLLDEAFDLYCRGLLIGFGPFWEHMLGYWRESLKRPEKVLFLKYE 245
Query: 246 DLKEDVNFHTKRIAEFVGIPFTQEEENNGVIENIIKLCSFESMKEIEGNQSGTISGDIEK 305
DLKED+ + K++A F+G+PFT+EEE GV++ I LCSFE++K++E N+S + + E
Sbjct: 246 DLKEDIETNLKKLASFLGLPFTEEEEQKGVVKAIADLCSFENLKKLEVNKSSKLIQNYEN 305
Query: 306 EFYFRKGEIGDWANYLSSSMVEKLSKVMEEKLNGSSLSFKV 346
F FRKGE+ D NYLS S VE+LS ++++KL GS L+F++
Sbjct: 306 RFLFRKGEVSDLVNYLSPSQVERLSALVDDKLAGSGLTFRL 346
>At2g03770 putative steroid sulfotransferase
Length = 324
Score = 286 bits (732), Expect = 1e-77
Identities = 144/330 (43%), Positives = 210/330 (63%), Gaps = 15/330 (4%)
Query: 18 QETTSKV---NKVSHDYNNLILSLPRENGCETQYMYFFHGFWCPSTLIQSVNSFQNNFHA 74
+ T SK+ +++ + L+ SLPR+ +Y+Y + GFW P L++ V Q +F A
Sbjct: 5 ETTLSKLLVESELVQECEELLSSLPRDRSVFAEYLYQYQGFWYPPNLLEGVLYSQKHFQA 64
Query: 75 KDSDIVVASIPKSGTTWLKGLAYAIVNRQHFTSLENNHPLLLFNPHELVPQFEVNLYGDK 134
+DSDIV+ASIPKSGTTWLK L +A+++RQ F + +HPLL NPH LV E +
Sbjct: 65 RDSDIVLASIPKSGTTWLKSLVFALIHRQEFQTPLVSHPLLDNNPHTLVTFIEGFHLHTQ 124
Query: 135 DGPLPQIDVSNMTEPRLFGTHMPFPSLPKSVKESNCKIIYICRNPFDTFVSYWIFINKIR 194
D T PR+F TH+P SLP+SVK+S+CK++Y CRNP D FVS W F+ +
Sbjct: 125 D-----------TSPRIFSTHIPVGSLPESVKDSSCKVVYCCRNPKDAFVSLWHFMKNL- 172
Query: 195 LRKSLTELTLEESFERYCKGICLFGPFWDNMLGYLKESIERPDRVLFLKYEDLKEDVNFH 254
+ K + T+EE +C+G ++GPFWD++L Y KES E P +V+F+ YE+++E
Sbjct: 173 IVKEMVGCTMEEMVRFFCRGSSIYGPFWDHVLQYWKESRENPKKVMFVMYEEMREQPQEW 232
Query: 255 TKRIAEFVGIPFTQEEENNGVIENIIKLCSFESMKEIEGNQSGTISGDIEKEFYFRKGEI 314
RIAEF+G FT+EE NGV+E+IIKLCS E++ ++E N+ G + +E + +FRKGEI
Sbjct: 233 VMRIAEFLGYSFTEEEIENGVLEDIIKLCSLENLSKLEVNEKGKLLNGMETKAFFRKGEI 292
Query: 315 GDWANYLSSSMVEKLSKVMEEKLNGSSLSF 344
G W + L+ + E++ K +EKL GS F
Sbjct: 293 GGWRDTLTPLLAEEIDKTTKEKLIGSDFRF 322
>At1g13420 unknown protein
Length = 331
Score = 285 bits (728), Expect = 3e-77
Identities = 142/337 (42%), Positives = 210/337 (62%), Gaps = 18/337 (5%)
Query: 8 NMKEDQSRDGQETTSKVNKVSHDYNNLILSLPRENGCETQYMYFFHGFWCPSTLIQSVNS 67
N+KE++ + + S + +LI SLP + C +Y + G W ++Q++ +
Sbjct: 9 NLKEEEEEEEENQ-------SEETKSLISSLPSDIDCSGTKLYKYQGCWYDKDILQAILN 61
Query: 68 FQNNFHAKDSDIVVASIPKSGTTWLKGLAYAIVNRQHFTSLENNHPLLLFNPHELVPQFE 127
F NF +++DI+VAS PKSGTTWLK L +A+ R TS +NHPLL NPHELVP E
Sbjct: 62 FNKNFQPQETDIIVASFPKSGTTWLKALTFALAQRSKHTS--DNHPLLTHNPHELVPYLE 119
Query: 128 VNLYGDKDGPLPQIDVSNM--TEPRLFGTHMPFPSLPKSVKESNCKIIYICRNPFDTFVS 185
++LY P D++ + + PRLF THM F +L +KES CKI+Y+CRN D VS
Sbjct: 120 LDLYLKSSKP----DLTKLPSSSPRLFSTHMSFDALKVPLKESPCKIVYVCRNVKDVLVS 175
Query: 186 YWIFINKIRLRKSLTELTLEESFERYCKGICLFGPFWDNMLGYLKESIERPDRVLFLKYE 245
W F N + L+LE FE C G+ L GP W+N+LGY + S+E P VLFL+YE
Sbjct: 176 LWCFENSM---SGENNLSLEALFESLCSGVNLCGPLWENVLGYWRGSLEDPKHVLFLRYE 232
Query: 246 DLKEDVNFHTKRIAEFVGIPFTQEEENNGVIENIIKLCSFESMKEIEGNQSGTISGDIEK 305
+LK + KR+AEF+ PFT+EEE++G ++ I++LCS ++ +E N++G++S +
Sbjct: 233 ELKTEPRVQIKRLAEFLDCPFTKEEEDSGGVDKILELCSLRNLSGLEINKTGSLSEGVSF 292
Query: 306 EFYFRKGEIGDWANYLSSSMVEKLSKVMEEKLNGSSL 342
+ +FRKGE+GDW +Y++ M K+ ++EEKL GS L
Sbjct: 293 KSFFRKGEVGDWKSYMTPEMENKIDMIVEEKLQGSGL 329
>At3g45070 sulfotransferase-like protein
Length = 323
Score = 282 bits (722), Expect = 2e-76
Identities = 134/327 (40%), Positives = 210/327 (63%), Gaps = 6/327 (1%)
Query: 19 ETTSKVNKVSHDYNNLILSLPRENGCETQYMYFFHGFWCPSTLIQSVNSFQNNFHAKDSD 78
E ++ ++ + LI SLP + + + + G W ++Q+V +FQ +F +D+D
Sbjct: 2 EMNLRIEDLNEETKTLISSLPSDKDFTGKTICKYQGCWYTHNVLQAVLNFQKSFKPQDTD 61
Query: 79 IVVASIPKSGTTWLKGLAYAIVNRQHFTSLENNHPLLLFNPHELVPQFEVNLYGDKDGPL 138
I+VAS PK GTTWLK L +A+++R S +++HPLL NPH LVP FE++LY + P
Sbjct: 62 IIVASFPKCGTTWLKALTFALLHRSKQPSHDDDHPLLSNNPHVLVPYFEIDLYLRSENP- 120
Query: 139 PQIDVSNMTE-PRLFGTHMPFPSLPKSVKESNCKIIYICRNPFDTFVSYWIFINKIRLRK 197
D++ + PRLF TH+P +L + +K S CKI+YI RN DT VSYW F K + +
Sbjct: 121 ---DLTKFSSSPRLFSTHVPSHTLQEGLKGSTCKIVYISRNVKDTLVSYWHFFTKKQTDE 177
Query: 198 SLTELTLEESFERYCKGICLFGPFWDNMLGYLKESIERPDRVLFLKYEDLKEDVNFHTKR 257
+ + E++FE +C+G+ +FGPFWD++L Y + S+E P+ VLF+K+E++K + K+
Sbjct: 178 KIIS-SFEDTFEMFCRGVSIFGPFWDHVLSYWRGSLEDPNHVLFMKFEEMKAEPRDQIKK 236
Query: 258 IAEFVGIPFTQEEENNGVIENIIKLCSFESMKEIEGNQSGTISGDIEKEFYFRKGEIGDW 317
AEF+G PFT+EEE +G ++ II LCS ++ +E N++G ++ E + +FRKGE+GDW
Sbjct: 237 FAEFLGCPFTKEEEESGSVDEIIDLCSLRNLSSLEINKTGKLNSGRENKMFFRKGEVGDW 296
Query: 318 ANYLSSSMVEKLSKVMEEKLNGSSLSF 344
NYL+ M K+ +++EKL S L F
Sbjct: 297 KNYLTPEMENKIDMIIQEKLQNSGLKF 323
>At2g03760 putative steroid sulfotransferase
Length = 326
Score = 272 bits (695), Expect = 2e-73
Identities = 141/312 (45%), Positives = 192/312 (61%), Gaps = 11/312 (3%)
Query: 34 LILSLPRENGCETQYMYFFHGFWCPSTLIQSVNSFQNNFHAKDSDIVVASIPKSGTTWLK 93
LI SLP+E G +Y F G W ++Q + Q F AKDSDI++ + PKSGTTWLK
Sbjct: 23 LISSLPKEKGWLVSEIYEFQGLWHTQAILQGILICQKRFEAKDSDIILVTNPKSGTTWLK 82
Query: 94 GLAYAIVNRQHF-TSLENNHPLLLFNPHELVPQFEVNLYGDKDGPLPQIDVSNMTEPRLF 152
L +A++NR F S NHPLL+ NPH LVP E Y P D S++ PRL
Sbjct: 83 ALVFALLNRHKFPVSSSGNHPLLVTNPHLLVPFLEGVYYES-----PDFDFSSLPSPRLM 137
Query: 153 GTHMPFPSLPKSVKESNCKIIYICRNPFDTFVSYWIFINKIRLRKSLTELTLEESFERYC 212
TH+ SLP+SVK S+CKI+Y CRNP D FVS W F K+ ++ + +E++ E +C
Sbjct: 138 NTHISHLSLPESVKSSSCKIVYCCRNPKDMFVSLWHFGKKLAPEET-ADYPIEKAVEAFC 196
Query: 213 KGICLFGPFWDNMLGYLKESIERPDRVLFLKYEDLKEDVNFHTKRIAEFVGIPFTQEEEN 272
+G + GPFWD++L Y S E P++VLF+ YE+LK+ KRIAEF+ F +EEE
Sbjct: 197 EGKFIGGPFWDHILEYWYASRENPNKVLFVTYEELKKQTEVEMKRIAEFLECGFIEEEE- 255
Query: 273 NGVIENIIKLCSFESMKEIEGNQSGTISGDIEKEFYFRKGEIGDWANYLSSSMVEKLSKV 332
+ I+KLCSFES+ +E N+ G + IE + +FRKGEIG W + LS S+ E++ +
Sbjct: 256 ---VREIVKLCSFESLSNLEVNKEGKLPNGIETKTFFRKGEIGGWRDTLSESLAEEIDRT 312
Query: 333 MEEKLNGSSLSF 344
+EEK GS L F
Sbjct: 313 IEEKFKGSGLKF 324
>At2g14920 putative steroid sulfotransferase
Length = 333
Score = 270 bits (690), Expect = 9e-73
Identities = 133/338 (39%), Positives = 208/338 (61%), Gaps = 14/338 (4%)
Query: 8 NMKEDQSRDGQETTSKVNKVSHDYNNLILSLPRENGCETQYMYFFHGFWCPSTLIQSVNS 67
N++E++ + +ET LI SLP E ++ + G+W ++QS+ +
Sbjct: 9 NLREEEEKPSEETKI-----------LISSLPWEIDYLGNKLFNYEGYWYSEDILQSIPN 57
Query: 68 FQNNFHAKDSDIVVASIPKSGTTWLKGLAYAIVNRQHFTSLENNHPLLLFNPHELVPQFE 127
F +++DI++AS KSGTTWLK L +A+V R + ++ HPLL NPHE+VP E
Sbjct: 58 IHTGFQPQETDIILASFYKSGTTWLKALTFALVQRSKHSLEDHQHPLLHHNPHEIVPNLE 117
Query: 128 VNLYGDKDGP-LPQIDVSNMTEPRLFGTHMPFPSLPKSVKESNCKIIYICRNPFDTFVSY 186
++LY P L + S+ + PRLF THM L +KE+ CKI+Y+CRN D VS
Sbjct: 118 LDLYLKSSKPDLTKFLSSSSSSPRLFSTHMSLDPLQVPLKENLCKIVYVCRNVKDVMVSV 177
Query: 187 WIFINKIRLRKSLTELTLEESFERYCKGICLFGPFWDNMLGYLKESIERPDRVLFLKYED 246
W F ++ ++ + +LE FE +C G+ L GPFWD+ L Y + S+E P LF++YED
Sbjct: 178 WYFRQSKKITRA-EDYSLEAIFESFCNGVTLHGPFWDHALSYWRGSLEDPKHFLFMRYED 236
Query: 247 LKEDVNFHTKRIAEFVGIPFTQEEENNGVIENIIKLCSFESMKEIEGNQSGTISGDIEKE 306
LK + KR+AEF+ PFT+EEE++G ++ I++LCS +++ +E N++ T S ++ +
Sbjct: 237 LKAEPRTQVKRLAEFLDCPFTKEEEDSGSVDKILELCSLSNLRSVEINKTRT-SSRVDFK 295
Query: 307 FYFRKGEIGDWANYLSSSMVEKLSKVMEEKLNGSSLSF 344
YFRKG++GDW +Y++ MV+K+ ++EEKL GS L F
Sbjct: 296 SYFRKGQVGDWKSYMTPEMVDKIDMIIEEKLKGSGLKF 333
>At2g03750 steroid sulfotransferase like protein
Length = 351
Score = 270 bits (690), Expect = 9e-73
Identities = 128/323 (39%), Positives = 202/323 (61%), Gaps = 6/323 (1%)
Query: 23 KVNKVSHDYNNLILSLPRENGCETQYMYFFHGFWCPSTLIQSVNSFQNNFHAKDSDIVVA 82
K + VS + NLI SLP + +Y + G W +Q+V Q +F +D+DI++A
Sbjct: 35 KDDNVSQETKNLITSLPSDKDFMGYGLYNYKGCWYYPNTLQAVLDVQKHFKPRDTDIILA 94
Query: 83 SIPKSGTTWLKGLAYAIVNRQHFTSLENNHPLLLFNPHELVPQFEVNLYGDKDGPLPQID 142
S+PK GTTWLK L +A+V+R+ + HPLLL NPH+LVP EV LY + P D
Sbjct: 95 SLPKGGTTWLKSLIFAVVHREKYRGTPQTHPLLLQNPHDLVPFLEVELYANSQIP----D 150
Query: 143 VSNMTEPRLFGTHMPFPSLPKSVKESNCKIIYICRNPFDTFVSYWIFINKIRLRKSLTEL 202
++ + P +F THM +L ++ ++ CK +Y+CR DTFVS W + N + R + +
Sbjct: 151 LAKYSSPMIFSTHMHLQALREATTKA-CKTVYVCRGIKDTFVSGWHYRNMLH-RTKMDQA 208
Query: 203 TLEESFERYCKGICLFGPFWDNMLGYLKESIERPDRVLFLKYEDLKEDVNFHTKRIAEFV 262
T E F+ YC+G+ L+GP+W+++L Y K S+E + VLF+KYE++ E+ KR+AEF+
Sbjct: 209 TFELMFDAYCRGVLLYGPYWEHVLSYWKGSLEAKENVLFMKYEEIIEEPRVQVKRLAEFL 268
Query: 263 GIPFTQEEENNGVIENIIKLCSFESMKEIEGNQSGTISGDIEKEFYFRKGEIGDWANYLS 322
PFT+EEE +G +E I+KLCS ++ +E N++GT ++ + +FRKGE+GDW N+L+
Sbjct: 269 ECPFTKEEEESGSVEEILKLCSLRNLSNLEVNKNGTTRIGVDSQVFFRKGEVGDWKNHLT 328
Query: 323 SSMVEKLSKVMEEKLNGSSLSFK 345
M + ++++ +L S L F+
Sbjct: 329 PQMAKTFDEIIDYRLGDSGLIFQ 351
>At3g45080 sulfotransferase-like protein
Length = 329
Score = 269 bits (688), Expect = 1e-72
Identities = 135/322 (41%), Positives = 204/322 (62%), Gaps = 7/322 (2%)
Query: 25 NKVSHDYNNLILSLPRENGCETQYMYFFHGFWCPSTLIQSVNSFQNNFHAKDSDIVVASI 84
+++S + LI SLP + + + G W ++Q V +FQ NF +D+DI+VAS
Sbjct: 13 DELSEESKTLISSLPSDKNSTGVNVCKYQGCWYTPPILQGVLNFQKNFKPQDTDIIVASF 72
Query: 85 PKSGTTWLKGLAYAIVNRQHFTSLENNHPLLLFNPHELVPQFEVNLYGDKDGPLPQIDVS 144
PK GTTWLK L +A+V R S +++HPLL NPH L P E+ LY + P D++
Sbjct: 73 PKCGTTWLKALTFALVRRSKHPSHDDHHPLLSDNPHVLSPSLEMYLYLCSENP----DLT 128
Query: 145 NMTEP-RLFGTHMPFPSLPKSVKESNCKIIYICRNPFDTFVSYWIFINKIRLRKSLTELT 203
+ RLF THMP +L + +K S CKI+Y+ RN DT VSYW F K + ++ +
Sbjct: 129 KFSSSSRLFSTHMPSHTLQEGLKGSTCKIVYMSRNVKDTLVSYWHFFCKKQTDDNIIS-S 187
Query: 204 LEESFERYCKGICLFGPFWDNMLGYLKESIERPDRVLFLKYEDLKEDVNFHTKRIAEFVG 263
+E++FE +C+G+ FGPFWD++L Y + S+E P+ VLF+K+E++KE+ KR+AEF+G
Sbjct: 188 VEDTFEMFCRGVNFFGPFWDHVLSYWRGSLEDPNHVLFMKFEEMKEEPREQIKRLAEFLG 247
Query: 264 IPFTQEEENNGVIENIIKLCSFESMKEIEGNQSGTI-SGDIEKEFYFRKGEIGDWANYLS 322
FT+EEE +G+++ II LCS ++ +E N++G + S E + +FRKGE+GDW NYL+
Sbjct: 248 CLFTKEEEESGLVDEIIDLCSLRNLSSLEINKTGKLHSTGRENKTFFRKGEVGDWKNYLT 307
Query: 323 SSMVEKLSKVMEEKLNGSSLSF 344
M K+ +++EKL S L F
Sbjct: 308 PEMENKIDMIIQEKLQNSGLKF 329
>At1g13430 unknown protein
Length = 351
Score = 264 bits (674), Expect = 6e-71
Identities = 134/353 (37%), Positives = 215/353 (59%), Gaps = 22/353 (6%)
Query: 6 LTNMKEDQSRDGQETTSKVNKVSHDYNNLILSLPRENGCETQYMYFFHGFWCPSTLIQSV 65
L N++E++ + +E S+ K+ LI SLP E ++ + G+W ++QS+
Sbjct: 7 LRNLREEEEEE-EENQSEETKI------LISSLPWEIDYLGNKLFKYQGYWYYEDVLQSI 59
Query: 66 NSFQNNFHAKDSDIVVASIPKSGTTWLKGLAYAIVNRQHFTSLENNHPLLLFNPHELVPQ 125
+ ++F +++DIVVAS KSGTTWLK L +A+V R + +++HPLL NPHE+VP
Sbjct: 60 PNIHSSFQPQETDIVVASFYKSGTTWLKALTFALVQRSKHSLEDHHHPLLSHNPHEIVPY 119
Query: 126 FEVNLYGDKDGP-LPQI--DVSNMTEPRLFGTHMPFPSLPKSVKESNCKIIYICRNPFDT 182
E++LY + P L + S+ + PRLF THM +L +K+S CK++Y+CRN D
Sbjct: 120 LELDLYLNSSKPDLTKFLSSSSSSSSPRLFSTHMSLDALKLPLKKSPCKVVYVCRNVKDV 179
Query: 183 FVSYWIFINKIRLRK-----------SLTELTLEESFERYCKGICLFGPFWDNMLGYLKE 231
VS W F+N + + + + FE +C G+ L GPFWD+ Y +
Sbjct: 180 LVSLWCFLNANKGVEWGDFSQNEKIIRAENYSFKAIFESFCNGVTLHGPFWDHAQSYWRG 239
Query: 232 SIERPDRVLFLKYEDLKEDVNFHTKRIAEFVGIPFTQEEENNGVIENIIKLCSFESMKEI 291
S+E P LF++YE+LK + KR+AEF+ PFT+EEE++G ++ I++LCS ++ +
Sbjct: 240 SLEDPKHFLFMRYEELKAEPRTQVKRLAEFLDCPFTKEEEDSGTVDKILELCSLSNLSSL 299
Query: 292 EGNQSGTISGDIEKEFYFRKGEIGDWANYLSSSMVEKLSKVMEEKLNGSSLSF 344
E N++G++ G ++ + YFRKG++GDW +Y++S MV K+ ++EEKL GS L F
Sbjct: 300 EINKTGSLGG-VDYKTYFRKGQVGDWKSYMTSEMVNKIDMIVEEKLKGSGLKF 351
>At5g43690 steroid sulfotransferase-like
Length = 331
Score = 261 bits (668), Expect = 3e-70
Identities = 132/322 (40%), Positives = 196/322 (59%), Gaps = 6/322 (1%)
Query: 25 NKVSHDYNNLILSLPRENGCETQYMYFFHGFWCPSTLIQSVNSFQNNFHAKDSDIVVASI 84
+ +S + LI SLP + + + G W +Q+V ++Q NF +D+DI++AS
Sbjct: 14 DNLSEETKTLISSLPTYQDSHVK-LCKYQGCWYYHNTLQAVINYQRNFQPQDTDIILASF 72
Query: 85 PKSGTTWLKGLAYAIVNR--QHFTSLENNHPLLLFNPHELVPQFEVNLYGDKDGPLPQID 142
PKSGTTWLK L+ AIV R Q F HPLL NPH +VP FE ++Y P +
Sbjct: 73 PKSGTTWLKALSVAIVERSKQPFDDDPLTHPLLSDNPHGIVPFFEFDMYLKTS--TPDLT 130
Query: 143 VSNMTEPRLFGTHMPFPSLPKSVKESNCKIIYICRNPFDTFVSYWIFINKIRLRKSLTEL 202
+ + PRLF THMP + + +K S CK++Y+CRN D +S W F +K ++
Sbjct: 131 KFSTSSPRLFSTHMPLHTFKEGLKGSPCKVVYMCRNIKDVLISDWHFRSKYS-NNEVSRS 189
Query: 203 TLEESFERYCKGICLFGPFWDNMLGYLKESIERPDRVLFLKYEDLKEDVNFHTKRIAEFV 262
TLE FE +C G+ +GPFWD+ L Y + S+E P VLF++YE++K + KR+AEF+
Sbjct: 190 TLESMFESFCGGVSFYGPFWDHALSYWRGSLENPKHVLFMRYEEMKTEPCVQVKRLAEFL 249
Query: 263 GIPFTQEEENNGVIENIIKLCSFESMKEIEGNQSGTISGDIEKEFYFRKGEIGDWANYLS 322
G PFT+EEE++G I +++LCS ++ +E N++G + + + YFRKGE+GDW N+L+
Sbjct: 250 GFPFTKEEEDSGSISKLLELCSLGNLSGLEVNKTGKTWMNYDYKSYFRKGEVGDWKNHLT 309
Query: 323 SSMVEKLSKVMEEKLNGSSLSF 344
M K+ ++EEKL GS L F
Sbjct: 310 PEMENKIDMIIEEKLKGSDLKF 331
>At1g74100 putative flavonol sulfotransferase
Length = 338
Score = 257 bits (656), Expect = 8e-69
Identities = 136/346 (39%), Positives = 206/346 (59%), Gaps = 25/346 (7%)
Query: 9 MKEDQSRDGQETT--SKVNKVSHDYNNLILSLPRENGCET-QYMYFFHGFWCPSTLIQSV 65
M+ +++G E ++ K Y + I +LP+ G + + + G W L++ +
Sbjct: 1 MESKTTQNGSEVVELTEFEKTQKKYQDFIATLPKSKGWRPDEILTQYGGHWWQECLLEGL 60
Query: 66 NSFQNNFHAKDSDIVVASIPKSGTTWLKGLAYAIVNRQHFTSLENNHPLLLFNPHELVPQ 125
+++F A+ +D +V S PK+GTTWLK L YAIVNR + N PLL NPHE VP
Sbjct: 61 FHAKDHFEARPTDFLVCSYPKTGTTWLKALTYAIVNRSRYDDAAN--PLLKRNPHEFVPY 118
Query: 126 FEVNLYGDKDGPLPQIDVSNMTEPRLFGTHMPFPSLPKSVKESNCKIIYICRNPFDTFVS 185
E++ P +DV + LF TH+P LP S+ S CK++YI R+P DTF+S
Sbjct: 119 VEIDF-----AFYPTVDVLQDRKNPLFSTHIPNGLLPDSIVNSGCKMVYIWRDPKDTFIS 173
Query: 186 YWIFINKIRLRKSLTELTLEESFERYCKGICLFGPFWDNMLGYLKESIERPDRVLFLKYE 245
W F++K + ++ +LE+SF+ +CKG+ ++GP+ D++LGY K E PDR+LFL+YE
Sbjct: 174 MWTFLHKEKSQEGQLA-SLEDSFDMFCKGLSVYGPYLDHVLGYWKAYQENPDRILFLRYE 232
Query: 246 DLKEDVNFHTKRIAEFVGIPFTQEEENNGVIENIIKLCSFESMKEIEGNQSGTISGDIEK 305
++ + KR+AEF+G FT EEE NGV E ++KLCSFE++K +E N+ GD E+
Sbjct: 233 TMRANPLPFVKRLAEFMGYGFTDEEEENGVAEKVVKLCSFETLKNLEANK-----GDKER 287
Query: 306 E---------FYFRKGEIGDWANYLSSSMVEKLSKVMEEKLNGSSL 342
E YFRKG++GDWANYL+ M ++ ++EEK + L
Sbjct: 288 EDRPAVYANSAYFRKGKVGDWANYLTPEMAARIDGLVEEKFKDTGL 333
>At1g18590 unknown protein
Length = 346
Score = 254 bits (650), Expect = 4e-68
Identities = 136/338 (40%), Positives = 198/338 (58%), Gaps = 15/338 (4%)
Query: 9 MKEDQSRDGQETTSKVNKVSHDYNNLILSLPRENGCETQYMYF-FHGFWCPSTLIQSVNS 67
+ E + S+ K Y +I +LP ++G + + + G W L++ +
Sbjct: 11 VSESNHELASSSPSEFEKNQKHYQEIIATLPHKDGWRPKDPFVEYGGHWWLQPLLEGLLH 70
Query: 68 FQNNFHAKDSDIVVASIPKSGTTWLKGLAYAIVNRQHFTSLENNHPLLLFNPHELVPQFE 127
Q F A+ +D V S PK+GTTWLK L +AI NR F N PLL NPHE VP E
Sbjct: 71 AQKFFKARPNDFFVCSYPKTGTTWLKALTFAIANRSKFDVSTN--PLLKRNPHEFVPYIE 128
Query: 128 VNLYGDKDGPL-PQIDVSNMTEPRLFGTHMPFPSLPKSVKESNCKIIYICRNPFDTFVSY 186
+ D P P +DV LF TH+P+ LP+SV +S CKI+YI R+P DTFVS
Sbjct: 129 I------DFPFFPSVDVLKDEGNTLFSTHIPYDLLPESVVKSGCKIVYIWRDPKDTFVSM 182
Query: 187 WIFINKIRLRKSLTELTLEESFERYCKGICLFGPFWDNMLGYLKESIERPDRVLFLKYED 246
W F +K R ++ +++EE+F++YC+G+ +GP+ D++LGY K PD++LFLKYE
Sbjct: 183 WTFAHKERSQQGPV-VSIEEAFDKYCQGLSAYGPYLDHVLGYWKAYQANPDQILFLKYET 241
Query: 247 LKEDVNFHTKRIAEFVGIPFTQEEENNGVIENIIKLCSFESMKEIEGNQSGTISGD---- 302
++ D + KR+AEF+G FT+EEE V+E ++KLCSFE++K +E N+ D
Sbjct: 242 MRADPLPYVKRLAEFMGYGFTKEEEEGNVVEKVVKLCSFETLKNLEANKGEKDREDRPAV 301
Query: 303 IEKEFYFRKGEIGDWANYLSSSMVEKLSKVMEEKLNGS 340
YFRKG++GDW NYL+ MV ++ +MEEK G+
Sbjct: 302 YANSAYFRKGKVGDWQNYLTPEMVARIDGLMEEKFKGT 339
>At1g74090 flavonol sulfotransferase like protein
Length = 350
Score = 252 bits (644), Expect = 2e-67
Identities = 140/340 (41%), Positives = 200/340 (58%), Gaps = 24/340 (7%)
Query: 14 SRDGQETTS-KVNKVSHDYNNLILSLPRENGCETQY-MYFFHGFWCPSTLIQSVNSFQNN 71
S D +T S + K Y +LI + P E G + + + G+W +L++ Q
Sbjct: 19 SHDETKTESTEFEKNQKRYQDLISTFPHEKGWRPKEPLIEYGGYWWLPSLLEGCIHAQEF 78
Query: 72 FHAKDSDIVVASIPKSGTTWLKGLAYAIVNRQHFTSLENNHPLLLFNPHELVPQFEVNLY 131
F A+ SD +V S PK+GTTWLK L +AI NR F ++++PLL NPHE VP E+
Sbjct: 79 FQARPSDFLVCSYPKTGTTWLKALTFAIANRSRFD--DSSNPLLKRNPHEFVPYIEI--- 133
Query: 132 GDKDGPL-PQIDVSNMTEPRLFGTHMPFPSLPKSVKESNCKIIYICRNPFDTFVSYWIFI 190
D P P++DV LF TH+P+ LP SV +S CK++YI R P DTF+S W F+
Sbjct: 134 ---DFPFFPEVDVLKDKGNTLFSTHIPYELLPDSVVKSGCKMVYIWREPKDTFISMWTFL 190
Query: 191 NKIRLRKSLTEL----TLEESFERYCKGICLFGPFWDNMLGYLKESIERPDRVLFLKYED 246
+K R TEL LEESF+ +C+G+ +GP+ +++L Y K E PDR+LFLKYE
Sbjct: 191 HKER-----TELGPVSNLEESFDMFCRGLSGYGPYLNHILAYWKAYQENPDRILFLKYET 245
Query: 247 LKEDVNFHTKRIAEFVGIPFTQEEENNGVIENIIKLCSFESMKEIEGNQSGTISGD---- 302
++ D + K +AEF+G FT EEE GV+E ++ LCSFE++K +E N+ D
Sbjct: 246 MRADPLPYVKSLAEFMGHGFTAEEEEKGVVEKVVNLCSFETLKNLEANKGEKDREDRPGV 305
Query: 303 IEKEFYFRKGEIGDWANYLSSSMVEKLSKVMEEKLNGSSL 342
YFRKG++GDW+NYL+ M ++ +MEEK G+ L
Sbjct: 306 YANSAYFRKGKVGDWSNYLTPEMAARIDGLMEEKFKGTGL 345
>At1g28170 steroid sulfotransferase, putative
Length = 326
Score = 252 bits (643), Expect = 2e-67
Identities = 128/330 (38%), Positives = 196/330 (58%), Gaps = 6/330 (1%)
Query: 16 DGQETTSKVNKVSHDYNNLILSLPRENGCETQYMYFFHGFWCPSTLIQSVNSFQNNFHAK 75
D + K+ + +LI SLP + ++ ++ + G W +Q V +FQ F +
Sbjct: 2 DETKIPKKLQSDDEENISLISSLPFDVDFDSTKLFKYQGCWYDDKTLQGVLNFQRGFEPQ 61
Query: 76 DSDIVVASIPKSGTTWLKGLAYAIVNRQHFTSLENNHPLLLFNPHELVPQFEVNLYGDKD 135
D+DI++AS PKSGTTWLK L A++ R ++HPLLL NPH LVP E+ L+ +
Sbjct: 62 DTDIIIASFPKSGTTWLKALTVALLERSKQKHSSDDHPLLLDNPHGLVPFLELRLFTETS 121
Query: 136 GPLPQIDVSNMTE-PRLFGTHMPFPSLPKSVKESNCKIIYICRNPFDTFVSYWIFINKIR 194
P D+++++ PRLF TH+ F +L +++K S CKI+Y+ RN D VS+W F N +
Sbjct: 122 KP----DLTSISSSPRLFSTHVAFQTLREALKNSPCKIVYVWRNVKDVLVSFWYF-NSAK 176
Query: 195 LRKSLTELTLEESFERYCKGICLFGPFWDNMLGYLKESIERPDRVLFLKYEDLKEDVNFH 254
L+ L+ FE +C+G+ +GP W+++L Y + S+E VLFLKYE+LK +
Sbjct: 177 LKIEEERSILDSMFESFCRGVINYGPSWEHVLNYWRASLEDSKNVLFLKYEELKTEPRVQ 236
Query: 255 TKRIAEFVGIPFTQEEENNGVIENIIKLCSFESMKEIEGNQSGTISGDIEKEFYFRKGEI 314
KR+AEF+ PFT EEE G +E I+ LCS ++K +E N++G + + +FRKGE+
Sbjct: 237 LKRLAEFLDCPFTVEEEERGSVEEILDLCSLRNLKNLEINKTGKTLRGADHKIFFRKGEV 296
Query: 315 GDWANYLSSSMVEKLSKVMEEKLNGSSLSF 344
GD N+L+ M + + + EEK GS L F
Sbjct: 297 GDSKNHLTPEMEKIIDMITEEKFEGSDLKF 326
>At4g26280 steroid sulfotransferase - like protein
Length = 314
Score = 244 bits (623), Expect = 5e-65
Identities = 125/317 (39%), Positives = 187/317 (58%), Gaps = 24/317 (7%)
Query: 27 VSHDYNNLILSLPRENGCETQYMYFFHGFWCPSTLIQSVNSFQNNFHAKDSDIVVASIPK 86
++ + LI SL E G + + + G W +Q V +FQ F +D+DI+VAS PK
Sbjct: 12 LTEETKTLISSLSSEKGYLGRNLCKYQGSWYYYNFLQGVLNFQRGFKPQDTDIIVASYPK 71
Query: 87 SGTTWLKGLAYAIVNRQHFTSLEN--NHPLLLFNPHELVPQFEVNLYGDKDGPLPQIDVS 144
SGT WLK L A+ R S ++ +HPLL NPH L+
Sbjct: 72 SGTLWLKALTVALFERTKNPSHDDPMSHPLLSNNPHNLL--------------------- 110
Query: 145 NMTEPRLFGTHMPFPSLPKSVKESNCKIIYICRNPFDTFVSYWIFINKIRLRKSLTELTL 204
+ + PRLF TH PF +L +VK+S CK++YICR+ D+ VS W + + L K L
Sbjct: 111 SSSSPRLFSTHTPFHTLQVAVKDSPCKVVYICRDAKDSLVSRWHIVCR-SLNKEEDRTIL 169
Query: 205 EESFERYCKGICLFGPFWDNMLGYLKESIERPDRVLFLKYEDLKEDVNFHTKRIAEFVGI 264
E FE +C G+CLFGPFWD++L Y K S+E+P +VLF++Y+++K D + K++AEF+G
Sbjct: 170 ESMFESFCSGVCLFGPFWDHILSYWKASLEKPKQVLFMRYDEIKTDPHGQLKKLAEFLGC 229
Query: 265 PFTQEEENNGVIENIIKLCSFESMKEIEGNQSGTISGDIEKEFYFRKGEIGDWANYLSSS 324
PF++EEE NG + I+++CS ++ +E N++G IE + +FRKG +GDW N+L+
Sbjct: 230 PFSKEEEKNGSLNKILEMCSLPNLSSLEVNKTGKSINGIEYKNHFRKGIVGDWKNHLTPE 289
Query: 325 MVEKLSKVMEEKLNGSS 341
M K+ +M+EKL S
Sbjct: 290 MGSKIDMIMKEKLKDYS 306
>At3g51210 putative protein
Length = 67
Score = 37.4 bits (85), Expect = 0.012
Identities = 22/61 (36%), Positives = 36/61 (58%), Gaps = 4/61 (6%)
Query: 282 LCSFESMKEIEGNQSGTISGDIEKEFYFRKGEIGDWANYLSSSM--VEKLSKVMEEKLNG 339
LC S +E+ + T++G K+F FR G++GDW N+LS ++ K+ ++EK G
Sbjct: 5 LCVALSGREVNKTRR-TVNGVDHKDF-FRDGKVGDWKNHLSVTLETENKIDMTIKEKFQG 62
Query: 340 S 340
S
Sbjct: 63 S 63
>At4g16960 disease resistance RPP5 like protein
Length = 1041
Score = 30.4 bits (67), Expect = 1.5
Identities = 13/42 (30%), Positives = 25/42 (58%)
Query: 2 ASTDLTNMKEDQSRDGQETTSKVNKVSHDYNNLILSLPRENG 43
A TD++N+ + R+G + V K+++D +N + LP+ G
Sbjct: 144 ALTDISNLAGEDLRNGPSEAAMVVKIANDVSNKLFPLPKGFG 185
>At4g16940 disease resistance RPP5 like protein
Length = 1103
Score = 30.4 bits (67), Expect = 1.5
Identities = 13/42 (30%), Positives = 25/42 (58%)
Query: 2 ASTDLTNMKEDQSRDGQETTSKVNKVSHDYNNLILSLPRENG 43
A TD++N+ + R+G + V K+++D +N + LP+ G
Sbjct: 137 ALTDISNLAGEDLRNGPSEAAMVVKIANDVSNKLFPLPKGFG 178
>At4g16860 disease resistance RPP5 like protein
Length = 1256
Score = 30.0 bits (66), Expect = 2.0
Identities = 13/42 (30%), Positives = 24/42 (56%)
Query: 2 ASTDLTNMKEDQSRDGQETTSKVNKVSHDYNNLILSLPRENG 43
A TD++N+ + R+G V K+++D +N + LP+ G
Sbjct: 142 ALTDISNLAGEDLRNGPTEAFMVKKIANDVSNKLFPLPKGFG 183
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.137 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,461,052
Number of Sequences: 26719
Number of extensions: 392519
Number of successful extensions: 995
Number of sequences better than 10.0: 31
Number of HSP's better than 10.0 without gapping: 24
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 921
Number of HSP's gapped (non-prelim): 32
length of query: 348
length of database: 11,318,596
effective HSP length: 100
effective length of query: 248
effective length of database: 8,646,696
effective search space: 2144380608
effective search space used: 2144380608
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC148349.13


