

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148347.6 + phase: 0
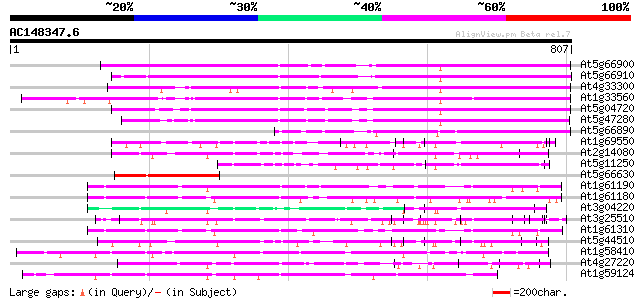
(807 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g66900 disease resistance protein-like 444 e-125
At5g66910 disease resistance protein-like 409 e-114
At4g33300 putative protein 359 4e-99
At1g33560 ADR1 (activated disease resistance) 343 3e-94
At5g04720 unknown protein 337 1e-92
At5g47280 disease resistance protein-like 332 4e-91
At5g66890 putative protein 244 1e-64
At1g69550 putative disease resistance protein 132 8e-31
At2g14080 putative disease resistance protein 132 1e-30
At5g11250 RPP1 disease resistance protein - like 120 4e-27
At5g66630 unknown protein 119 5e-27
At1g61190 110 2e-24
At1g61180 105 1e-22
At3g04220 putative disease resistance protein 104 2e-22
At3g25510 unknown protein 103 5e-22
At1g61310 similar to disease resistance protein 103 5e-22
At5g44510 disease resistance protein-like 100 3e-21
At1g58410 unknown protein 100 4e-21
At4g27220 putative protein 99 7e-21
At1g59124 PRM1 homolog (RF45) 99 1e-20
>At5g66900 disease resistance protein-like
Length = 809
Score = 444 bits (1143), Expect = e-125
Identities = 264/683 (38%), Positives = 392/683 (56%), Gaps = 36/683 (5%)
Query: 131 PPFKRPFDVPENPKFTVGLDIPFSKLKMELIRDGSSTLVLTGLGGLGKTTLATKLCWDQE 190
P F+ VP+ K VGLD P +LK L+ D TLV++ G GKTTL ++LC D +
Sbjct: 153 PVFRDLCSVPKLDKVIVGLDWPLGELKKRLLDDSVVTLVVSAPPGCGKTTLVSRLCDDPD 212
Query: 191 VNGKFKENIIFVTFSKTPMLKTIVERIIEHCGYPVPEFQSDEDAVNKLELLLKKV-EGSP 249
+ GKFK +I F S TP + IV+ +++H GY F++D A L LL+++ E P
Sbjct: 213 IKGKFK-HIFFNVVSNTPNFRVIVQNLLQHNGYNALTFENDSQAEVGLRKLLEELKENGP 271
Query: 250 LLLVLDDVWPTSESLVKKLQFEISDFKILVTSRVSFPRFRTTCILKPLAHEDAVTLFHHY 309
+LLVLDDVW ++S ++K Q ++ ++KILVTSR FP F + LKPL +DA L H+
Sbjct: 272 ILLVLDDVWRGADSFLQKFQIKLPNYKILVTSRFDFPSFDSNYRLKPLEDDDARALLIHW 331
Query: 310 AQMEKNSSDIIDKNLVEKVVRSCKGLPLTIKVIATSLRNRPDDLWRKIVMELSQGHSILD 369
A N+S ++L++K+++ C G P+ I+V+ SL+ R + W+ V S+G IL
Sbjct: 332 ASRPCNTSPDEYEDLLQKILKRCNGFPIVIEVVGVSLKGRSLNTWKGQVESWSEGEKILG 391
Query: 370 SN-TELLTRLQKIFDVLEDNPTIMECFMDIALFPEDHRIPVAALVDMWAKLYKLDDNGIQ 428
+L LQ FD L+ P + ECF+D+ F ED +I + ++DMW +LY + +
Sbjct: 392 KPYPTVLECLQPSFDALD--PNLKECFLDMGSFLEDQKIRASVIIDMWVELYGKGSSILY 449
Query: 429 AMEIINKLGIMNLANVIIPRKNASDTDNNNYNNHFIILHDILRDLGIYQSTKQPFEQRKR 488
+ L NL ++ N + ++ YN+ + HDILR+L I QS + +RKR
Sbjct: 450 MY--LEDLASQNLLKLVPLGTN--EHEDGFYNDFLVTQHDILRELAICQSEFKENLERKR 505
Query: 489 LIIDINKNRSELAEKQQSLLTRILSKVMRLCIKQNPQQLAAHILSVSTDEACASDWSQMQ 548
L ++I +N C+ + A +LS+STD+ +S W +M
Sbjct: 506 LNLEILENTFP-----------------DWCLNT----INASLLSISTDDLFSSKWLEMD 544
Query: 549 PTQVEVLILNLHTKQYSFPESIKKMSKLKVLIITNYSFHPCELNNFELLGSLQNLEKIRL 608
VE L+LNL + Y+ P I M KLKVL ITN+ F+P L+NF L SL NL++IRL
Sbjct: 545 CPNVEALVLNLSSSDYALPSFISGMKKLKVLTITNHGFYPARLSNFSCLSSLPNLKRIRL 604
Query: 609 ERILVPSFGT----LKNLKKLSLYMCNTILAF-EKGSILISDAFPNLEELNIDYCKDLVV 663
E++ + L +LKKLSL MC+ F + I++S+A L+E++IDYC DL
Sbjct: 605 EKVSITLLDIPQLQLSSLKKLSLVMCSFGEVFYDTEDIVVSNALSKLQEIDIDYCYDLDE 664
Query: 664 LQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGKLLNLK 723
L I +I+SLK L++TNC+KLS LP+ IG L LE+L S +L +P + L NL+
Sbjct: 665 LPYWISEIVSLKTLSITNCNKLSQLPEAIGNLSRLEVLRLCSSMNLSELPEATEGLSNLR 724
Query: 724 HLDISNCISLSSLPEEFGNLCNLKNLDMASCASIELPFSVVNLQNLKTITCDEETAATWE 783
LDIS+C+ L LP+E G L NLK + M C+ ELP SV NL+NL+ + CDEET WE
Sbjct: 725 FLDISHCLGLRKLPQEIGKLQNLKKISMRKCSGCELPESVTNLENLE-VKCDEETGLLWE 783
Query: 784 DFQHMLPNMKIEVLHVDVNLNWL 806
+ + N++++ ++ NLN L
Sbjct: 784 RLKPKMRNLRVQEEEIEHNLNLL 806
>At5g66910 disease resistance protein-like
Length = 815
Score = 409 bits (1052), Expect = e-114
Identities = 249/669 (37%), Positives = 381/669 (56%), Gaps = 36/669 (5%)
Query: 147 VGLDIPFSKLKMELIRDGSSTLVLTGLGGLGKTTLATKLCWDQEVNGKFKENIIFVTFSK 206
VGLD P +LK +L+ +S +V++G G GKTTL TKLC D E+ G+FK+ I + S
Sbjct: 173 VGLDWPLVELKKKLL--DNSVVVVSGPPGCGKTTLVTKLCDDPEIEGEFKK-IFYSVVSN 229
Query: 207 TPMLKTIVERIIEHCGYPVPEFQSDEDAVNKLELLLKKV-EGSPLLLVLDDVWPTSESLV 265
TP + IV+ +++ G F D A L LL+++ + +LLVLDDVW SE L+
Sbjct: 230 TPNFRAIVQNLLQDNGCGAITFDDDSQAETGLRDLLEELTKDGRILLVLDDVWQGSEFLL 289
Query: 266 KKLQFEISDFKILVTSRVSFPRFRTTCILKPLAHEDAVTLFHHYAQMEKNSSDIIDKNLV 325
+K Q ++ D+KILVTS+ F T L PL +E A +L +A ++S ++L+
Sbjct: 290 RKFQIDLPDYKILVTSQFDFTSLWPTYHLVPLKYEYARSLLIQWASPPLHTSPDEYEDLL 349
Query: 326 EKVVRSCKGLPLTIKVIATSLRNRPDDLWRKIVMELSQGHSIL-DSNTELLTRLQKIFDV 384
+K+++ C G PL I+V+ SL+ + LW+ V S+G +IL ++N + RLQ F+V
Sbjct: 350 QKILKRCNGFPLVIEVVGISLKGQALYLWKGQVESWSEGETILGNANPTVRQRLQPSFNV 409
Query: 385 LEDNPTIMECFMDIALFPEDHRIPVAALVDMWAKLYKLDDNGIQA-MEIINKLGIMNLAN 443
L+ P + ECFMD+ F +D +I + ++D+W +LY + M +N+L NL
Sbjct: 410 LK--PHLKECFMDMGSFLQDQKIRASLIIDIWMELYGRGSSSTNKFMLYLNELASQNLLK 467
Query: 444 VIIPRKNASDTDNNNYNNHFIILHDILRDLGIYQSTKQPFEQRKRLIIDINKNRSELAEK 503
++ + ++ YN + H+ILR+L I+QS +P QRK+L ++I ++
Sbjct: 468 LV--HLGTNKREDGFYNELLVTQHNILRELAIFQSELEPIMQRKKLNLEIREDNFPDE-- 523
Query: 504 QQSLLTRILSKVMRLCIKQNPQQLAAHILSVSTDEACASDWSQMQPTQVEVLILNLHTKQ 563
C+ Q + A +LS+ TD+ +S W +M VE L+LN+ +
Sbjct: 524 ---------------CLNQ---PINARLLSIYTDDLFSSKWLEMDCPNVEALVLNISSLD 565
Query: 564 YSFPESIKKMSKLKVLIITNYSFHPCELNNFELLGSLQNLEKIRLERILVPSFGT----L 619
Y+ P I +M KLKVL I N+ F+P L+NF L SL NL++IR E++ V L
Sbjct: 566 YALPSFIAEMKKLKVLTIANHGFYPARLSNFSCLSSLPNLKRIRFEKVSVTLLDIPQLQL 625
Query: 620 KNLKKLSLYMCNTILAF-EKGSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLN 678
+LKKLS +MC+ F + I +S A NL+E++IDYC DL L I +++SLK L+
Sbjct: 626 GSLKKLSFFMCSFGEVFYDTEDIDVSKALSNLQEIDIDYCYDLDELPYWIPEVVSLKTLS 685
Query: 679 VTNCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPE 738
+TNC+KLS LP+ IG L LE+L SC +L +P + +L NL+ LDIS+C+ L LP+
Sbjct: 686 ITNCNKLSQLPEAIGNLSRLEVLRMCSCMNLSELPEATERLSNLRSLDISHCLGLRKLPQ 745
Query: 739 EFGNLCNLKNLDMASCASIELPFSVVNLQNLKTITCDEETAATWEDFQHMLPNMKIEVLH 798
E G L L+N+ M C+ ELP SV L+NL+ + CDE T WE + N+++
Sbjct: 746 EIGKLQKLENISMRKCSGCELPDSVRYLENLE-VKCDEVTGLLWERLMPEMRNLRVHTEE 804
Query: 799 VDVNLNWLL 807
+ NL LL
Sbjct: 805 TEHNLKLLL 813
>At4g33300 putative protein
Length = 855
Score = 359 bits (921), Expect = 4e-99
Identities = 228/705 (32%), Positives = 371/705 (52%), Gaps = 66/705 (9%)
Query: 141 ENPKFTVGLDIPFSKLKMELIRDGSSTLVLTGLGGLGKTTLATKLCWDQEVNGKFKENII 200
++ KF VGL++ K+K + ++G+GG+GKTTLA +L D EV F+ I+
Sbjct: 176 DSEKFGVGLELGKVKVKKMMFESQGGVFGISGMGGVGKTTLAKELQRDHEVQCHFENRIL 235
Query: 201 FVTFSKTPMLKTIVERI---IEHC--GYPVPEFQSDEDAVNKLELLLKKVEGSPLLLVLD 255
F+T S++P+L+ + E I + C G PVP+ D KL ++LD
Sbjct: 236 FLTVSQSPLLEELRELIWGFLSGCEAGNPVPDCNFPFDGARKL-------------VILD 282
Query: 256 DVWPTSESLVKKLQFEISDFKILVTSRVSFPRFRTTCILKPLAHEDAVTLFHHYAQMEKN 315
DVW T+++L + F+ LV SR + T ++ L+ ++A++LF A +K+
Sbjct: 283 DVW-TTQALDRLTSFKFPGCTTLVVSRSKLTEPKFTYDVEVLSEDEAISLFCLCAFGQKS 341
Query: 316 -----SSDIIDKNLVE-----------KVVRSCKGLPLTIKVIATSLRNRPDDLWRKIVM 359
D++ ++ ++ +V CKGLPL +KV SL +P+ W+ ++
Sbjct: 342 IPLGFCKDLVKQHNIQSFSILRVLCLAQVANECKGLPLALKVTGASLNGKPEMYWKGVLQ 401
Query: 360 ELSQGHSILDSN-TELLTRLQKIFDVLEDNPTIMECFMDIALFPEDHRIPVAALVDMWAK 418
LS+G DS+ + LL +++ D L+ T +CF+D+ FPED +IP+ L+++W +
Sbjct: 402 RLSKGEPADDSHESRLLRQMEASLDNLDQ--TTKDCFLDLGAFPEDRKIPLDVLINIWIE 459
Query: 419 LYKLDDNGIQAMEIINKLGIMNLANVIIPRKNASDTDNNNYNNH---FIILHDILRDLGI 475
L+ +D+ A I+ L NL + + Y +H F+ HD+LRDL +
Sbjct: 460 LHDIDEGN--AFAILVDLSHKNLLTL-----GKDPRLGSLYASHYDIFVTQHDVLRDLAL 512
Query: 476 YQSTKQPFEQRKRLII---------DINKNRSELAEKQ-QSLLTRILSKVMRLCIKQNPQ 525
+ S +RKRL++ D +N E Q S+ T +++ + +
Sbjct: 513 HLSNAGKVNRRKRLLMPKRELDLPGDWERNNDEHYIAQIVSIHTGKCFSTLQIFLVLSCN 572
Query: 526 QLAAHILSVSTDEACASDWSQMQPTQVEVLILNLHTKQYSFPESIKKMSKLKVLIITNYS 585
H + E W M+ + E+LILN + +Y P I KMS+LKVL+I N
Sbjct: 573 NHQDH----NVGEMNEMQWFDMEFPKAEILILNFSSDKYVLPPFISKMSRLKVLVIINNG 628
Query: 586 FHPCELNNFELLGSLQNLEKIRLERILVPSFGT----LKNLKKLSLYMCNTILAFEKGSI 641
P L++F + L L + LER+ VP LKNL K+SL +C +F++ +
Sbjct: 629 MSPAVLHDFSIFAHLSKLRSLWLERVHVPQLSNSTTPLKNLHKMSLILCKINKSFDQTGL 688
Query: 642 LISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELL 701
++D FP L +L ID+C DLV L + IC + SL L++TNC +L LP+++ KL+ LE+L
Sbjct: 689 DVADIFPKLGDLTIDHCDDLVALPSSICGLTSLSCLSITNCPRLGELPKNLSKLQALEIL 748
Query: 702 SFSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMASCASIELPF 761
+C +L+ +P I +L LK+LDIS C+SLS LPEE G L L+ +DM C + P
Sbjct: 749 RLYACPELKTLPGEICELPGLKYLDISQCVSLSCLPEEIGKLKKLEKIDMRECCFSDRPS 808
Query: 762 SVVNLQNLKTITCDEETAATWEDFQHMLPNMKIEVLHVDVNLNWL 806
S V+L++L+ + CD + A WE+ + +P +KIE +L+WL
Sbjct: 809 SAVSLKSLRHVICDTDVAFMWEEVEKAVPGLKIEAAEKCFSLDWL 853
>At1g33560 ADR1 (activated disease resistance)
Length = 787
Score = 343 bits (879), Expect = 3e-94
Identities = 242/821 (29%), Positives = 416/821 (50%), Gaps = 83/821 (10%)
Query: 18 VLNKISQIVETARKHQSARQLLRSILKDLTLVVQDIKQYNEHL-DHPREEIKTLLEENDA 76
+L ++ + T + + L ++++D+ +++I+ L +H + ++ E +
Sbjct: 16 LLKLLALVANTVYSCKGIAERLITMIRDVQPTIREIQYSGAELSNHHQTQLGVFYEILEK 75
Query: 77 EESAC----KCSSENDSYVVDENQSLIVNDVEET----------LYKAREILEL-LNYET 121
C +C+ N +V N+ + D+E+ L+ E+ L +N +
Sbjct: 76 ARKLCEKVLRCNRWNLKHVYHANK---MKDLEKQISRFLNSQILLFVLAEVCHLRVNGDR 132
Query: 122 FEHKFNEAKPPFKRPFDVPE---------NPKFTVGLDIPFSKLKMELIRDGSSTLV-LT 171
E + PE +P+ L++ K+K + + + L ++
Sbjct: 133 IERNMDRLLTERNDSLSFPETMMEIETVSDPEIQTVLELGKKKVKEMMFKFTDTHLFGIS 192
Query: 172 GLGGLGKTTLATKLCWDQEVNGKFKENIIFVTFSKTPMLKTIVERIIEHCGYPVPEFQSD 231
G+ G GKTTLA +L D +V G FK ++F+T S++P + + E C + EF D
Sbjct: 193 GMSGSGKTTLAIELSKDDDVRGLFKNKVLFLTVSRSPNFENL-----ESC---IREFLYD 244
Query: 232 EDAVNKLELLLKKVEGSPLLLVLDDVWPTSESLVKKLQFEISDFKILVTSRVSFPRFRTT 291
KL ++LDDVW T ESL +L +I LV SR RTT
Sbjct: 245 GVHQRKL-------------VILDDVW-TRESL-DRLMSKIRGSTTLVVSRSKLADPRTT 289
Query: 292 CILKPLAHEDAVTLFHHYAQMEKNSSDIIDKNLVEKVVRSCKGLPLTIKVIATSLRNRPD 351
++ L ++A++L A +K+ +K LV++VV CKGLPL++KV+ SL+N+P+
Sbjct: 290 YNVELLKKDEAMSLLCLCAFEQKSPPSPFNKYLVKQVVDECKGLPLSLKVLGASLKNKPE 349
Query: 352 DLWRKIVMELSQGHSILDSN-TELLTRLQKIFDVLEDNPTIMECFMDIALFPEDHRIPVA 410
W +V L +G + +++ + + +++ + L+ P I +CF+D+ FPED +IP+
Sbjct: 350 RYWEGVVKRLLRGEAADETHESRVFAHMEESLENLD--PKIRDCFLDMGAFPEDKKIPLD 407
Query: 411 ALVDMWAKLYKLDDNGIQAMEIINKLGIMNLANVIIPRKNASDTDNN-NYNNHFIILHDI 469
L +W + + +D+ A + +L NL ++ N D + Y + F+ HD+
Sbjct: 408 LLTSVWVERHDIDEE--TAFSFVLRLADKNLLTIV---NNPRFGDVHIGYYDVFVTQHDV 462
Query: 470 LRDLGIYQSTKQPFEQRKRLIIDINKNRSELAEKQQSLLTRILSKVMRLCIKQNPQQLAA 529
LRDL ++ S + +D+N+ L K + +L R K + A
Sbjct: 463 LRDLALHMSNR----------VDVNRRERLLMPKTEPVLPREWEK-------NKDEPFDA 505
Query: 530 HILSVSTDEACASDWSQMQPTQVEVLILNLHTKQYSFPESIKKMSKLKVLIITNYSFHPC 589
I+S+ T E +W M + EVLILN + Y P I KMS+L+VL+I N P
Sbjct: 506 KIVSLHTGEMDEMNWFDMDLPKAEVLILNFSSDNYVLPPFIGKMSRLRVLVIINNGMSPA 565
Query: 590 ELNNFELLGSLQNLEKIRLERILVPSFGT----LKNLKKLSLYMCNTILAFEKGSILISD 645
L+ F + +L L + L+R+ VP + LKNL K+ L C +F + S IS
Sbjct: 566 RLHGFSIFANLAKLRSLWLKRVHVPELTSCTIPLKNLHKIHLIFCKVKNSFVQTSFDISK 625
Query: 646 AFPNLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSFSS 705
FP+L +L ID+C DL+ L++ I I SL L++TNC ++ LP+++ +++LE L +
Sbjct: 626 IFPSLSDLTIDHCDDLLELKS-IFGITSLNSLSITNCPRILELPKNLSNVQSLERLRLYA 684
Query: 706 CTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMASCASIELPFSVVN 765
C +L ++P + +L LK++DIS C+SL SLPE+FG L +L+ +DM C+ + LP SV
Sbjct: 685 CPELISLPVEVCELPCLKYVDISQCVSLVSLPEKFGKLGSLEKIDMRECSLLGLPSSVAA 744
Query: 766 LQNLKTITCDEETAATWEDFQHMLPNMKIEVLHVDVNLNWL 806
L +L+ + CDEET++ WE + ++P + IEV ++WL
Sbjct: 745 LVSLRHVICDEETSSMWEMVKKVVPELCIEVAKKCFTVDWL 785
>At5g04720 unknown protein
Length = 811
Score = 337 bits (865), Expect = 1e-92
Identities = 216/668 (32%), Positives = 352/668 (52%), Gaps = 46/668 (6%)
Query: 147 VGLDIPFSKLKMELIR--DGSSTLVLTGLGGLGKTTLATKLCWDQEVNGKFKENIIFVTF 204
VGLD+ K+K L + DG + ++G+ G GKTTLA +L D+EV G F ++F+T
Sbjct: 180 VGLDLGKRKVKEMLFKSIDGERLIGISGMSGSGKTTLAKELARDEEVRGHFGNKVLFLTV 239
Query: 205 SKTPMLKTIVERIIEHCGYPVPEFQSDEDAVNKLELLLKKVEGSPLLLVLDDVWPTSESL 264
S++P L E + H + +++ A + S L++LDDVW T ESL
Sbjct: 240 SQSPNL----EELRAHIWGFLTSYEAGVGAT---------LPESRKLVILDDVW-TRESL 285
Query: 265 VKKLQFEISDFKILVTSRVSFPRFRTTCILKPLAHEDAVTLFHHYAQMEKNSSDIIDKNL 324
+ + I LV SR R T ++ L +A LF +K ++L
Sbjct: 286 DQLMFENIPGTTTLVVSRSKLADSRVTYDVELLNEHEATALFCLSVFNQKLVPSGFSQSL 345
Query: 325 VEKVVRSCKGLPLTIKVIATSLRNRPDDLWRKIVMELSQGHSILDSN-TELLTRLQKIFD 383
V++VV CKGLPL++KVI SL+ RP+ W V LS+G +++ + + +++ +
Sbjct: 346 VKQVVGECKGLPLSLKVIGASLKERPEKYWEGAVERLSRGEPADETHESRVFAQIEATLE 405
Query: 384 VLEDNPTIMECFMDIALFPEDHRIPVAALVDMWAKLYKLDDNGIQAMEIINKLGIMNLAN 443
L+ P +CF+ + FPED +IP+ L+++ +L+ L+D A +I L NL
Sbjct: 406 NLD--PKTRDCFLVLGAFPEDKKIPLDVLINVLVELHDLED--ATAFAVIVDLANRNLLT 461
Query: 444 VII-PRKNASDTDNNNYNNHFIILHDILRDLGIYQSTKQPFEQRKRLIIDINKNRSELAE 502
++ PR T +Y + F+ HD+LRD+ + S R+RL++
Sbjct: 462 LVKDPRFGHMYT---SYYDIFVTQHDVLRDVALRLSNHGKVNNRERLLMP---------- 508
Query: 503 KQQSLLTRILSKVMRLCIKQNPQQLAAHILSVSTDEACASDWSQMQPTQVEVLILNLHTK 562
K++S+L R + N + A ++S+ T E DW M+ + EVLIL+ +
Sbjct: 509 KRESMLPREWER-------NNDEPYKARVVSIHTGEMTQMDWFDMELPKAEVLILHFSSD 561
Query: 563 QYSFPESIKKMSKLKVLIITNYSFHPCELNNFELLGSLQNLEKIRLERILVPSFGT---- 618
+Y P I KM KL L+I N P L++F + +L L+ + L+R+ VP +
Sbjct: 562 KYVLPPFIAKMGKLTALVIINNGMSPARLHDFSIFTNLAKLKSLWLQRVHVPELSSSTVP 621
Query: 619 LKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLN 678
L+NL KLSL C + ++ + I+ FP L +L ID+C DL+ L + IC I SL ++
Sbjct: 622 LQNLHKLSLIFCKINTSLDQTELDIAQIFPKLSDLTIDHCDDLLELPSTICGITSLNSIS 681
Query: 679 VTNCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPE 738
+TNC ++ LP+++ KL+ L+LL +C +L ++P I +L LK++DIS C+SLSSLPE
Sbjct: 682 ITNCPRIKELPKNLSKLKALQLLRLYACHELNSLPVEICELPRLKYVDISQCVSLSSLPE 741
Query: 739 EFGNLCNLKNLDMASCASIELPFSVVNLQNLKTITCDEETAATWEDFQHMLPNMKIEVLH 798
+ G + L+ +D C+ +P SVV L +L+ + CD E WE Q + +++E
Sbjct: 742 KIGKVKTLEKIDTRECSLSSIPNSVVLLTSLRHVICDREALWMWEKVQKAVAGLRVEAAE 801
Query: 799 VDVNLNWL 806
+ +WL
Sbjct: 802 KSFSRDWL 809
>At5g47280 disease resistance protein-like
Length = 623
Score = 332 bits (852), Expect = 4e-91
Identities = 214/651 (32%), Positives = 352/651 (53%), Gaps = 41/651 (6%)
Query: 161 IRDGSSTLVLTGLGGLGKTTLATKLCWDQEVNGKFKENIIFVTFSKTPMLKTIVERIIEH 220
+ D + + ++G+ G GKT LA +L D+EV G F ++F+T S++P L+ + I
Sbjct: 5 LNDEARIIGISGMIGSGKTILAKELARDEEVRGHFANRVLFLTVSQSPNLEELRSLI--- 61
Query: 221 CGYPVPEFQSDEDAVNKLELLLKKVEGSPLLLVLDDVWPTSESLVKKLQFEISDFKILVT 280
+F + +A L + V + L++LDDV T ESL +L F I LV
Sbjct: 62 -----RDFLTGHEA-GFGTALPESVGHTRKLVILDDV-RTRESL-DQLMFNIPGTTTLVV 113
Query: 281 SRVSFPRFRTTCILKPLAHEDAVTLFHHYAQMEKNSSDIIDKNLVEKVVRSCKGLPLTIK 340
S+ RTT ++ L DA +LF A +K+ K+LV++VV KGLPL++K
Sbjct: 114 SQSKLVDPRTTYDVELLNEHDATSLFCLSAFNQKSVPSGFSKSLVKQVVGESKGLPLSLK 173
Query: 341 VIATSLRNRPDDLWRKIVMELSQGHSILDSN-TELLTRLQKIFDVLEDNPTIMECFMDIA 399
V+ SL +RP+ W V LS+G + +++ +++ +++ + L+ P ECF+D+
Sbjct: 174 VLGASLNDRPETYWAIAVERLSRGEPVDETHESKVFAQIEATLENLD--PKTKECFLDMG 231
Query: 400 LFPEDHRIPVAALVDMWAKLYKLDDNGIQAMEIINKLGIMNLANVII-PRKNASDTDNNN 458
FPE +IPV L++M K++ L+D A +++ L NL ++ P A T +
Sbjct: 232 AFPEGKKIPVDVLINMLVKIHDLEDAA--AFDVLVDLANRNLLTLVKDPTFVAMGT---S 286
Query: 459 YNNHFIILHDILRDLGIYQSTKQPFEQRKRLIIDINKNRSELAEKQQSLLTRILSKVMRL 518
Y + F+ HD+LRD+ ++ + + +R RL++ K++++L +
Sbjct: 287 YYDIFVTQHDVLRDVALHLTNRGKVSRRDRLLMP----------KRETMLPSEWER---- 332
Query: 519 CIKQNPQQLAAHILSVSTDEACASDWSQMQPTQVEVLILNLHTKQYSFPESIKKMSKLKV 578
N + A ++S+ T E DW M + EVLI+N + Y P I KM L+V
Sbjct: 333 ---SNDEPYNARVVSIHTGEMTEMDWFDMDFPKAEVLIVNFSSDNYVLPPFIAKMGMLRV 389
Query: 579 LIITNYSFHPCELNNFELLGSLQNLEKIRLERILVPSFGT----LKNLKKLSLYMCNTIL 634
+I N P L++F + SL NL + LER+ VP + LKNL KL L +C
Sbjct: 390 FVIINNGTSPAHLHDFPIPTSLTNLRSLWLERVHVPELSSSMIPLKNLHKLYLIICKINN 449
Query: 635 AFEKGSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGK 694
+F++ +I I+ FP L ++ IDYC DL L + IC I SL +++TNC + LP++I K
Sbjct: 450 SFDQTAIDIAQIFPKLTDITIDYCDDLAELPSTICGITSLNSISITNCPNIKELPKNISK 509
Query: 695 LENLELLSFSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMASC 754
L+ L+LL +C +L+++P I +L L ++DIS+C+SLSSLPE+ GN+ L+ +DM C
Sbjct: 510 LQALQLLRLYACPELKSLPVEICELPRLVYVDISHCLSLSSLPEKIGNVRTLEKIDMREC 569
Query: 755 ASIELPFSVVNLQNLKTITCDEETAATWEDFQHMLPNMKIEVLHVDVNLNW 805
+ +P S V+L +L +TC E W++ + +P ++IE N+ W
Sbjct: 570 SLSSIPSSAVSLTSLCYVTCYREALWMWKEVEKAVPGLRIEATEKWFNMTW 620
>At5g66890 putative protein
Length = 415
Score = 244 bits (623), Expect = 1e-64
Identities = 156/435 (35%), Positives = 237/435 (53%), Gaps = 41/435 (9%)
Query: 382 FDVLEDNPTIMECFMDIALFPEDHRIPVAALVDMWAKLYKLDDNGIQAMEIINKLGIMNL 441
FD L N + ECF+D+A F ED RI + ++D+W+ Y G + M + L NL
Sbjct: 9 FDALPHN--LRECFLDMASFLEDQRIIASTIIDLWSASY-----GKEGMNNLQDLASRNL 61
Query: 442 ANVIIPRKNASDTDNNNYNNHFIILHDILRDLGIYQSTKQPFE--QRKRLIIDINKNRSE 499
++ +N + ++ YN + ++LR+ I Q K+ +RKRL ++I N
Sbjct: 62 LKLLPIGRN--EYEDGFYNELLVKQDNVLREFAINQCLKESSSIFERKRLNLEIQDN--- 116
Query: 500 LAEKQQSLLTRILSKVMRLCIKQNPQQ---LAAHILSVSTDEACASDWSQMQPTQVEVLI 556
K C+ NP+Q + A + S+STD++ AS W +M VE L+
Sbjct: 117 --------------KFPNWCL--NPKQPIVINASLFSISTDDSFASSWFEMDCPNVEALV 160
Query: 557 LNLHTKQYSFPESIKKMSKLKVLIITNYSFHPCELNNFELLGSLQNLEKIRLERILV--- 613
LN+ + Y+ P I M +LKV+II N+ P +L N L SL NL++IR E++ +
Sbjct: 161 LNISSSNYALPNFIATMKELKVVIIINHGLEPAKLTNLSCLSSLPNLKRIRFEKVSISLL 220
Query: 614 --PSFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQTGICDI 671
P G LK+L+KLSL+ C+ + A + +S+ +L+E+ IDYC +L L I +
Sbjct: 221 DIPKLG-LKSLEKLSLWFCHVVDALNELED-VSETLQSLQEIEIDYCYNLDELPYWISQV 278
Query: 672 ISLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGKLLNLKHLDISNCI 731
+SLKKL+VTNC+KL + + IG L +LE L SSC L +P +I +L NL+ LD+S
Sbjct: 279 VSLKKLSVTNCNKLCRVIEAIGDLRDLETLRLSSCASLLELPETIDRLDNLRFLDVSGGF 338
Query: 732 SLSSLPEEFGNLCNLKNLDMASCASIELPFSVVNLQNLKTITCDEETAATWEDFQHMLPN 791
L +LP E G L L+ + M C ELP SV NL+NL+ + CDE+TA W+ + + N
Sbjct: 339 QLKNLPLEIGKLKKLEKISMKDCYRCELPDSVKNLENLE-VKCDEDTAFLWKILKPEMKN 397
Query: 792 MKIEVLHVDVNLNWL 806
+ I + NLN L
Sbjct: 398 LTITEEKTEHNLNLL 412
>At1g69550 putative disease resistance protein
Length = 1398
Score = 132 bits (332), Expect = 8e-31
Identities = 86/221 (38%), Positives = 123/221 (54%), Gaps = 23/221 (10%)
Query: 567 PESIKKMSKLKVLIITNYSFHPCELNNFELLGSLQNLEKIRLERI--------LVPSFGT 618
P SI + LK+L + S + E+ S+ NL ++L + L S G
Sbjct: 827 PSSIGNLISLKILYLKRIS------SLVEIPSSIGNLINLKLLNLSGCSSLVELPSSIGN 880
Query: 619 LKNLKKLSLYMCNTI--LAFEKGSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKK 676
L NLKKL L C+++ L G+++ NL+EL + C LV L + I ++I+LK
Sbjct: 881 LINLKKLDLSGCSSLVELPLSIGNLI------NLQELYLSECSSLVELPSSIGNLINLKT 934
Query: 677 LNVTNCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSL 736
LN++ C L LP IG L NL+ L S C+ L +P+SIG L+NLK LD+S C SL L
Sbjct: 935 LNLSECSSLVELPSSIGNLINLQELYLSECSSLVELPSSIGNLINLKKLDLSGCSSLVEL 994
Query: 737 PEEFGNLCNLKNLDMASCAS-IELPFSVVNLQNLKTITCDE 776
P GNL NLK L+++ C+S +ELP S+ NL NL+ + E
Sbjct: 995 PLSIGNLINLKTLNLSECSSLVELPSSIGNLINLQELYLSE 1035
Score = 121 bits (304), Expect = 1e-27
Identities = 75/184 (40%), Positives = 110/184 (59%), Gaps = 15/184 (8%)
Query: 597 LGSLQNLEKIRLERI-----LVPSFGTLKNLKKLSLYMCNTILAFEK--GSILISDAFPN 649
+G+L NL+ + L L S G L NL++L L C++++ G+++ N
Sbjct: 926 IGNLINLKTLNLSECSSLVELPSSIGNLINLQELYLSECSSLVELPSSIGNLI------N 979
Query: 650 LEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTDL 709
L++L++ C LV L I ++I+LK LN++ C L LP IG L NL+ L S C+ L
Sbjct: 980 LKKLDLSGCSSLVELPLSIGNLINLKTLNLSECSSLVELPSSIGNLINLQELYLSECSSL 1039
Query: 710 EAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMASCAS-IELPFSVVNLQN 768
+P+SIG L+NLK LD+S C SL LP GNL NLK L+++ C+S +ELP S+ NL N
Sbjct: 1040 VELPSSIGNLINLKKLDLSGCSSLVELPLSIGNLINLKTLNLSGCSSLVELPSSIGNL-N 1098
Query: 769 LKTI 772
LK +
Sbjct: 1099 LKKL 1102
Score = 121 bits (303), Expect = 2e-27
Identities = 97/307 (31%), Positives = 144/307 (46%), Gaps = 29/307 (9%)
Query: 476 YQSTKQPFEQRKRLIIDINKNRSELAEKQQSLLTRILSKVMRLCIKQNPQQLAAHILSVS 535
Y T P + + ++ I SEL + + + + KVM L + ++L +++
Sbjct: 656 YPMTSLPSKFNLKFLVKIILKHSELEKLWEGIQPLVNLKVMDLRYSSHLKELPNLSTAIN 715
Query: 536 TDEACASDWSQMQPTQVEVLILNLHTKQYSFPESIKKMSKLKVLIITNYSFH---PCELN 592
E SD S + P SI + +K L I S P +
Sbjct: 716 LLEMVLSDCSSL----------------IELPSSIGNATNIKSLDIQGCSSLLKLPSSIG 759
Query: 593 NFELLGSLQNLEKIRLERILVPSFGTLKNLKKLSLYMCNTILAFEK--GSILISDAFPNL 650
N L L + L L S G L NL +L L C++++ G+++ NL
Sbjct: 760 NLITLPRLDLMGCSSLVE-LPSSIGNLINLPRLDLMGCSSLVELPSSIGNLI------NL 812
Query: 651 EELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTDLE 710
E C L+ L + I ++ISLK L + L +P IG L NL+LL+ S C+ L
Sbjct: 813 EAFYFHGCSSLLELPSSIGNLISLKILYLKRISSLVEIPSSIGNLINLKLLNLSGCSSLV 872
Query: 711 AIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMASCAS-IELPFSVVNLQNL 769
+P+SIG L+NLK LD+S C SL LP GNL NL+ L ++ C+S +ELP S+ NL NL
Sbjct: 873 ELPSSIGNLINLKKLDLSGCSSLVELPLSIGNLINLQELYLSECSSLVELPSSIGNLINL 932
Query: 770 KTITCDE 776
KT+ E
Sbjct: 933 KTLNLSE 939
Score = 120 bits (301), Expect = 3e-27
Identities = 181/708 (25%), Positives = 293/708 (40%), Gaps = 123/708 (17%)
Query: 147 VGLDIPFSKLKMELIRDGSS---TLVLTGLGGLGKTTLATKLC------WDQEVNGKFKE 197
+G+ K+K L D + T+ ++G G+GK+T+A L + V KFK
Sbjct: 253 IGMKAHIEKMKQLLCLDSTDERRTVGISGPSGIGKSTIARVLHNQISDGFQMSVFMKFKP 312
Query: 198 NIIFVTFSKTPMLKTIVERIIEHCGYPVPEFQSDEDA-VNKLELLLKKVEGSPLLLVLDD 256
+ S +K +E+ + + + ED +++L V G +L+VLD
Sbjct: 313 SYTRPICSDDHDVKLQLEQQF------LAQLINQEDIKIHQLGTAQNFVMGKKVLIVLDG 366
Query: 257 VWPTSESLVKKLQFEIS-----DFKILVTSRVS--FPRFRTTCILK---PLAHEDAVTLF 306
V + LV+ L + +I++T++ F+ I P HE A+ +F
Sbjct: 367 V----DQLVQLLAMPKAVCLGPGSRIIITTQDQQLLKAFQIKHIYNVDFPPDHE-ALQIF 421
Query: 307 HHYAQMEKNSSDIIDKNLVEKVVRSCKGLPLTIKVIATSLRNRPDDLWRKIVMELSQGHS 366
+A + D +K L KV R LPL ++V+ + R + W+ EL +
Sbjct: 422 CIHAFGHDSPDDGFEK-LATKVTRLAGNLPLGLRVMGSHFRGMSKEDWKG---ELPRLRI 477
Query: 367 ILDSNTELLTRLQKIFDVLEDNPTIMECFMDIALFPEDHRIPVAALVDMWAKLYKLDDNG 426
LD E+ + L+ +DVL+D + F+ IA F D I D + G
Sbjct: 478 RLDG--EIGSILKFSYDVLDDED--KDLFLHIACFFNDEGID-HTFEDTLRHKFSNVQRG 532
Query: 427 IQAMEIINKLGIMNLANVIIPRKNASDTDNNNYNNHFIIL-HDILRDLGIYQSTKQPF-- 483
+Q V++ R S+ +N + L +I+R+ +Y+ K+ F
Sbjct: 533 LQ---------------VLVQRSLISEDLTQPMHNLLVQLGREIVRNQSVYEPGKRQFLV 577
Query: 484 -----------EQRKRLIIDIN---------KNRSELAEKQQSLLTRIL---SKVMRLCI 520
+I IN N S+ + S L + RL +
Sbjct: 578 DGKEICEVLTSHTGSESVIGINFEVYWSMDELNISDRVFEGMSNLQFFRFDENSYGRLHL 637
Query: 521 KQNPQQLAAHILSVSTDEACASDWSQMQPTQVEVLILNLHTKQYSFPESIKKMSKLKVLI 580
Q L + + D + + V I+ H++ E I+ + LKV+
Sbjct: 638 PQGLNYLPPKLRILHWDYYPMTSLPSKFNLKFLVKIILKHSELEKLWEGIQPLVNLKVMD 697
Query: 581 ITNYSFHPCELNNFELLGSLQNLEKIRLERI-----LVPSFGTLKNLKKLSLYMCNTILA 635
+ YS H EL N L + NL ++ L L S G N+K L + C+++L
Sbjct: 698 L-RYSSHLKELPN---LSTAINLLEMVLSDCSSLIELPSSIGNATNIKSLDIQGCSSLLK 753
Query: 636 FEK--GSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIG 693
G+++ L L++ C LV L + I ++I+L +L++ C L LP IG
Sbjct: 754 LPSSIGNLI------TLPRLDLMGCSSLVELPSSIGNLINLPRLDLMGCSSLVELPSSIG 807
Query: 694 KLENLELLSFSSC------------------------TDLEAIPTSIGKLLNLKHLDISN 729
L NLE F C + L IP+SIG L+NLK L++S
Sbjct: 808 NLINLEAFYFHGCSSLLELPSSIGNLISLKILYLKRISSLVEIPSSIGNLINLKLLNLSG 867
Query: 730 CISLSSLPEEFGNLCNLKNLDMASCAS-IELPFSVVNLQNLKTITCDE 776
C SL LP GNL NLK LD++ C+S +ELP S+ NL NL+ + E
Sbjct: 868 CSSLVELPSSIGNLINLKKLDLSGCSSLVELPLSIGNLINLQELYLSE 915
Score = 115 bits (289), Expect = 7e-26
Identities = 87/251 (34%), Positives = 129/251 (50%), Gaps = 29/251 (11%)
Query: 556 ILNLHTKQYS-------FPESIKKMSKLKVLIITNYSFHPCELNNFELLGSLQNLEKIRL 608
++NL T S P SI + L+ L ++ S EL + +G+L NL+K+ L
Sbjct: 1001 LINLKTLNLSECSSLVELPSSIGNLINLQELYLSECS-SLVELPSS--IGNLINLKKLDL 1057
Query: 609 ERI-----LVPSFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVV 663
L S G L NLK L+L C++++ S NL++L++ C LV
Sbjct: 1058 SGCSSLVELPLSIGNLINLKTLNLSGCSSLVELPS-----SIGNLNLKKLDLSGCSSLVE 1112
Query: 664 LQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGKLLNLK 723
L + I ++I+LKKL+++ C L LP IG L NL+ L S C+ L +P+SIG L+NL+
Sbjct: 1113 LPSSIGNLINLKKLDLSGCSSLVELPLSIGNLINLQELYLSECSSLVELPSSIGNLINLQ 1172
Query: 724 HLDISNCISLSSLPEEFGNLCNLKNLDMASCASI----ELPFSVVNL-----QNLKTITC 774
L +S C SL LP GNL NLK LD+ C + +LP S+ L ++L+T+ C
Sbjct: 1173 ELYLSECSSLVELPSSIGNLINLKKLDLNKCTKLVSLPQLPDSLSVLVAESCESLETLAC 1232
Query: 775 DEETAATWEDF 785
W F
Sbjct: 1233 SFPNPQVWLKF 1243
>At2g14080 putative disease resistance protein
Length = 1215
Score = 132 bits (331), Expect = 1e-30
Identities = 168/658 (25%), Positives = 282/658 (42%), Gaps = 92/658 (13%)
Query: 147 VGLDIPFSKLKMELIRDGSSTLVLT--GLGGLGKTTLATKLCWDQEVNGKFKENIIFVTF 204
VG+ KL++ L D ++ G G+GKTT+ L +++ F+ +I
Sbjct: 231 VGMGAHMEKLELLLCLDSCEVRMIGIWGPPGIGKTTIVRFLY--NQLSSSFELSIFMENI 288
Query: 205 --------------SKTPMLKTIVERIIEHCGYPVPEFQSDEDAVNKLELLLKKVEGSPL 250
+K + + + +I++H +P L +L +++ +
Sbjct: 289 KTMHTILASSDDYSAKLILQRQFLSKILDHKDIEIPH----------LRVLQERLYNKKV 338
Query: 251 LLVLDDVWPTSE--SLVKKLQFEISDFKILVTSR----VSFPRFRTTCILKPLAHEDAVT 304
L+VLDDV + + +L K+ ++ +IL+T++ + R + +DA+
Sbjct: 339 LVVLDDVDQSVQLDALAKETRWFGPRSRILITTQDRKLLKAHRINNIYKVDLPNSDDALQ 398
Query: 305 LFHHYAQMEKNSSDIIDKNLVEKVVRSCKGLPLTIKVIATSLRNRPDDLWRKIVMELSQG 364
+F YA +K D K L KV PL ++V+ + R WRK E+ +
Sbjct: 399 IFCMYAFGQKTPYDGFYK-LARKVTWLVGNFPLGLRVVGSYFREMSKQEWRK---EIPRL 454
Query: 365 HSILDSNTELLTRLQKIFDVLEDNPTIMECFMDIALFPEDHRIPVAALVDMWAKLYKLDD 424
+ LD E + + +D L D + F+ IA F I KL+D
Sbjct: 455 RARLDGKIESVLKFS--YDALCDEDK--DLFLHIACFFNHESIE------------KLED 498
Query: 425 N-GIQAMEIINKLGIMNLANVIIPRKNASDTDNNNYNNHFIILHDILRDLGIYQSTKQPF 483
G ++I + ++ ++I + N++F+ +HD L LG KQ
Sbjct: 499 FLGKTFLDIAQRFHVLAEKSLI------------SINSNFVEMHDSLAQLGKEIVRKQSV 546
Query: 484 EQ--RKRLIIDINKNRSELAEKQQSLLTRILSKVMRLCIKQNPQQLAAHILSVSTDEACA 541
+ +++ ++D LA+ + I + L + +N + ++S E
Sbjct: 547 REPGQRQFLVDARDISEVLADDTAGGRSVI---GIYLDLHRNDD-----VFNIS--EKAF 596
Query: 542 SDWSQMQPTQVEVLILNLHTKQYSFPESIKKMSKLKVLIITNYSFHPCELNNF--ELLGS 599
S +Q +V+ NL P + +S+ L+ Y C + F E L
Sbjct: 597 EGMSNLQFLRVKNFG-NLFPAIVCLPHCLTYISRKLRLLDWMYFPMTCFPSKFNPEFLVE 655
Query: 600 LQNLEKIRLERILVPSFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCK 659
L N+ +LE+ L L+NLK++ L+ + S + NLE LN++ C
Sbjct: 656 L-NMWGSKLEK-LWEEIQPLRNLKRMDLFSSKNLKELPDLS-----SATNLEVLNLNGCS 708
Query: 660 DLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGKL 719
LV L I + L KL ++ C L LP IG NL+ + FS C +L +P+SIG
Sbjct: 709 SLVELPFSIGNATKLLKLELSGCSSLLELPSSIGNAINLQTIDFSHCENLVELPSSIGNA 768
Query: 720 LNLKHLDISNCISLSSLPEEFGNLCNLKNLDMASCASI-ELPFSVVNLQNLKT--ITC 774
NLK LD+S C SL LP GN NLK L + C+S+ ELP S+ N NLK +TC
Sbjct: 769 TNLKELDLSCCSSLKELPSSIGNCTNLKKLHLICCSSLKELPSSIGNCTNLKELHLTC 826
Score = 45.8 bits (107), Expect = 1e-04
Identities = 57/222 (25%), Positives = 92/222 (40%), Gaps = 47/222 (21%)
Query: 552 VEVLILNLHTKQYSFPESIKKMSKLKVLIITNYSFHPCELNNFELLGSLQNLEKIRLERI 611
+E LIL P I K + LK+L N + C + +G+L L ++RL
Sbjct: 843 LEKLILAGCESLVELPSFIGKATNLKIL---NLGYLSCLVELPSFIGNLHKLSELRLRGC 899
Query: 612 ----LVPSFGTLKNLKKLSLYMCNTILAFEKGSILISD----------------AFPNLE 651
++P+ L+ L +L L C + F S I ++P LE
Sbjct: 900 KKLQVLPTNINLEFLNELDLTDCILLKTFPVISTNIKRLHLRGTQIEEVPSSLRSWPRLE 959
Query: 652 ELNIDYCKDL----------VVLQTGICDI----------ISLKKLNVTNCHKLSSLPQD 691
+L + Y ++L VL+ +I L++L ++ C KL SLPQ
Sbjct: 960 DLQMLYSENLSEFSHVLERITVLELSDINIREMTPWLNRITRLRRLKLSGCGKLVSLPQ- 1018
Query: 692 IGKLENLELLSFSSCTDLEAIPTSIGKLLNLKHLDISNCISL 733
++L +L +C LE + S N+K LD +NC+ L
Sbjct: 1019 --LSDSLIILDAENCGSLERLGCSFNNP-NIKCLDFTNCLKL 1057
>At5g11250 RPP1 disease resistance protein - like
Length = 1189
Score = 120 bits (300), Expect = 4e-27
Identities = 135/504 (26%), Positives = 233/504 (45%), Gaps = 69/504 (13%)
Query: 300 EDAVTLFHHYAQMEKNSSDIIDKNLVEKVVRSCKGLPLTIKVIATSLRNRPDDLWRKIVM 359
E+A+ +F YA + + D +NL KV+ LPL ++++ + R + W+K +
Sbjct: 400 EEALQIFCMYAFGQNSPKDGF-QNLAWKVINLAGNLPLGLRIMGSYFRGMSREEWKKSLP 458
Query: 360 ELSQGHSILDSNTELLTRLQKIFDVLEDNPTIMECFMDIALFPEDHRIPVAALVDMWAKL 419
L S LD++ + + + +D L+D + F+ IA F I + L + AK
Sbjct: 459 RLE---SSLDADIQSILKFS--YDALDDEDKNL--FLHIACFFNGKEIKI--LEEHLAKK 509
Query: 420 YKLDDNGIQAMEIINKLGIMNLANVIIPRKNASDTDNNNYNNHFIILH---DILRDLGIY 476
+ +E+ +L NV+ + S ++ H ++ +I+R+ I+
Sbjct: 510 F---------VEVRQRL------NVLAEKSLISFSNWGTIEMHKLLAKLGGEIVRNQSIH 554
Query: 477 QSTKQPFEQRKRLIIDINKNRSE------------LAEKQQSLLTRILS-----KVMRLC 519
+ ++ F I D+ + + E++ + R+ + +R
Sbjct: 555 EPGQRQFLFDGEEICDVLNGDAAGSKSVIGIDFHYIIEEEFDMNERVFEGMSNLQFLRFD 614
Query: 520 IKQNPQQLAAHILSVSTDEACASDWSQMQPT------QVEVLI-LNL-HTKQYSFPESIK 571
+ QL+ LS + + DW T VE LI LNL H+K E +K
Sbjct: 615 CDHDTLQLSRG-LSYLSRKLQLLDWIYFPMTCLPSTVNVEFLIELNLTHSKLDMLWEGVK 673
Query: 572 KMSKLKVLIITNYSFHPCELNNFELLGSLQNLEKIRLER----ILVPS-FGTLKNLKKLS 626
+ L+ + ++ YS + EL + L + NL K+ L I +PS G NL+ L
Sbjct: 674 PLHNLRQMDLS-YSVNLKELPD---LSTAINLRKLILSNCSSLIKLPSCIGNAINLEDLD 729
Query: 627 LYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLS 686
L C++++ DA NL++L + YC +LV L + I + I+L++L++ C L
Sbjct: 730 LNGCSSLVELPS----FGDAI-NLQKLLLRYCSNLVELPSSIGNAINLRELDLYYCSSLI 784
Query: 687 SLPQDIGKLENLELLSFSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNL 746
LP IG NL +L + C++L +P+SIG +NL+ LD+ C L LP GN NL
Sbjct: 785 RLPSSIGNAINLLILDLNGCSNLLELPSSIGNAINLQKLDLRRCAKLLELPSSIGNAINL 844
Query: 747 KNLDMASCAS-IELPFSVVNLQNL 769
+NL + C+S +ELP S+ N NL
Sbjct: 845 QNLLLDDCSSLLELPSSIGNATNL 868
Score = 99.0 bits (245), Expect = 9e-21
Identities = 63/185 (34%), Positives = 103/185 (55%), Gaps = 11/185 (5%)
Query: 598 GSLQNLEKIRLERI-----LVPSFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEE 652
G NL+K+ L L S G NL++L LY C++++ I +A NL
Sbjct: 743 GDAINLQKLLLRYCSNLVELPSSIGNAINLRELDLYYCSSLIRLPSS---IGNAI-NLLI 798
Query: 653 LNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTDLEAI 712
L+++ C +L+ L + I + I+L+KL++ C KL LP IG NL+ L C+ L +
Sbjct: 799 LDLNGCSNLLELPSSIGNAINLQKLDLRRCAKLLELPSSIGNAINLQNLLLDDCSSLLEL 858
Query: 713 PTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMASCASIE-LPFSVVNLQNLKT 771
P+SIG NL ++++SNC +L LP GNL L+ L + C+ +E LP + +NL++L
Sbjct: 859 PSSIGNATNLVYMNLSNCSNLVELPLSIGNLQKLQELILKGCSKLEDLPIN-INLESLDI 917
Query: 772 ITCDE 776
+ ++
Sbjct: 918 LVLND 922
>At5g66630 unknown protein
Length = 702
Score = 119 bits (299), Expect = 5e-27
Identities = 66/151 (43%), Positives = 95/151 (62%), Gaps = 2/151 (1%)
Query: 152 PFSKLKMELIRDGSSTLVLTGLGGLGKTTLATKLCWDQEVNGKFKENIIFVTFSKTPMLK 211
P +LK L DG T+V++ LGKTTL TKLC D +V KFK+ I F++ SK P ++
Sbjct: 175 PLMELKKMLFEDGVVTVVVSAPYALGKTTLVTKLCHDADVKEKFKQ-IFFISVSKFPNVR 233
Query: 212 TIVERIIEHCGYPVPEFQSDEDAVNKLELLLKKV-EGSPLLLVLDDVWPTSESLVKKLQF 270
I +++EH G E+++D DA+ ++ LLK++ +LLVLDDVW ESL++K
Sbjct: 234 LIGHKLLEHIGCKANEYENDLDAMLYIQQLLKQLGRNGSILLVLDDVWAEEESLLQKFLI 293
Query: 271 EISDFKILVTSRVSFPRFRTTCILKPLAHED 301
++ D+KILVTSR F F T LKPL ++
Sbjct: 294 QLPDYKILVTSRFEFTSFGPTFHLKPLIDDE 324
>At1g61190
Length = 967
Score = 110 bits (276), Expect = 2e-24
Identities = 163/715 (22%), Positives = 303/715 (41%), Gaps = 96/715 (13%)
Query: 113 ILELLNYETFEHKFNEAKPPFKRPFDVPENP-KFTVGLDIPFSKLKMELIRDGSSTLVLT 171
+LE + E F+E P R +V E P + T+G + K L+ DG + L
Sbjct: 121 LLEEVTKLKSEGNFDEVSQPPPRS-EVEERPTQPTIGQEEMLKKAWNRLMEDGVGIMGLH 179
Query: 172 GLGGLGKTTLATKLCWDQEVNGKFKENIIFVTFSKTPMLKTIVERIIE--HCGYPVPEFQ 229
G+GG+GKTTL K+ G + +I++ S+ L + E I E H + + +
Sbjct: 180 GMGGVGKTTLFKKIHNKFAETGGTFDIVIWIVVSQGAKLSKLQEDIAEKLHLCDDLWKNK 239
Query: 230 SDEDAVNKLELLLKKVEGSPLLLVLDDVWPTS--ESLVKKLQFEISDFKILVTSR----V 283
++ D + +LK G +L+LDD+W E++ E++ K+ T+R
Sbjct: 240 NESDKATDIHRVLK---GKRFVLMLDDIWEKVDLEAIGIPYPSEVNKCKVAFTTRDQKVC 296
Query: 284 SFPRFRTTCILKPLAHEDAVTLFHHYAQMEKNSSDIIDKNLVEKVVRSCKGLPLTIKVIA 343
+K L EDA LF + SD + L +V + C+GLPL + I
Sbjct: 297 GQMGDHKPMQVKCLEPEDAWELFKNKVGDNTLRSDPVIVGLAREVAQKCRGLPLALSCIG 356
Query: 344 TSLRNRPD-DLWRKIVMELSQGHS-ILDSNTELLTRLQKIFDVLEDNPTIMECFMDIALF 401
++ ++ W + L++ + D ++L L+ +D LED I CF+ ALF
Sbjct: 357 ETMASKTMVQEWEHAIDVLTRSAAEFSDMQNKILPILKYSYDSLEDEH-IKSCFLYCALF 415
Query: 402 PEDHRIPVAALVDMWAKLYKLDDNGIQAMEIINKLGIMNLANVIIPRKNASDTDNNNYNN 461
PED +I L++ W + ++ Q ++ G L +I R N D
Sbjct: 416 PEDDKIDTKTLINKWICEGFIGED--QVIKRARNKGYEMLGTLI--RANLLTNDRGFVKW 471
Query: 462 HFIILHDILRDLGIYQSTKQPFEQRKRLIIDINKNRSELAEKQQSLLTRILSKVMRLCIK 521
H +++HD++R++ ++ ++ +Q++ ++ E+ + + R +S +M
Sbjct: 472 H-VVMHDVVREMALWIASDFG-KQKENYVVRARVGLHEIPKVKDWGAVRRMSLMMN---- 525
Query: 522 QNPQQLAAHILSVSTDEACASDWSQMQPTQVEVLILNLHTKQYSFPESIKKMSKLKVLII 581
I ++ + C+ ++ L L + + E I+ M KL VL +
Sbjct: 526 --------EIEEITCESKCS---------ELTTLFLQSNQLKNLSGEFIRYMQKLVVLDL 568
Query: 582 TNYSFHPCELNNF-ELLGSLQNLEKIRLERILVPSFGT-LKNLKKLSLYMCNTILAFEKG 639
+ H + N E + L +L+ + L + LK LKKL
Sbjct: 569 S----HNPDFNELPEQISGLVSLQYLDLSWTRIEQLPVGLKELKKLIF------------ 612
Query: 640 SILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLNV--TNCHKLSSLPQDIGKLEN 697
LN+ + + L + +GI ++SL+ L++ +N H +S+ +++ +LEN
Sbjct: 613 -------------LNLCFTERLCSI-SGISRLLSLRWLSLRESNVHGDASVLKELQQLEN 658
Query: 698 LELLSFSSCTDLEAIPTSIGKLLNL--------KHLDISNCISLSSL-----PEEFGNLC 744
L+ L + +L ++ + KL+++ K D+S S+ +L + +
Sbjct: 659 LQDLRITESAELISLDQRLAKLISVLRIEGFLQKPFDLSFLASMENLYGLLVENSYFSEI 718
Query: 745 NLKNLDMASCASI-----ELPFSVVNLQNLKTITCDEETAATWEDFQHMLPNMKI 794
N+K + + +S ++P NL L + C TW F L N+ I
Sbjct: 719 NIKCRESETESSYLHINPKIP-CFTNLTGLIIMKCHSMKDLTWILFAPNLVNLDI 772
>At1g61180
Length = 889
Score = 105 bits (261), Expect = 1e-22
Identities = 178/789 (22%), Positives = 335/789 (41%), Gaps = 142/789 (17%)
Query: 113 ILELLNYETFEHKFNEAKPPFKRPFDVPENP-KFTVGLDIPFSKLKMELIRDGSSTLVLT 171
+LE + E F+E P R +V E P + T+G + K L+ DG + L
Sbjct: 120 LLEEVKKLNSEGNFDEVSQPPPRS-EVEERPTQPTIGQEDMLEKAWNRLMEDGVGIMGLH 178
Query: 172 GLGGLGKTTLATKLCWD-QEVNGKFKENIIFVTFSKTPMLKTIVERIIE--HCGYPVPEF 228
G+GG+GKTTL K+ E+ G F + +I++ SK M+ + E I E H + +
Sbjct: 179 GMGGVGKTTLFKKIHNKFAEIGGTF-DIVIWIVVSKGVMISKLQEDIAEKLHLCDDLWKN 237
Query: 229 QSDEDAVNKLELLLKKVEGSPLLLVLDDVWPTS--ESLVKKLQFEISDFKILVTSRVSFP 286
+++ D + +LK G +L+LDD+W E++ E++ K+ T+R
Sbjct: 238 KNESDKATDIHRVLK---GKRFVLMLDDIWEKVDLEAIGIPYPSEVNKCKVAFTTRS--- 291
Query: 287 RFRTTC---------ILKPLAHEDAVTLFHHYAQMEKNSSDIIDKNLVEKVVRSCKGLPL 337
R C + L EDA LF + SSD + L +V + C+GLPL
Sbjct: 292 --REVCGEMGDHKPMQVNCLEPEDAWELFKNKVGDNTLSSDPVIVELAREVAQKCRGLPL 349
Query: 338 TIKVIATSLRNRP--DDLWRKIVMELSQGHSILDSNTELLTRLQKIFDVLEDNPTIMECF 395
+ VI ++ ++ + I + + D ++L L+ +D L D I CF
Sbjct: 350 ALNVIGETMSSKTMVQEWEHAIHVFNTSAAEFSDMQNKILPILKYSYDSLGDE-HIKSCF 408
Query: 396 MDIALFPEDHRIPVAALVDMWA--------KLYKLDDNGIQAMEIINKLGIMNLANVIIP 447
+ ALFPED I L+D W ++ K N AM LG + AN++
Sbjct: 409 LYCALFPEDGEIYNEKLIDYWICEGFIGEDQVIKRARNKGYAM-----LGTLTRANLL-- 461
Query: 448 RKNASDTDNNNYNNHFIILHDILRDLGIYQSTKQPFEQRKRLII-------DINKNRSEL 500
++ ++HD++R++ ++ ++ +Q++ ++ +I K +
Sbjct: 462 ---------TKVGTYYCVMHDVVREMALWIASDFG-KQKENFVVQAGVGLHEIPKVKDWG 511
Query: 501 AEKQQSLLTRIL---------SKVMRLCIKQN-----PQQLAAHILS-VSTDEACASDWS 545
A ++ SL+ + S++ L ++ N P ++ V D + D++
Sbjct: 512 AVRKMSLMDNDIEEITCESKCSELTTLFLQSNKLKNLPGAFIRYMQKLVVLDLSYNRDFN 571
Query: 546 QMQPTQVEVLI----LNL-HTKQYSFPESIKKMSKLKVLIITNYSFHPCELNNFELLGSL 600
++ P Q+ L+ L+L +T P +K++ KL L +T Y+ C ++ L SL
Sbjct: 572 KL-PEQISGLVSLQFLDLSNTSIEHMPIGLKELKKLTFLDLT-YTDRLCSISGISRLLSL 629
Query: 601 QNL----EKIRLERILVPSFGTLKNLKKLSLYMCNTILAFEKG-SILISD---------- 645
+ L K+ + ++ L+NL++L++ + +++ ++ + LIS+
Sbjct: 630 RLLRLLGSKVHGDASVLKELQQLQNLQELAITVSAELISLDQRLAKLISNLCIEGFLQKP 689
Query: 646 -------AFPNLEELNID-------YCKD------LVVLQTGICDIISLKKLNVTNCHKL 685
+ NL L ++ C++ + + I +L +L + CH +
Sbjct: 690 FDLSFLASMENLSSLRVENSYFSEIKCRESETESSYLRINPKIPCFTNLSRLEIMKCHSM 749
Query: 686 SSL------PQDIGKL-----ENLELLSFSSCTDLEAIPTSIGKLLNLKHLDISNCISLS 734
L P + L E E+++ T+L TSI L L+ L + N L
Sbjct: 750 KDLTWILFAPNLVVLLIEDSREVGEIINKEKATNL----TSITPFLKLEWLILYNLPKLE 805
Query: 735 SLPEEFGNLCNLKNLDMASCASI-ELPFSVVNLQNLKTITC------DEETAATWED--- 784
S+ L +D+++C + +LP + ++ ++ ++E WED
Sbjct: 806 SIYWSPLPFPVLLTMDVSNCPKLRKLPLNATSVSKVEEFEIHMYPPPEQENELEWEDDDT 865
Query: 785 FQHMLPNMK 793
LP++K
Sbjct: 866 KNRFLPSIK 874
>At3g04220 putative disease resistance protein
Length = 896
Score = 104 bits (260), Expect = 2e-22
Identities = 154/704 (21%), Positives = 281/704 (39%), Gaps = 121/704 (17%)
Query: 112 EILELLNYETFEHKFNEAKPPFKRPFDVPENPKFTVGLDIPFSKLK--MELIRDGSSTLV 169
+I E+LN+ T F++ +G+ K+K +++ D T+
Sbjct: 220 DISEMLNHSTPSRDFDDL-----------------IGMGDHMEKMKPLLDIDSDEMKTIG 262
Query: 170 LTGLGGLGKTTLATKLCWDQEVNGKFKENIIFVTFSKTPMLKTIVERIIEHCGYP---VP 226
+ G G+GKTT+A L + + KF+ ++ + + + E +
Sbjct: 263 IWGPPGVGKTTIARSLY--NQHSDKFQLSVFMESIKTAYTIPACSDDYYEKLQLQQRFLS 320
Query: 227 EFQSDEDA-VNKLELLLKKVEGSPLLLVLDDVWPT--SESLVKKLQFEISDFKILVTSR- 282
+ + E+ + L + +++ +L+V+DDV + ++L K+ + +I++T++
Sbjct: 321 QITNQENVQIPHLGVAQERLNDKKVLVVIDDVNQSVQVDALAKENDWLGPGSRIIITTQD 380
Query: 283 --------------VSFPRFRTTCILKPLAHEDAVTLFHHYAQMEKNSSDIIDKNLVEKV 328
V +P + E+A+ +F +A +K+ D ++ L ++V
Sbjct: 381 RGILRAHGIEHIYEVDYPNY-----------EEALQIFCMHAFGQKSPYDGFEE-LAQQV 428
Query: 329 VRSCKGLPLTIKVIATSLRNRPDDLWRKIVMELSQGHSILDSNTELLTRLQKIFDVLEDN 388
LPL +KV+ + R W M L + + LD E + +L +D L
Sbjct: 429 TTLSGRLPLGLKVMGSYFRGMTKQEW---TMALPRVRTHLDGKIESILKLS--YDAL--- 480
Query: 389 PTIMECFMDIALFPEDHRIPVAALVDMWAKLYKLDDNGIQAMEIINKLGIMNLANVIIPR 448
C +D +LF A + DD + ++ K + ++
Sbjct: 481 -----CDVDKSLFLH------------LACSFHNDDTELVEQQLGKKFSDLRQGLHVLAE 523
Query: 449 KNASDTDNNNYNNHFIILHDILRDLGIYQSTKQPFEQRKRLIIDINKNRSELAEKQQSLL 508
K+ D H ++L + R++ QS +P +++ ++D R L + S
Sbjct: 524 KSLIHMDLRLIRMH-VLLAQLGREIVRKQSIHEP--GQRQFLVDATDIREVLTDDTGS-- 578
Query: 509 TRILSKVMRLCIKQNPQQLAAHILSVSTDEACASDWSQMQPTQVEVLILNLHTKQY---- 564
V+ + N + I E S +Q ++ + + H Y
Sbjct: 579 ----RSVIGIDFDFNTMEKELDI-----SEKAFRGMSNLQFIRIYGDLFSRHGVYYFGGR 629
Query: 565 ------------SFPESIKKM-SKLKVLIITNYSFH--PCELNNFELLGSLQNLEKIRLE 609
FP + + KL++L + P E + L+ K+
Sbjct: 630 GHRVSLDYDSKLHFPRGLDYLPGKLRLLHWQQFPMTSLPSEFHAEFLVKLCMPYSKLEK- 688
Query: 610 RILVPSFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQTGIC 669
L L+NL+ L L + S + L+ L+I+ C LV L + I
Sbjct: 689 --LWEGIQPLRNLEWLDLTCSRNLKELPDLSTATN-----LQRLSIERCSSLVKLPSSIG 741
Query: 670 DIISLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGKLLNLKHLDISN 729
+ +LKK+N+ C L LP G L NL+ L C+ L +PTS G L N++ L+
Sbjct: 742 EATNLKKINLRECLSLVELPSSFGNLTNLQELDLRECSSLVELPTSFGNLANVESLEFYE 801
Query: 730 CISLSSLPEEFGNLCNLKNLDMASCAS-IELPFSVVNLQNLKTI 772
C SL LP FGNL NL+ L + C+S +ELP S NL NL+ +
Sbjct: 802 CSSLVKLPSTFGNLTNLRVLGLRECSSMVELPSSFGNLTNLQVL 845
Score = 104 bits (260), Expect = 2e-22
Identities = 70/212 (33%), Positives = 109/212 (51%), Gaps = 18/212 (8%)
Query: 568 ESIKKMSKLKVLIITNYSFHPCELNNFEL--LGSLQNLEKIRLERI-----LVPSFGTLK 620
E I+ + L+ L +T C N EL L + NL+++ +ER L S G
Sbjct: 691 EGIQPLRNLEWLDLT------CSRNLKELPDLSTATNLQRLSIERCSSLVKLPSSIGEAT 744
Query: 621 NLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLNVT 680
NLKK++L C +++ ++ NL+EL++ C LV L T ++ +++ L
Sbjct: 745 NLKKINLRECLSLVELPSSFGNLT----NLQELDLRECSSLVELPTSFGNLANVESLEFY 800
Query: 681 NCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEF 740
C L LP G L NL +L C+ + +P+S G L NL+ L++ C +L LP F
Sbjct: 801 ECSSLVKLPSTFGNLTNLRVLGLRECSSMVELPSSFGNLTNLQVLNLRKCSTLVELPSSF 860
Query: 741 GNLCNLKNLDMASCASIELPFSVVNLQNLKTI 772
NL NL+NLD+ C+S+ LP S N+ LK +
Sbjct: 861 VNLTNLENLDLRDCSSL-LPSSFGNVTYLKRL 891
Score = 87.8 bits (216), Expect = 2e-17
Identities = 54/163 (33%), Positives = 83/163 (50%), Gaps = 11/163 (6%)
Query: 597 LGSLQNLEKIRLERILV-----PSFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLE 651
+G NL+KI L L SFG L NL++L L C++++ ++ N+E
Sbjct: 740 IGEATNLKKINLRECLSLVELPSSFGNLTNLQELDLRECSSLVELPTSFGNLA----NVE 795
Query: 652 ELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTDLEA 711
L C LV L + ++ +L+ L + C + LP G L NL++L+ C+ L
Sbjct: 796 SLEFYECSSLVKLPSTFGNLTNLRVLGLRECSSMVELPSSFGNLTNLQVLNLRKCSTLVE 855
Query: 712 IPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMASC 754
+P+S L NL++LD+ +C SL LP FGN+ LK L C
Sbjct: 856 LPSSFVNLTNLENLDLRDCSSL--LPSSFGNVTYLKRLKFYKC 896
Score = 36.6 bits (83), Expect = 0.058
Identities = 29/92 (31%), Positives = 37/92 (39%), Gaps = 24/92 (26%)
Query: 709 LEAIPTSIGKLLNLKHLD-----------------------ISNCISLSSLPEEFGNLCN 745
LE + I L NL+ LD I C SL LP G N
Sbjct: 686 LEKLWEGIQPLRNLEWLDLTCSRNLKELPDLSTATNLQRLSIERCSSLVKLPSSIGEATN 745
Query: 746 LKNLDMASCAS-IELPFSVVNLQNLKTITCDE 776
LK +++ C S +ELP S NL NL+ + E
Sbjct: 746 LKKINLRECLSLVELPSSFGNLTNLQELDLRE 777
Score = 34.7 bits (78), Expect = 0.22
Identities = 24/81 (29%), Positives = 38/81 (46%)
Query: 550 TQVEVLILNLHTKQYSFPESIKKMSKLKVLIITNYSFHPCELNNFELLGSLQNLEKIRLE 609
T + VL L + P S ++ L+VL + S ++F L +L+NL+
Sbjct: 816 TNLRVLGLRECSSMVELPSSFGNLTNLQVLNLRKCSTLVELPSSFVNLTNLENLDLRDCS 875
Query: 610 RILVPSFGTLKNLKKLSLYMC 630
+L SFG + LK+L Y C
Sbjct: 876 SLLPSSFGNVTYLKRLKFYKC 896
>At3g25510 unknown protein
Length = 1981
Score = 103 bits (256), Expect = 5e-22
Identities = 83/251 (33%), Positives = 122/251 (48%), Gaps = 41/251 (16%)
Query: 550 TQVEVLILNLHTKQYSFPESIKKMSKLKVLIITNYSFHPC----ELNNF----------- 594
T +E LIL P + K+ KL+VL + H C EL +F
Sbjct: 687 TNLEELILKYCVSLVKVPSCVGKLGKLQVLCL-----HGCTSILELPSFTKNVTGLQSLD 741
Query: 595 --------ELLGS------LQNLEKIRLERILVP-SFGTLKNLKKLSLYMCNTILAFEKG 639
EL S LQNL+ L + +P S NLKK L C++++
Sbjct: 742 LNECSSLVELPSSIGNAINLQNLDLGCLRLLKLPLSIVKFTNLKKFILNGCSSLVELP-- 799
Query: 640 SILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLE 699
+ +A NL+ L++ C LV L + I + I+L+ L+++NC L LP IG NLE
Sbjct: 800 --FMGNA-TNLQNLDLGNCSSLVELPSSIGNAINLQNLDLSNCSSLVKLPSFIGNATNLE 856
Query: 700 LLSFSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMASCAS-IE 758
+L C+ L IPTSIG + NL LD+S C SL LP GN+ L+ L++ +C++ ++
Sbjct: 857 ILDLRKCSSLVEIPTSIGHVTNLWRLDLSGCSSLVELPSSVGNISELQVLNLHNCSNLVK 916
Query: 759 LPFSVVNLQNL 769
LP S + NL
Sbjct: 917 LPSSFGHATNL 927
Score = 101 bits (252), Expect = 1e-21
Identities = 152/664 (22%), Positives = 291/664 (42%), Gaps = 70/664 (10%)
Query: 158 MELIRDGSSTLVLTGLGGLGKTTLATKLCWDQEVNGKFKENIIFVTFSK---TPMLKTIV 214
+ L D + + G G+GKTT+A L +V+ F+ + I V + +P L
Sbjct: 216 LRLDLDDVRMIGIWGPPGIGKTTIARFLL--SQVSKSFQLSTIMVNIKECYPSPCLDEYS 273
Query: 215 ERIIEHCGYPVPEFQSDEDAVNKLELLLKKVEGSPLLLVLDDVWPTSE--SLVKKLQFEI 272
++ + + L + ++++ + LVLDDV + +L K+ ++
Sbjct: 274 VQLQLQNKMLSKMINQKDIMIPHLGVAQERLKDKKVFLVLDDVDQLGQLDALAKETRWFG 333
Query: 273 SDFKILVTSR----VSFPRFRTTCILKPLAHEDAVTLFHHYAQMEKNSSDIIDKNLVEKV 328
+I++T+ + R ++ + ++A +F +A +K+ + + L +V
Sbjct: 334 PGSRIIITTENLRLLMAHRINHIYKVEFSSTDEAFQIFCMHAFGQKHPYNGFYE-LSREV 392
Query: 329 VRSCKGLPLTIKVIATSLRNRPDDLWRKIVMELSQGHSILDSNTE--LLTRLQKIFDVLE 386
GLPL +KV+ +SLR W++ + L + LD E L+ + + +
Sbjct: 393 TELAGGLPLGLKVMGSSLRGMSKQEWKRTLPRL---RTCLDGKIESILMFSYEALSHEDK 449
Query: 387 DNPTIMECFMDIALFPEDHRIPVAALVDMWAKLYKLDDNGIQAMEIINKLGIMNLANVII 446
D + CF + + + +D+ LY L + + + I M+ V +
Sbjct: 450 DLFLCIACFFNYQKIKKVEKHLADRFLDVRQGLYVLAEKSL--IHIGTGATEMHTLLVQL 507
Query: 447 PRKNASDTDNNNYNNHFIILHDILRDLGIYQSTKQPFEQRKRLI---IDINKNRSELAEK 503
R+ A N+ ++ + R++ + + + +R+I D++KN E+
Sbjct: 508 GREIAHTQSTNDPRKSLFLVDE--REI-CEALSDETMDSSRRIIGMDFDLSKNGEEVTNI 564
Query: 504 QQSLLTRILSKVMRLCIKQNPQQLAAHILSV----STDEACASDWS---------QMQPT 550
+ L R+ + I+ + + A H ++ S+D CA + Q Q
Sbjct: 565 SEKGLQRMSNLQF---IRFDGRSCARHSSNLTVVRSSDNNCAHPDTVNALQDLNYQFQEI 621
Query: 551 QV-----------------EVLI-LNLHTKQ-YSFPESIKKMSKLKVLIITNYSFHPCEL 591
++ E L+ LN+ + ++ E K + LK + + +YS EL
Sbjct: 622 RLLHWINFRRLCLPSTFNPEFLVELNMPSSTCHTLWEGSKALRNLKWMDL-SYSISLKEL 680
Query: 592 NNFELLGSLQNL-EKIRLERILVPS-FGTLKNLKKLSLYMCNTILAFEKGSILISDAFPN 649
+ +L+ L K + + VPS G L L+ L L+ C +IL +
Sbjct: 681 PDLSTATNLEELILKYCVSLVKVPSCVGKLGKLQVLCLHGCTSILELPS----FTKNVTG 736
Query: 650 LEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTDL 709
L+ L+++ C LV L + I + I+L+ L++ C +L LP I K NL+ + C+ L
Sbjct: 737 LQSLDLNECSSLVELPSSIGNAINLQNLDL-GCLRLLKLPLSIVKFTNLKKFILNGCSSL 795
Query: 710 EAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMASCAS-IELPFSVVNLQN 768
+P +G NL++LD+ NC SL LP GN NL+NLD+++C+S ++LP + N N
Sbjct: 796 VELP-FMGNATNLQNLDLGNCSSLVELPSSIGNAINLQNLDLSNCSSLVKLPSFIGNATN 854
Query: 769 LKTI 772
L+ +
Sbjct: 855 LEIL 858
Score = 99.8 bits (247), Expect = 6e-21
Identities = 78/259 (30%), Positives = 123/259 (47%), Gaps = 33/259 (12%)
Query: 567 PESIKKMSKLKVLIITNYSF--------HPCELNNFEL------------LGSLQNLEKI 606
P SI K + LK I+ S + L N +L +G+ NL+ +
Sbjct: 775 PLSIVKFTNLKKFILNGCSSLVELPFMGNATNLQNLDLGNCSSLVELPSSIGNAINLQNL 834
Query: 607 RLER----ILVPSF-GTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDL 661
L + +PSF G NL+ L L C++++ ++ NL L++ C L
Sbjct: 835 DLSNCSSLVKLPSFIGNATNLEILDLRKCSSLVEIPTSIGHVT----NLWRLDLSGCSSL 890
Query: 662 VVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGKLLN 721
V L + + +I L+ LN+ NC L LP G NL L S C+ L +P+SIG + N
Sbjct: 891 VELPSSVGNISELQVLNLHNCSNLVKLPSSFGHATNLWRLDLSGCSSLVELPSSIGNITN 950
Query: 722 LKHLDISNCISLSSLPEEFGNLCNLKNLDMASCASIELPFSVVNLQNLKTITCDEETAAT 781
L+ L++ NC +L LP GNL L L +A C +E S +NL++L+ + D +
Sbjct: 951 LQELNLCNCSNLVKLPSSIGNLHLLFTLSLARCQKLEALPSNINLKSLERL--DLTDCSQ 1008
Query: 782 WEDFQHMLPNMKIEVLHVD 800
++ F + N IE L++D
Sbjct: 1009 FKSFPEISTN--IECLYLD 1025
Score = 68.2 bits (165), Expect = 2e-11
Identities = 57/187 (30%), Positives = 88/187 (46%), Gaps = 15/187 (8%)
Query: 592 NNFELLGSLQNLEKIRLERILVPSFGTLKNLKKLSLYMCNTILAFEKGSILIS-DAFP-- 648
N+ ELLG L N+ + E + F +K + +Y+ + + L+ D FP
Sbjct: 1777 NSAELLGEL-NISERAFEGMSNLKFLRIKCDRSDKMYLPRGLKYISRKLRLLEWDRFPLT 1835
Query: 649 ---------NLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLE 699
L ELN+ + K LV L G + +LK +N+ + L LP D NL+
Sbjct: 1836 CLPSNFCTEYLVELNMRHSK-LVKLWEGNLSLGNLKWMNLFHSKNLKELP-DFSTATNLQ 1893
Query: 700 LLSFSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMASCASIEL 759
L C+ L +P SIG NL+ L + C SL LP GNL L+N+ + C+ +E+
Sbjct: 1894 TLILCGCSSLVELPYSIGSANNLQKLHLCRCTSLVELPASIGNLHKLQNVTLKGCSKLEV 1953
Query: 760 PFSVVNL 766
+ +NL
Sbjct: 1954 VPTNINL 1960
Score = 55.5 bits (132), Expect = 1e-07
Identities = 140/627 (22%), Positives = 259/627 (40%), Gaps = 98/627 (15%)
Query: 124 HKFNEAKPPFKRPFDVPENPKFTVGLDIPFSKLKMELIRDGSSTLVLT--GLGGLGKTTL 181
+K N++ P R FD VG+ ++++ L D ++ G G+GKTT+
Sbjct: 1410 NKLNKSTP--SRDFDE------LVGMGAHMERMELLLCLDSDEVRMIGIWGPSGIGKTTI 1461
Query: 182 ATKLCW---DQEVNGKFKENIIFVTF----------SKTPMLKTIVERIIEHCGYPVPEF 228
A L D F ENI + + +K + + +II H VP
Sbjct: 1462 ARFLFSQFSDSFELSAFMENIKELMYRKPVCSDDYSAKLHLQNQFMSQIINHMDVEVPHL 1521
Query: 229 QSDEDAVNKLELLLKKVEGSPLLLVLDDVWPTSE--SLVKKLQFEISDFKILVTSRVS-- 284
E+ +N KKV L+VLD++ + + ++ K+ ++ +I++T++
Sbjct: 1522 GVVENRLND-----KKV-----LIVLDNIDQSMQLDAIAKETRWFGHGSRIIITTQDQKL 1571
Query: 285 FPRFRTTCILK---PLAHEDAVTLFHHYAQMEKNSSDIIDKNLVEKVVRSCKGLPLTIKV 341
I K P HE A +F A +K D + +E V LPL ++V
Sbjct: 1572 LKAHGINHIYKVDYPSTHE-ACQIFCMSAVGKKFPKDEFQELALE-VTNLLGNLPLGLRV 1629
Query: 342 IATSLRNRPDDLWRKIVMELSQGHSILDSNTELLTRLQKIFDVL--EDNPTIMECFMDIA 399
+ + R W + L + + LDSN + + + +D L ED + F+ IA
Sbjct: 1630 MGSHFRGMSKQEW---INALPRLRTHLDSNIQSILKFS--YDALCREDK----DLFLHIA 1680
Query: 400 LFPEDHRIPVAALVDMWAKLYKLDDNGIQAMEIINKLGIMNLANVIIPRKNASDTDNNNY 459
+ RI ++ A L + Q ++ + ++++ I N +
Sbjct: 1681 CTFNNKRIE-----NVEAHLTHKFLDTKQRFHVLAEKSLISIEEGWIKMHNLLELLGRE- 1734
Query: 460 NNHFIILHDILRDLGIYQSTKQPFEQRKRLIIDINKNRSELAEKQQSLLTRILSKVMRLC 519
I+ H+ ++S ++P +++ ++D ++ E +LT +
Sbjct: 1735 ----IVCHE-------HESIREP--GKRQFLVDAR----DICE----VLTDDTGSKSVVG 1773
Query: 520 IKQNPQQLAAHILSVSTDEACASDWSQMQPTQVEVLILNLHTKQYSFPESIKKMS-KLKV 578
I N +L + + E S ++ +++ + K Y P +K +S KL++
Sbjct: 1774 IYFNSAELLGEL---NISERAFEGMSNLKFLRIKC---DRSDKMY-LPRGLKYISRKLRL 1826
Query: 579 LIITNYSFHPCELNNF--ELLGSLQNLEKIRLERILVPSFGTLKNLKKLSLYMCNTILAF 636
L + C +NF E L L N+ +L ++ + +L NLK ++L+ +
Sbjct: 1827 LEWDRFPL-TCLPSNFCTEYLVEL-NMRHSKLVKLWEGNL-SLGNLKWMNLFHSKNLKEL 1883
Query: 637 EKGSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLE 696
S NL+ L + C LV L I +L+KL++ C L LP IG L
Sbjct: 1884 PDFSTAT-----NLQTLILCGCSSLVELPYSIGSANNLQKLHLCRCTSLVELPASIGNLH 1938
Query: 697 NLELLSFSSCTDLEAIPTSIGKLLNLK 723
L+ ++ C+ LE +PT+I +L++K
Sbjct: 1939 KLQNVTLKGCSKLEVVPTNINLILDVK 1965
Score = 51.6 bits (122), Expect = 2e-06
Identities = 30/99 (30%), Positives = 48/99 (48%), Gaps = 1/99 (1%)
Query: 649 NLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTD 708
NL+ +N+ + K+L L +L+ L + C L LP IG NL+ L CT
Sbjct: 1868 NLKWMNLFHSKNLKELPD-FSTATNLQTLILCGCSSLVELPYSIGSANNLQKLHLCRCTS 1926
Query: 709 LEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLK 747
L +P SIG L L+++ + C L +P + ++K
Sbjct: 1927 LVELPASIGNLHKLQNVTLKGCSKLEVVPTNINLILDVK 1965
Score = 43.1 bits (100), Expect = 6e-04
Identities = 52/212 (24%), Positives = 87/212 (40%), Gaps = 45/212 (21%)
Query: 567 PESIKKMSKLKVLIITNYSFH---PCELNNFELLGSLQNLEKIRLERILVPSFGTLKNLK 623
P SI ++ L+ L + N S P + N LL +L +LE + PS LK+L+
Sbjct: 942 PSSIGNITNLQELNLCNCSNLVKLPSSIGNLHLLFTLSLARCQKLEAL--PSNINLKSLE 999
Query: 624 KLSLYMCNTILAFEKGS---------------------------ILISDAFPNLEELN-- 654
+L L C+ +F + S +L F L+E +
Sbjct: 1000 RLDLTDCSQFKSFPEISTNIECLYLDGTAVEEVPSSIKSWSRLTVLHMSYFEKLKEFSHV 1059
Query: 655 ------IDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTD 708
+++ +D+ + I +I L L + C KL SLPQ E+L +++ C
Sbjct: 1060 LDIITWLEFGEDIQEVAPWIKEISRLHGLRLYKCRKLLSLPQ---LPESLSIINAEGCES 1116
Query: 709 LEAIPTSIGKLLNLKHLDISNCISLSSLPEEF 740
LE + S L+L L+ + C L+ +F
Sbjct: 1117 LETLDCSYNNPLSL--LNFAKCFKLNQEARDF 1146
>At1g61310 similar to disease resistance protein
Length = 925
Score = 103 bits (256), Expect = 5e-22
Identities = 170/730 (23%), Positives = 312/730 (42%), Gaps = 118/730 (16%)
Query: 113 ILELLNYETFEHKFNEAKPPFKRPFDVPENP-KFTVGLDIPFSKLKMELIRDGSSTLVLT 171
+LE + E F+E P R +V E P + T+G + K L+ DG + L
Sbjct: 122 LLEEVKILKSEGNFDEVSQPPPRS-EVEERPTQPTIGQEEMLEKAWNRLMEDGVGIMGLH 180
Query: 172 GLGGLGKTTLATKLCWD-QEVNGKFKENIIFVTFSKTPMLKTIVERIIE--HCGYPVPEF 228
G+GG+GKTTL K+ E+ G F + +I++ S+ L + E I E H + +
Sbjct: 181 GMGGVGKTTLFKKIHNKFAEIGGTF-DIVIWIVVSQGAKLSKLQEDIAEKLHLCDDLWKN 239
Query: 229 QSDEDAVNKLELLLKKVEGSPLLLVLDDVWPTS--ESLVKKLQFEISDFKILVTSRVSFP 286
+++ D + +LK G +L+LDD+W E++ E++ K+ T+R
Sbjct: 240 KNESDKATDIHRVLK---GKRFVLMLDDIWEKVDLEAIGIPYPSEVNKCKVAFTTRS--- 293
Query: 287 RFRTTC---------ILKPLAHEDAVTLFHHYAQMEKNSSDIIDKNLVEKVVRSCKGLPL 337
R C + L EDA LF + SSD + L +V + C+GLPL
Sbjct: 294 --REVCGEMGDHKPMQVNCLEPEDAWELFKNKVGDNTLSSDPVIVGLAREVAQKCRGLPL 351
Query: 338 TIKVIATSLRNRP-DDLWRKIVMELSQGHSILDS-NTELLTRLQKIFDVLEDNPTIMECF 395
+ VI ++ ++ W + L++ + ++L L+ +D L D I CF
Sbjct: 352 ALNVIGETMASKTMVQEWEYAIDVLTRSAAEFSGMENKILPILKYSYDSLGDE-HIKSCF 410
Query: 396 MDIALFPEDHRIPVAALVDMWAKLYKLDDNGIQAMEIINK---------LGIMNLANVII 446
+ ALFPED +I L+D KL G + + K LG + AN++
Sbjct: 411 LYCALFPEDGQIYTETLID------KLICEGFIGEDQVIKRARNKGYAMLGTLTRANLLT 464
Query: 447 PRKNASDTDNNNYNNHFIILHDILRDLGIYQSTKQPFEQRKRLIIDINKNRSELAEKQQS 506
+ + ++HD++R++ ++ ++ +Q++ ++ + E+ E
Sbjct: 465 KVGTELANLLTKVSIYHCVMHDVVREMALWIASDFG-KQKENFVVQASAGLHEIPE---- 519
Query: 507 LLTRILSKVMRLCIKQNPQQLAAHILSVSTDEACASDWSQMQPTQVEVLILNLHTKQYSF 566
+ V R+ + +N I ++ + C +++ L L + +
Sbjct: 520 --VKDWGAVRRMSLMRN------EIEEITCESKC---------SELTTLFLQSNQLKNLS 562
Query: 567 PESIKKMSKLKVLIIT-NYSFH--PCELNNFELLGSLQNLEKIRLERILVPSFGTLKNLK 623
E I+ M KL VL ++ N F+ P +++ L L +L R+E++ V LK LK
Sbjct: 563 GEFIRYMQKLVVLDLSDNRDFNELPEQISGLVSLQYL-DLSFTRIEQLPV----GLKELK 617
Query: 624 KLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLNV--TN 681
KL+ L++ Y L + +GI ++SL+ L++ +
Sbjct: 618 KLTF-------------------------LDLAYTARLCSI-SGISRLLSLRVLSLLGSK 651
Query: 682 CHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGKLLNL--------KHLDISNCIS- 732
H +S+ +++ +LENL+ L+ + +L ++ + K++++ K D+S S
Sbjct: 652 VHGDASVLKELQQLENLQDLAITLSAELISLDQRLAKVISILGIEGFLQKPFDLSFLASM 711
Query: 733 --LSSLPEEFGNLCNLKNLDMASCASI-----ELPFSVVNLQNLKTITCDEETAATWEDF 785
LSSL + +K + + +S ++P NL L + C TW F
Sbjct: 712 ENLSSLWVKNSYFSEIKCRESETDSSYLHINPKIP-CFTNLSRLDIVKCHSMKDLTWILF 770
Query: 786 QHMLPNMKIE 795
L + IE
Sbjct: 771 APNLVVLFIE 780
>At5g44510 disease resistance protein-like
Length = 1187
Score = 100 bits (249), Expect = 3e-21
Identities = 166/719 (23%), Positives = 300/719 (41%), Gaps = 118/719 (16%)
Query: 127 NEAKPPFKRPFDVPENPKFT--------VGLDIPFSKLK--MELIRDGSSTLVLTGLGGL 176
NEA K DV + FT VG++ +++ ++L + + + G G+
Sbjct: 186 NEADMIIKISKDVSDVLSFTPSKDFDEFVGIEAHTTEITSLLQLDLEEVRMIGIWGPAGI 245
Query: 177 GKTTLATKL---CWDQEVNGKFKENIIF--------VTFSKTPMLKTIVERIIEHCGYPV 225
GKTT++ L + Q G +NI +K + K ++ ++I V
Sbjct: 246 GKTTISRVLYNKLFHQFQLGAIIDNIKVRYPRPCHDEYSAKLQLQKELLSQMINQKDMVV 305
Query: 226 PEFQSDEDAVNKLELLLKKVEGSPLLLVLDDV--WPTSESLVKKLQ-FEISDFKILVTSR 282
P L + ++++ +LLVLDDV +++ K +Q F + I+VT
Sbjct: 306 PH----------LGVAQERLKDKKVLLVLDDVDGLVQLDAMAKDVQWFGLGSRIIVVTQD 355
Query: 283 VSFPRFRTTCILKPL---AHEDAVTLFHHYAQMEKNSSDIIDKNLVEKVVRSCKGLPLTI 339
+ + + + ++A+ +F YA EK S + + + V LPL +
Sbjct: 356 LKLLKAHGIKYIYKVDFPTSDEALEIFCMYAFGEK-SPKVGFEQIARTVTTLAGKLPLGL 414
Query: 340 KVIATSLRNRPDDLWRKIVMELSQGHSILDSNTELLTRLQKIFDVLEDNPTIMECFMDIA 399
+V+ + LR W K + L + LD + E + + ++ L + + F+ I
Sbjct: 415 RVMGSYLRRMSKQEWAKSIPRL---RTSLDDDIESVLKFS--YNSLAEQEK--DLFLHIT 467
Query: 400 LFPEDHRIPVAAL------VDMWAKLYKLDDNGIQAMEIINKLGIMNLANVIIPRKNASD 453
F RI + VDM L L D + ++ LG + + N+++
Sbjct: 468 CFFRRERIETLEVFLAKKSVDMRQGLQILADKSLLSLN----LGNIEMHNLLVQ------ 517
Query: 454 TDNNNYNNHFIILHDILRDLGIYQSTKQPF-------------EQRKRLIIDINKNRSEL 500
+ DI+R I++ K+ F + R +I I+ S +
Sbjct: 518 -----------LGLDIVRKQSIHKPGKRQFLVDTEDICEVLTDDTGTRTLIGIDLELSGV 566
Query: 501 AEKQQSLLTRILSKVMRLCIKQ--NPQQLAAHI-------LSVSTDEACASDWSQMQPTQ 551
E ++ R ++ L + +P H LS + + W + T
Sbjct: 567 IEGVINISERAFERMCNLQFLRFHHPYGDRCHDILYLPQGLSHISRKLRLLHWERYPLTC 626
Query: 552 V------EVLI-LNLHT----KQYSFPESIKKMSKLKVLIITNYSFHPCELNNFELLGSL 600
+ E L+ +N+ K + E I+ + + + N EL +F +L
Sbjct: 627 LPPKFNPEFLVKINMRDSMLEKLWDGNEPIRNLKWMDLSFCVNLK----ELPDFSTATNL 682
Query: 601 QNLEKIR-LERILVPS-FGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYC 658
Q L I L + +PS G NL +L L C++++ ++ NL++L ++ C
Sbjct: 683 QELRLINCLSLVELPSSIGNATNLLELDLIDCSSLVKLPSSIGNLT----NLKKLFLNRC 738
Query: 659 KDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGK 718
LV L + ++ SLK+LN++ C L +P IG + NL+ + C+ L +P+SIG
Sbjct: 739 SSLVKLPSSFGNVTSLKELNLSGCSSLLEIPSSIGNIVNLKKVYADGCSSLVQLPSSIGN 798
Query: 719 LLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMASCAS-IELPF--SVVNLQNLKTITC 774
NLK L + NC SL P NL L++L+++ C S ++LP +V+NLQ+L C
Sbjct: 799 NTNLKELHLLNCSSLMECPSSMLNLTRLEDLNLSGCLSLVKLPSIGNVINLQSLYLSDC 857
Score = 90.5 bits (223), Expect = 3e-18
Identities = 49/128 (38%), Positives = 75/128 (58%), Gaps = 2/128 (1%)
Query: 649 NLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTD 708
NL+ +++ +C +L L +L++L + NC L LP IG NL L C+
Sbjct: 658 NLKWMDLSFCVNLKELPD-FSTATNLQELRLINCLSLVELPSSIGNATNLLELDLIDCSS 716
Query: 709 LEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMASCASI-ELPFSVVNLQ 767
L +P+SIG L NLK L ++ C SL LP FGN+ +LK L+++ C+S+ E+P S+ N+
Sbjct: 717 LVKLPSSIGNLTNLKKLFLNRCSSLVKLPSSFGNVTSLKELNLSGCSSLLEIPSSIGNIV 776
Query: 768 NLKTITCD 775
NLK + D
Sbjct: 777 NLKKVYAD 784
Score = 86.3 bits (212), Expect = 6e-17
Identities = 66/212 (31%), Positives = 104/212 (48%), Gaps = 40/212 (18%)
Query: 597 LGSLQNLEKIRLERI-----LVPSFGTLKNLKKLSLYMCNTILAFEK--GSILISDAFPN 649
+G+L NL+K+ L R L SFG + +LK+L+L C+++L G+I+ N
Sbjct: 724 IGNLTNLKKLFLNRCSSLVKLPSSFGNVTSLKELNLSGCSSLLEIPSSIGNIV------N 777
Query: 650 LEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQD------------------ 691
L+++ D C LV L + I + +LK+L++ NC L P
Sbjct: 778 LKKVYADGCSSLVQLPSSIGNNTNLKELHLLNCSSLMECPSSMLNLTRLEDLNLSGCLSL 837
Query: 692 -----IGKLENLELLSFSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNL 746
IG + NL+ L S C+ L +P +I NL L + C +L LP N+ NL
Sbjct: 838 VKLPSIGNVINLQSLYLSDCSSLMELPFTIENATNLDTLYLDGCSNLLELPSSIWNITNL 897
Query: 747 KNLDMASCASI-ELPFSV---VNLQNLKTITC 774
++L + C+S+ ELP V +NLQ+L + C
Sbjct: 898 QSLYLNGCSSLKELPSLVENAINLQSLSLMKC 929
Score = 81.3 bits (199), Expect = 2e-15
Identities = 57/177 (32%), Positives = 90/177 (50%), Gaps = 17/177 (9%)
Query: 567 PESIKKMSKLKVLIITNYSFHPCELNNFELLGSLQNLEKIR-------LERILVPSFGTL 619
P SI + LK L + N S + E S+ NL ++ L + +PS G +
Sbjct: 793 PSSIGNNTNLKELHLLNCS------SLMECPSSMLNLTRLEDLNLSGCLSLVKLPSIGNV 846
Query: 620 KNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLNV 679
NL+ L L C++++ I +A NL+ L +D C +L+ L + I +I +L+ L +
Sbjct: 847 INLQSLYLSDCSSLMELP---FTIENA-TNLDTLYLDGCSNLLELPSSIWNITNLQSLYL 902
Query: 680 TNCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSL 736
C L LP + NL+ LS C+ L +P+SI ++ NL +LD+SNC SL L
Sbjct: 903 NGCSSLKELPSLVENAINLQSLSLMKCSSLVELPSSIWRISNLSYLDVSNCSSLLEL 959
Score = 80.1 bits (196), Expect = 5e-15
Identities = 64/237 (27%), Positives = 109/237 (45%), Gaps = 37/237 (15%)
Query: 550 TQVEVLILNLHTKQYSFPESIKKMSKLKVLIITNYSFHPCELNNFELLGSLQNLEKIRLE 609
T ++ L LN + P S ++ LK L ++ S L +G++ NL+K+ +
Sbjct: 728 TNLKKLFLNRCSSLVKLPSSFGNVTSLKELNLSGCS---SLLEIPSSIGNIVNLKKVYAD 784
Query: 610 RI-----LVPSFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVL 664
L S G NLK+L L C++++ + ++ LE+LN+ C LV L
Sbjct: 785 GCSSLVQLPSSIGNNTNLKELHLLNCSSLMECPSSMLNLT----RLEDLNLSGCLSLVKL 840
Query: 665 QTGICDIISLKKL----------------NVTN--------CHKLSSLPQDIGKLENLEL 700
+ I ++I+L+ L N TN C L LP I + NL+
Sbjct: 841 PS-IGNVINLQSLYLSDCSSLMELPFTIENATNLDTLYLDGCSNLLELPSSIWNITNLQS 899
Query: 701 LSFSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMASCASI 757
L + C+ L+ +P+ + +NL+ L + C SL LP + NL LD+++C+S+
Sbjct: 900 LYLNGCSSLKELPSLVENAINLQSLSLMKCSSLVELPSSIWRISNLSYLDVSNCSSL 956
>At1g58410 unknown protein
Length = 899
Score = 100 bits (248), Expect = 4e-21
Identities = 189/824 (22%), Positives = 350/824 (41%), Gaps = 93/824 (11%)
Query: 11 LTTRFHDVLNKISQIVETARKHQSARQLLRSILKDLTLVVQDIKQYNEHLDHPREEIKTL 70
L R ++ + + + +S LL+S LKD K +E + H EEIK +
Sbjct: 11 LWDRLSQEYDQFKGVEDQVTELKSNLNLLKSFLKDADAK----KHISEMVRHCVEEIKDI 66
Query: 71 L-------------EENDAEESACKCSSENDSYVVDENQ-SLIVNDVEETLYKAREILEL 116
+ E+ + + K S ++D + + + + + + K + ++
Sbjct: 67 VYDTEDIIETFILKEKVEMKRGIMKRIKRFASTIMDRRELASDIGGISKRISKVIQDMQS 126
Query: 117 LNYETF-------EHKFNEAKPPFKRPFDVPENPKFTVGLDIPFSKLKMELI-RDGSSTL 168
+ H E + + F F VG++ KL L+ +D +
Sbjct: 127 FGVQQIITDGSRSSHPLQERQREMRHTFSRDSENDF-VGMEANVKKLVGYLVEKDDYQIV 185
Query: 169 VLTGLGGLGKTTLATKLCWDQEVNGKFKENIIFVT----FSKTPMLKTIVERIIEHCGYP 224
LTG+GGLGKTTLA ++ V +F + +V+ F++ + +TI++ +
Sbjct: 186 SLTGMGGLGKTTLARQVFNHDVVKDRF-DGFAWVSVSQEFTRISVWQTILQNLTSK--ER 242
Query: 225 VPEFQSDEDAVNKLELLLKKVEGSPLLLVLDDVWPTSE-SLVKKLQFEISDFKILVTSRV 283
E Q+ ++A + + L + +E S L+VLDD+W + L+K + +K+L+TSR
Sbjct: 243 KDEIQNMKEA-DLHDDLFRLLESSKTLIVLDDIWKEEDWDLIKPIFPPKKGWKVLLTSRT 301
Query: 284 SFPRFR--TTCI-LKP--LAHEDAVTLFHHYAQMEKNSSDI-ID---KNLVEKVVRSCKG 334
R TT I KP L+ D+ TLF A K++S+ +D +N+ +K+++ C G
Sbjct: 302 ESIAMRGDTTYISFKPKCLSIPDSWTLFQSIAMPRKDTSEFKVDEEMENMGKKMIKHCGG 361
Query: 335 LPLTIKVIATSLRNRPD-DLWRKIVMELSQGHSILDSNTELLTRLQKIFDV-LEDNPTIM 392
L L +KV+ L + W+++ + G I++ + + + + V E+ P +
Sbjct: 362 LSLAVKVLGGLLAAKYTLHDWKRLSENI--GSHIVERTSGNNSSIDHVLSVSFEELPNYL 419
Query: 393 E-CFMDIALFPEDHRIPVAALVDMWAKLYKLDDNGIQAMEIINKLGIMNLANVIIPRKNA 451
+ CF+ +A FPEDH I V L WA + + E I G + ++
Sbjct: 420 KHCFLYLAHFPEDHEIDVEKLHYYWA-AEGISERRRYDGETIRDTGDSYIEELVRRNMVI 478
Query: 452 SDTDNNNYNNHFIILHDILRDLGIYQSTKQPFEQRKRLIIDINKNRSELAEKQQSLLTR- 510
S+ D LHD++R++ ++++ ++ F + I N S + Q +R
Sbjct: 479 SERDVMTSRFETCRLHDMMREICLFKAKEENF-------LQIVSNHSPTSNPQTLGASRR 531
Query: 511 -ILSKVMRLCIKQNPQQLAAHILSVSTDEACASDW--SQMQPTQVEVL-ILNLHTKQY-- 564
+L L +++ L V D+ W S T+V++L +L+L ++
Sbjct: 532 FVLHNPTTLHVERYKNNPKLRSLVVVYDDIGNRRWMLSGSIFTRVKLLRVLDLVQAKFKG 591
Query: 565 -SFPESIKKMSKLKVLII--TNYSFHPCELNNFELLGSLQNLEKIRLERILVPS-FGTLK 620
P I K+ L+ L + S P L N LL L + I VP+ F ++
Sbjct: 592 GKLPSDIGKLIHLRYLSLKDAKVSHLPSSLRNLVLLIYLD--IRTDFTDIFVPNVFMGMR 649
Query: 621 NLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLNV- 679
L+ L L EK + +S NLE+L + ++ + + D+ + +L
Sbjct: 650 ELRYLEL----PRFMHEKTKLELS----NLEKL--EALENFSTKSSSLEDLRGMVRLRTL 699
Query: 680 -------TNCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGKLLNLKHLDISNCIS 732
T+ LS+ + LEN +++ + + + LK L +S I
Sbjct: 700 VIILSEGTSLQTLSASVCGLRHLENFKIMENAGVNRM-GEERMVLDFTYLKKLTLS--IE 756
Query: 733 LSSLPEEFGNLCNLKNLDMASCASIELPFSVV-NLQNLKTITCD 775
+ LP+ +L LD++ C E P ++ L LK ++ D
Sbjct: 757 MPRLPKIQHLPSHLTVLDLSYCCLEEDPMPILEKLLELKDLSLD 800
Score = 39.3 bits (90), Expect = 0.009
Identities = 28/106 (26%), Positives = 51/106 (47%), Gaps = 1/106 (0%)
Query: 684 KLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNL 743
K LP DIGKL +L LS + +P+S+ L+ L +LDI + +P F +
Sbjct: 590 KGGKLPSDIGKLIHLRYLSLKDAK-VSHLPSSLRNLVLLIYLDIRTDFTDIFVPNVFMGM 648
Query: 744 CNLKNLDMASCASIELPFSVVNLQNLKTITCDEETAATWEDFQHML 789
L+ L++ + + NL+ L+ + +++ ED + M+
Sbjct: 649 RELRYLELPRFMHEKTKLELSNLEKLEALENFSTKSSSLEDLRGMV 694
>At4g27220 putative protein
Length = 919
Score = 99.4 bits (246), Expect = 7e-21
Identities = 142/571 (24%), Positives = 242/571 (41%), Gaps = 95/571 (16%)
Query: 155 KLKMELIRDGSSTLVLTGLGGLGKTTLATKLCWDQEVNGKFKEN--IIFVTFSKTPMLKT 212
KLK L + + + G+GG+GKTTL L D ++ +I+VT SK LK
Sbjct: 124 KLKDCLKKKNVQKIGVWGMGGVGKTTLVRTLNNDLLKYAATQQFALVIWVTVSKDFDLKR 183
Query: 213 IVERIIEHCGYPVPEFQSDEDAVNKLELLLKKVEGSPLLLVLDDVWPTSE--SLVKKLQF 270
+ I + G Q ++ + E L ++ LL+LDDVW + L L
Sbjct: 184 VQMDIAKRLGKRFTREQMNQLGLTICERL---IDLKNFLLILDDVWHPIDLDQLGIPLAL 240
Query: 271 EIS-DFKILVTSR--------VSFPRFRTTCILKPLAHEDAVTLFHHYAQMEKNSSDIID 321
E S D K+++TSR ++ + C L ++A LF H NS ++
Sbjct: 241 ERSKDSKVVLTSRRLEVCQQMMTNENIKVAC----LQEKEAWELFCHNVGEVANSDNV-- 294
Query: 322 KNLVEKVVRSCKGLPLTIKVIATSLRNRPD-DLWRKIVMELSQGHSILDSNTELLTRLQK 380
K + + V C GLPL I I +LR +P ++W+ + L + +D+ ++ L+
Sbjct: 295 KPIAKDVSHECCGLPLAIITIGRTLRGKPQVEVWKHTLNLLKRSAPSIDTEEKIFGTLKL 354
Query: 381 IFDVLEDNPTIMECFMDIALFPEDHRIPVAALVDMWAKLYKLDDNGIQAMEIINKLGIMN 440
+D L+DN + CF+ ALFPED+ I V+ L+ W LD G E +MN
Sbjct: 355 SYDFLQDN--MKSCFLFCALFPEDYSIKVSELIMYWVAEGLLD--GQHHYE-----DMMN 405
Query: 441 LANVIIPRKNASDTDNNNYNNHFIILHDILRDLGIYQSTKQPFEQRKRLIIDINKNRSEL 500
++ R S + + + +HD++RD I+ + Q + ++ + E
Sbjct: 406 EGVTLVERLKDSCLLEDGDSCDTVKMHDVVRDFAIWFMSSQ--GEGFHSLVMAGRGLIEF 463
Query: 501 AEKQQSLLTRILSKVMRLCIKQNP-QQLAAHILSVSTDEACASDWSQMQPTQVEVLIL-- 557
+ + +S V R+ + N ++L +++ VE L+L
Sbjct: 464 PQ------DKFVSSVQRVSLMANKLERLPNNVIE-----------------GVETLVLLL 500
Query: 558 --NLHTKQYSFPES-IKKMSKLKVLIITNYSFHPCELNNFELLGSLQNLEKIRLERILVP 614
N H K+ P ++ L++L +L +R+ R L
Sbjct: 501 QGNSHVKE--VPNGFLQAFPNLRIL----------------------DLSGVRI-RTLPD 535
Query: 615 SFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQTGICDIISL 674
SF L +L+ L L C + L+ F +L E I L G+ + SL
Sbjct: 536 SFSNLHSLRSLVLRNCKKLRNLPSLESLVKLQFLDLHESAIR------ELPRGLEALSSL 589
Query: 675 KKLNVTNCHKLSSLPQ-DIGKLENLELLSFS 704
+ + V+N ++L S+P I +L +LE+L +
Sbjct: 590 RYICVSNTYQLQSIPAGTILQLSSLEVLDMA 620
Score = 47.4 bits (111), Expect = 3e-05
Identities = 37/135 (27%), Positives = 64/135 (47%), Gaps = 9/135 (6%)
Query: 646 AFPNLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSFSS 705
AFPNL L++ + + L ++ SL+ L + NC KL +LP LE+L L F
Sbjct: 516 AFPNLRILDLSGVR-IRTLPDSFSNLHSLRSLVLRNCKKLRNLP----SLESLVKLQFLD 570
Query: 706 CTD--LEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMASCASIELPFSV 763
+ + +P + L +L+++ +SN L S+P G + L +L++ A + +
Sbjct: 571 LHESAIRELPRGLEALSSLRYICVSNTYQLQSIPA--GTILQLSSLEVLDMAGSAYSWGI 628
Query: 764 VNLQNLKTITCDEET 778
+ T DE T
Sbjct: 629 KGEEREGQATLDEVT 643
Score = 45.8 bits (107), Expect = 1e-04
Identities = 63/254 (24%), Positives = 106/254 (40%), Gaps = 55/254 (21%)
Query: 554 VLILNLHTKQYSFPESIKKMSKLKVLIITNYSFHP------------CELNNFELLGSLQ 601
+ +L++ + Y F K+++K + L S P ++N + LQ
Sbjct: 653 IKLLDVLSFSYEFDSLTKRLTKFQFLFSPIRSVSPPGTGEGCLAISDVNVSNASIGWLLQ 712
Query: 602 NLEKIRL----------ERILVPSFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLE 651
++ + L E ++ S + +K LS++ + L+ G D FPNLE
Sbjct: 713 HVTSLDLNYCEGLNGMFENLVTKSKSSFVAMKALSIHYFPS-LSLASGCESQLDLFPNLE 771
Query: 652 ELNIDYCK-DLVVLQTGICD--IISLKKLNVTNCHKLSSLPQD---IGKLENLELLSFSS 705
EL++D + + G + LK L V+ C +L L D G L NL+ + S
Sbjct: 772 ELSLDNVNLESIGELNGFLGMRLQKLKLLQVSGCRQLKRLFSDQILAGTLPNLQEIKVVS 831
Query: 706 CTDLEAIPTSIGKLLNLKHLDISNC----------ISLSSLPEEFGNLCN-------LKN 748
C LE +L N + + C I L LP + +LCN L++
Sbjct: 832 CLRLE-------ELFNFSSVPVDFCAESLLPKLTVIKLKYLP-QLRSLCNDRVVLESLEH 883
Query: 749 LDMASCASIE-LPF 761
L++ SC S++ LPF
Sbjct: 884 LEVESCESLKNLPF 897
Score = 32.7 bits (73), Expect = 0.83
Identities = 26/91 (28%), Positives = 49/91 (53%), Gaps = 5/91 (5%)
Query: 683 HKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGKLL-NLKHLDISNCISLSSLPEEFG 741
+KL LP ++ + +L + ++ +P + NL+ LD+S + + +LP+ F
Sbjct: 480 NKLERLPNNVIEGVETLVLLLQGNSHVKEVPNGFLQAFPNLRILDLSG-VRIRTLPDSFS 538
Query: 742 NLCNLKNLDMASCASI-ELPF--SVVNLQNL 769
NL +L++L + +C + LP S+V LQ L
Sbjct: 539 NLHSLRSLVLRNCKKLRNLPSLESLVKLQFL 569
>At1g59124 PRM1 homolog (RF45)
Length = 1155
Score = 98.6 bits (244), Expect = 1e-20
Identities = 161/727 (22%), Positives = 315/727 (43%), Gaps = 82/727 (11%)
Query: 19 LNKISQIVE--TARKHQSARQLLRSILKDLTLVVQDIKQYNEHLDHPREEIKT-LLEEND 75
LN +S ++ A+KH SA ++++ ++++ ++ D + I+T +LE+N
Sbjct: 38 LNMLSSFLKDANAKKHTSA--VVKNCVEEIKEIIYD----------GEDTIETFVLEQNL 85
Query: 76 AEESACKCSSENDSYVVDENQ--SLIVNDVEETLYKAREILELLNYET------FEHKFN 127
+ S K S + ++ + + +L + + + K ++ + ++
Sbjct: 86 GKTSGIKKSIRRLACIIPDRRRYALGIGGLSNRISKVIRDMQSFGVQQAIVDGGYKQPQG 145
Query: 128 EAKPPFKRPFDVPENPKFTVGLDIPFSKLKMELIRDGSSTLV-LTGLGGLGKTTLATKLC 186
+ + ++ F ++ F VGL+ KL L+ + + +V +TG+GGLGKTTLA ++
Sbjct: 146 DKQREMRQKFSKDDDSDF-VGLEANVKKLVGYLVDEANVQVVSITGMGGLGKTTLAKQVF 204
Query: 187 WDQEVNGKFKENIIFVTFSKTPMLKTIVERIIEHCGYPVPEFQSDEDAVNKLE-LLLKKV 245
++V +F + + +V S+ + ++I+ E + E + L+ L++ +
Sbjct: 205 NHEDVKHQF-DGLSWVCVSQDFTRMNVWQKILRDLKPKEEEKKIMEMTQDTLQGELIRLL 263
Query: 246 EGSPLLLVLDDVWPTSE-SLVKKLQFEISDFKILVTSR---------VSFPRFRTTCILK 295
E S L+VLDD+W + L+K + +K+L+TSR S+ F+ C
Sbjct: 264 ETSKSLIVLDDIWEKEDWELIKPIFPPTKGWKVLLTSRNESVAMRRNTSYINFKPEC--- 320
Query: 296 PLAHEDAVTLFHHYAQMEKNSSDI-ID---KNLVEKVVRSCKGLPLTIKVIATSLRNR-P 350
L ED+ TLF A K++++ ID + L + +++ C GLPL I+V+ L +
Sbjct: 321 -LTTEDSWTLFQRIALPMKDAAEFKIDEEKEELGKLMIKHCGGLPLAIRVLGGMLAEKYT 379
Query: 351 DDLWRK----IVMELSQGHSIL--DSNTELLTRLQKIFDVLEDNPTIMECFMDIALFPED 404
WR+ I L G + D+N L F+ L + CF+ +A FPED
Sbjct: 380 SHDWRRLSENIGSHLVGGRTNFNDDNNNTCNNVLSLSFEELPS--YLKHCFLYLAHFPED 437
Query: 405 HRIPVAALVDMWAKLYKLDDNGIQAMEIINKLGIMNLANVIIPRKNASDTDNNNYNNHFI 464
+ I V L WA E I +G + + ++ S+ D
Sbjct: 438 YEIKVENLSYYWAAEGIFQPRHYDG-ETIRDVGDVYIEELVRRNMVISERDVKTSRFETC 496
Query: 465 ILHDILRDLGIYQSTKQPFEQRKRLIIDINKNRSELAEKQQSLLTR--ILSKVMRLCIKQ 522
LHD++R++ + ++ ++ F + I +R A Q ++ +R + L +++
Sbjct: 497 HLHDMMREVCLLKAKEENF-------LQITSSRPSTANLQSTVTSRRFVYQYPTTLHVEK 549
Query: 523 ---NPQQLAAHILSVSTDEACASDWSQMQPTQVEVLILNLHTKQYSFPESIKKMSKLKVL 579
NP+ A ++++ + S +++++ +V LI + K I K+ L+ L
Sbjct: 550 DINNPKLRALVVVTLGSWNLAGSSFTRLELLRVLDLI-EVKIKGGKLASCIGKLIHLRYL 608
Query: 580 II--TNYSFHPCELNNFELLGSLQNLEKIRLERILVPSFGTLKNLKKLSLYM-CNTILAF 636
+ + P L N +LL L NL + ++ L+ L+L
Sbjct: 609 SLEYAEVTHIPYSLGNLKLLIYL-NLASFGRSTFVPNVLMGMQELRYLALPSDMGRKTKL 667
Query: 637 EKGSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHK--LSSLPQDIGK 694
E +++ + N N +C ++ L LN+ + L +L IG
Sbjct: 668 ELSNLVKLETLENFSTENSSL--------EDLCGMVRLSTLNIKLIEETSLETLAASIGG 719
Query: 695 LENLELL 701
L+ LE L
Sbjct: 720 LKYLEKL 726
Score = 35.4 bits (80), Expect = 0.13
Identities = 36/115 (31%), Positives = 56/115 (48%), Gaps = 12/115 (10%)
Query: 597 LGSLQNLEKIRLE------RILVPSFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNL 650
LG L L++++L RI+V S G L+KLS+Y L + I+ + P L
Sbjct: 857 LGRLVYLKELQLGFRTFSGRIMVCSGGGFPQLQKLSIYR----LEEWEEWIVEQGSMPFL 912
Query: 651 EELNIDYCKDLVVLQTGICDIISLKKLNVTNCHK--LSSLPQDIGKLENLELLSF 703
L ID C L L G+ I SLK L ++ K LS ++ K++++ + F
Sbjct: 913 HTLYIDDCPKLKKLPDGLQFIYSLKNLKISERWKERLSEGGEEYYKVQHIPSVEF 967
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.136 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,566,161
Number of Sequences: 26719
Number of extensions: 765037
Number of successful extensions: 6651
Number of sequences better than 10.0: 457
Number of HSP's better than 10.0 without gapping: 371
Number of HSP's successfully gapped in prelim test: 88
Number of HSP's that attempted gapping in prelim test: 2665
Number of HSP's gapped (non-prelim): 2102
length of query: 807
length of database: 11,318,596
effective HSP length: 107
effective length of query: 700
effective length of database: 8,459,663
effective search space: 5921764100
effective search space used: 5921764100
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 64 (29.3 bits)
Medicago: description of AC148347.6


