

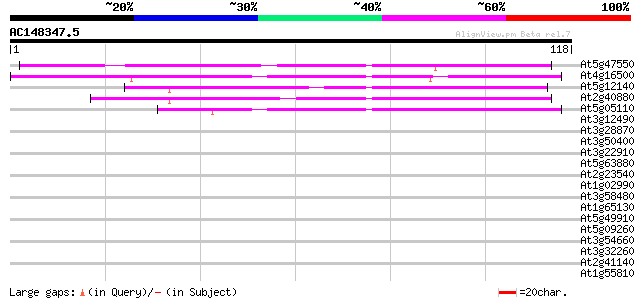
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148347.5 - phase: 0
(118 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g47550 unknown protein 73 2e-14
At4g16500 cysteine proteinase inhibitor like protein 63 2e-11
At5g12140 cystatin (emb|CAA03929.1) 45 8e-06
At2g40880 putative cysteine proteinase inhibitor B (cystatin B) 42 5e-05
At5g05110 cysteine proteinase inhibitor-like protein 42 6e-05
At3g12490 unknown protein 37 0.002
At3g28870 hypothetical protein 28 0.96
At3g50400 unknown protein 27 1.6
At3g22910 calmodulin-stimulated calcium-ATPase, putative 26 3.6
At5g63880 unknown protein 25 4.7
At2g23540 putative GDSL-motif lipase/hydrolase 25 4.7
At1g02990 overlap with bases 73023-78341 of BAC clone F10O3 (AC0... 25 4.7
At3g58480 putative protein 25 6.2
At1g65130 hypothetical protein 25 6.2
At5g49910 heat shock protein 70 (Hsc70-7) 25 8.1
At5g09260 unknown protein 25 8.1
At3g54660 Gluthatione reductase, chloroplast precursor 25 8.1
At3g32260 hypothetical protein 25 8.1
At2g41140 CDPK-related kinase 1 (CRK1) 25 8.1
At1g55810 uracil phosphoribosyltransferase 1, putative 25 8.1
>At5g47550 unknown protein
Length = 122
Score = 73.2 bits (178), Expect = 2e-14
Identities = 47/113 (41%), Positives = 67/113 (58%), Gaps = 9/113 (7%)
Query: 3 LESMVLVLVVLLAFTATKQAIPIGNLSPINNINDPKVIDVANFAVKEYNNRRRKPEEKLR 62
L S+V+VL+ L A A + +G SPI+N+ DP+V+++ FAV EYN R E L+
Sbjct: 10 LLSLVVVLLPLYASAAAR----VGGWSPISNVTDPQVVEIGEFAVSEYNKR---SESGLK 62
Query: 63 LWKVIKGESQIVADGVNYRLTLSATK-VYTSNTYEAIVLEWSLQHLRNLTSFK 114
V+ GE+Q+V+ G NYRL ++A S Y AIV + RNLTSF+
Sbjct: 63 FETVVSGETQVVS-GTNYRLKVAANDGDGVSKNYLAIVWDKPWMKFRNLTSFE 114
>At4g16500 cysteine proteinase inhibitor like protein
Length = 117
Score = 63.2 bits (152), Expect = 2e-11
Identities = 39/124 (31%), Positives = 74/124 (59%), Gaps = 15/124 (12%)
Query: 1 MRLESMVLVLVVLLAFTATKQAIP----IGNLSPINNINDPKVIDVANFAVKEYNNRRRK 56
M ++S++ + ++LL + + + +G+ PI N++DP V+ VA +A++E+N ++
Sbjct: 1 MMMKSLICLSLILLPLVSVVEGLGGGGGLGSRKPIKNVSDPDVVAVAKYAIEEHN---KE 57
Query: 57 PEEKLRLWKVIKGESQIVADGVNYRLTLSAT----KVYTSNTYEAIVLEWSLQHLRNLTS 112
+EKL KV++G +Q+V+ G Y L ++A K+ YEA+V+E H ++L S
Sbjct: 58 SKEKLVFVKVVEGTTQVVS-GTKYDLKIAAKDGGGKI---KNYEAVVVEKLWLHSKSLES 113
Query: 113 FKLI 116
FK +
Sbjct: 114 FKAL 117
>At5g12140 cystatin (emb|CAA03929.1)
Length = 101
Score = 44.7 bits (104), Expect = 8e-06
Identities = 32/90 (35%), Positives = 49/90 (53%), Gaps = 5/90 (5%)
Query: 25 IGNLSPIN-NINDPKVIDVANFAVKEYNNRRRKPEEKLRLWKVIKGESQIVADGVNYRLT 83
+G + I+ N ND +V +A FAV E+N E RL + ++Q+VA G + LT
Sbjct: 10 VGGVRDIDANANDLQVESLARFAVDEHNKNENLTLEYKRL---LGAKTQVVA-GTMHHLT 65
Query: 84 LSATKVYTSNTYEAIVLEWSLQHLRNLTSF 113
+ T+ YEA VLE + ++L+ L SF
Sbjct: 66 VEVADGETNKVYEAKVLEKAWENLKQLESF 95
>At2g40880 putative cysteine proteinase inhibitor B (cystatin B)
Length = 125
Score = 42.0 bits (97), Expect = 5e-05
Identities = 28/98 (28%), Positives = 51/98 (51%), Gaps = 5/98 (5%)
Query: 18 ATKQAIPIGNLSPIN-NINDPKVIDVANFAVKEYNNRRRKPEEKLRLWKVIKGESQIVAD 76
+T++ + +G + + N N ++ +A FA++E+N ++ K L K++K Q+VA
Sbjct: 28 STEKTMMLGGVHDLRGNQNSGEIESLARFAIQEHNKQQNKI---LEFKKIVKAREQVVA- 83
Query: 77 GVNYRLTLSATKVYTSNTYEAIVLEWSLQHLRNLTSFK 114
G Y LTL A + + +EA V + + L FK
Sbjct: 84 GTMYHLTLEAKEGDQTKNFEAKVWVKPWMNFKQLQEFK 121
>At5g05110 cysteine proteinase inhibitor-like protein
Length = 232
Score = 41.6 bits (96), Expect = 6e-05
Identities = 33/86 (38%), Positives = 44/86 (50%), Gaps = 5/86 (5%)
Query: 32 NNINDPKVID-VANFAVKEYNNRRRKPEEKLRLWKVIKGESQIVADGVNYRLTLSATKVY 90
N+ N K ID +A FAV+E+N R+ L L +V+K Q+VA G YRLTL +
Sbjct: 53 NDWNGGKEIDDIALFAVQEHN---RRENAVLELARVLKATEQVVA-GKLYRLTLEVIEAG 108
Query: 91 TSNTYEAIVLEWSLQHLRNLTSFKLI 116
YEA V + + L FK I
Sbjct: 109 EKKIYEAKVWVKPWMNFKQLQEFKNI 134
>At3g12490 unknown protein
Length = 201
Score = 36.6 bits (83), Expect = 0.002
Identities = 27/82 (32%), Positives = 39/82 (46%), Gaps = 4/82 (4%)
Query: 33 NINDPKVIDVANFAVKEYNNRRRKPEEKLRLWKVIKGESQIVADGVNYRLTLSATKVYTS 92
N N +V +A FAV E+N +K L +V+K + Q+VA G + LTL +
Sbjct: 13 NQNSGEVESLARFAVDEHN---KKENALLEFARVVKAKEQVVA-GTLHHLTLEILEAGQK 68
Query: 93 NTYEAIVLEWSLQHLRNLTSFK 114
YEA V + + L FK
Sbjct: 69 KLYEAKVWVKPWLNFKELQEFK 90
>At3g28870 hypothetical protein
Length = 355
Score = 27.7 bits (60), Expect = 0.96
Identities = 18/60 (30%), Positives = 30/60 (50%), Gaps = 8/60 (13%)
Query: 33 NINDPKVIDVANFAVKEYNNRRRKPEEKLRLWKVIKGESQIVADGVNYRLTLSATKVYTS 92
N+NDPK+ K +N + + E K +L +++K + RLT S +VYT+
Sbjct: 66 NLNDPKMRPRIRIRSKLVDNPKDRLEAKQKLTELLK--------ALEERLTRSELRVYTN 117
>At3g50400 unknown protein
Length = 374
Score = 26.9 bits (58), Expect = 1.6
Identities = 13/38 (34%), Positives = 21/38 (55%), Gaps = 9/38 (23%)
Query: 25 IGNLSPI---------NNINDPKVIDVANFAVKEYNNR 53
+GN++PI N +ND + +D+AN +YN R
Sbjct: 226 VGNVAPIGCIPYQKSINQLNDKQCVDLANKLAIQYNAR 263
>At3g22910 calmodulin-stimulated calcium-ATPase, putative
Length = 1017
Score = 25.8 bits (55), Expect = 3.6
Identities = 22/78 (28%), Positives = 32/78 (40%), Gaps = 14/78 (17%)
Query: 16 FTATKQAIPIGNLSPINNINDPKVIDVANFAVKEYNNR-----------RRKPEEKLRLW 64
FTA A+ G L+P + +N V++ F R R P +KL +
Sbjct: 673 FTARAIAVECGILTPEDEMNSEAVLEGEKFRNYTQEERLEKVERIKVMARSSPFDKLLMV 732
Query: 65 KVIKGESQIVA---DGVN 79
K +K +VA DG N
Sbjct: 733 KCLKELGHVVAVTGDGTN 750
>At5g63880 unknown protein
Length = 219
Score = 25.4 bits (54), Expect = 4.7
Identities = 20/86 (23%), Positives = 36/86 (41%), Gaps = 4/86 (4%)
Query: 34 INDPKVIDVANFAVKEYNNRRRKPEEKLRLWKVIKGESQIVADGVNY----RLTLSATKV 89
+ P++ +V + RR+ + + +L KVI+ E Q D + R L+ K
Sbjct: 6 VKKPQITEVDRAILSLKTQRRKLGQYQQKLEKVIEAEKQAARDLIREKRKDRALLALRKK 65
Query: 90 YTSNTYEAIVLEWSLQHLRNLTSFKL 115
T V +W + + LT +L
Sbjct: 66 RTQEELLKQVDQWVINVEQQLTDIEL 91
>At2g23540 putative GDSL-motif lipase/hydrolase
Length = 387
Score = 25.4 bits (54), Expect = 4.7
Identities = 14/46 (30%), Positives = 24/46 (51%), Gaps = 9/46 (19%)
Query: 25 IGNLSPI---------NNINDPKVIDVANFAVKEYNNRRRKPEEKL 61
IGN+ PI N +++ + +D+AN +YN R + E+L
Sbjct: 240 IGNVGPIGCIPYQKTINQLDENECVDLANKLANQYNVRLKSLLEEL 285
>At1g02990 overlap with bases 73023-78341 of BAC clone F10O3
(AC006550). Genes in this region are annotated in the
record for clone F10O3, AC006550
Length = 1306
Score = 25.4 bits (54), Expect = 4.7
Identities = 18/45 (40%), Positives = 23/45 (51%), Gaps = 1/45 (2%)
Query: 51 NNRRRKPEEKLRLWKVIKGES-QIVADGVNYRLTLSATKVYTSNT 94
N RR + E + L + GES +DG N TL+A K SNT
Sbjct: 1170 NIRRCRYELQAALQVIPSGESPSFASDGENSNHTLTAEKFALSNT 1214
>At3g58480 putative protein
Length = 575
Score = 25.0 bits (53), Expect = 6.2
Identities = 17/56 (30%), Positives = 27/56 (47%), Gaps = 8/56 (14%)
Query: 44 NFAVKEYNNRRRKPEEKLRLWK-VIKGE------SQIVADGVNYRLTLSATKVYTS 92
NF++K + K+R+WK V + E + ++ DG Y+ L KVY S
Sbjct: 54 NFSIKPLTFMKEDDRYKMRIWKPVCENEHAKEFLALLLGDG-TYQAALKLQKVYRS 108
>At1g65130 hypothetical protein
Length = 1088
Score = 25.0 bits (53), Expect = 6.2
Identities = 17/59 (28%), Positives = 28/59 (46%), Gaps = 6/59 (10%)
Query: 36 DPKVIDVANFAVKEYNNRRRKPEEKLRLWKVI---KGESQI---VADGVNYRLTLSATK 88
D +ID N ++E RR PE++ L+ + K E I D + +L+L A +
Sbjct: 531 DAALIDANNLCIREDERRRNIPEDQWTLYASLLCDKCEENIRRHAGDSLTTKLSLCAVQ 589
>At5g49910 heat shock protein 70 (Hsc70-7)
Length = 718
Score = 24.6 bits (52), Expect = 8.1
Identities = 23/90 (25%), Positives = 37/90 (40%), Gaps = 11/90 (12%)
Query: 17 TATKQAIPIGNLSPINNINDPKVIDVANFAVKEYNNRRRKPEEKLRLWKVIKGESQI--- 73
TATK A I L + IN+P +A RK E + ++ + G +
Sbjct: 228 TATKDAGRIAGLEVLRIINEPTAASLA-------YGFERKSNETILVFDLGGGTFDVSVL 280
Query: 74 -VADGVNYRLTLSATKVYTSNTYEAIVLEW 102
V DGV L+ S + ++ V++W
Sbjct: 281 EVGDGVFEVLSTSGDTHLGGDDFDKRVVDW 310
>At5g09260 unknown protein
Length = 216
Score = 24.6 bits (52), Expect = 8.1
Identities = 20/86 (23%), Positives = 35/86 (40%), Gaps = 4/86 (4%)
Query: 34 INDPKVIDVANFAVKEYNNRRRKPEEKLRLWKVIKGESQIVADGVNY----RLTLSATKV 89
+ PK+ +V + RR+ + + +L KVI+ E Q D + R L+ K
Sbjct: 6 VKKPKITEVDRAILSLKTQRRKLGQYQQQLEKVIEAEKQAARDLIREKRKDRALLALKKK 65
Query: 90 YTSNTYEAIVLEWSLQHLRNLTSFKL 115
T V +W + + L +L
Sbjct: 66 RTQEELLKQVDQWLINVEQQLADIEL 91
>At3g54660 Gluthatione reductase, chloroplast precursor
Length = 565
Score = 24.6 bits (52), Expect = 8.1
Identities = 12/39 (30%), Positives = 23/39 (58%)
Query: 61 LRLWKVIKGESQIVADGVNYRLTLSATKVYTSNTYEAIV 99
+R KV++G + V D V +++L + +T + EAI+
Sbjct: 290 IRQKKVLRGFDEDVRDFVGEQMSLRGIEFHTEESPEAII 328
>At3g32260 hypothetical protein
Length = 309
Score = 24.6 bits (52), Expect = 8.1
Identities = 12/42 (28%), Positives = 23/42 (54%), Gaps = 4/42 (9%)
Query: 44 NFAVKEYNNRRRKPEEKLRLWKVIKGESQIVADGVNYRLTLS 85
N A+++Y ++R +RLW++ K ++ DG R+ S
Sbjct: 16 NPALEQYKIKKRV----VRLWRLFKSIEMVLVDGEGTRIHAS 53
>At2g41140 CDPK-related kinase 1 (CRK1)
Length = 576
Score = 24.6 bits (52), Expect = 8.1
Identities = 21/79 (26%), Positives = 33/79 (41%), Gaps = 4/79 (5%)
Query: 16 FTATKQAIPIGNLSPINNINDPKVIDVANFAVKEYNNRRRKPEEKLRLWKVIKGESQIVA 75
F A +A P +P +++ V V K+Y R + W V E +I +
Sbjct: 336 FRAVLKAEPNFEEAPWPSLSPEAVDFVKRLLNKDYRKRLTAAQALCHPWLVGSHELKIPS 395
Query: 76 DGVNYRLTLSATKVYTSNT 94
D + Y+L KVY +T
Sbjct: 396 DMIIYKL----VKVYIMST 410
>At1g55810 uracil phosphoribosyltransferase 1, putative
Length = 466
Score = 24.6 bits (52), Expect = 8.1
Identities = 15/64 (23%), Positives = 29/64 (44%), Gaps = 5/64 (7%)
Query: 28 LSPINNINDPKVIDVANFAVKEYNN-----RRRKPEEKLRLWKVIKGESQIVADGVNYRL 82
LS + + + +D+ N+ K Y N RR P + + L ++ V D +N ++
Sbjct: 110 LSSMEKLRKGQAVDIPNYDFKSYKNNVFPPRRVNPSDVIILEGILIFHDPRVRDLMNMKI 169
Query: 83 TLSA 86
+ A
Sbjct: 170 FVDA 173
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.134 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,329,721
Number of Sequences: 26719
Number of extensions: 82805
Number of successful extensions: 260
Number of sequences better than 10.0: 21
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 14
Number of HSP's that attempted gapping in prelim test: 247
Number of HSP's gapped (non-prelim): 21
length of query: 118
length of database: 11,318,596
effective HSP length: 94
effective length of query: 24
effective length of database: 8,807,010
effective search space: 211368240
effective search space used: 211368240
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 52 (24.6 bits)
Medicago: description of AC148347.5


