

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148345.6 + phase: 0
(570 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
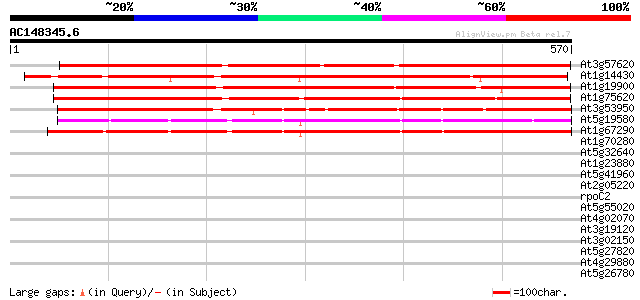
Score E
Sequences producing significant alignments: (bits) Value
At3g57620 putative protein 697 0.0
At1g14430 hypothetical protein 598 e-171
At1g19900 unknown protein 567 e-162
At1g75620 unknown protein 565 e-161
At3g53950 unknown protein 434 e-122
At5g19580 putative protein 431 e-121
At1g67290 unknown protein 427 e-120
At1g70280 NHL repeat-containing protein 35 0.15
At5g32640 putative protein 34 0.19
At1g23880 unknown protein 32 1.2
At5g41960 unknown protein 31 2.1
At2g05220 40S ribosomal protein S17 31 2.1
rpoC2 -chloroplast genome- RNA polymerase beta' subunit-2 30 2.8
At5g55020 putative transcription factor MYB120 (MYB120) 30 2.8
At4g02070 G/T DNA mismatch repair enzyme 30 2.8
At3g19120 unknown protein 30 3.6
At3g02150 unknown protein 30 3.6
At5g27820 putative protein 30 4.7
At4g29880 putative protein 30 4.7
At5g26780 glycine hydroxymethyltransferase - like protein 29 6.2
>At3g57620 putative protein
Length = 547
Score = 697 bits (1799), Expect = 0.0
Identities = 336/523 (64%), Positives = 412/523 (78%), Gaps = 17/523 (3%)
Query: 51 WELIQPTIGISAMHMQLSHNNKIIIFDRTDFGPSNLPLSNGRCRMDPFDTALKIDCTAHS 110
WE++ P+IGISAMHMQL HN +I+FDRTDFG SN+ L G CR DP DTA K DC+AHS
Sbjct: 37 WEMLLPSIGISAMHMQLLHNGMVIMFDRTDFGTSNVSLPGGICRYDPTDTAEKFDCSAHS 96
Query: 111 VLYDIATNTFRSLTVQTDTWCSSGSVLSNGTLVQTGGFNDGERRIRMFTPC-FNENCDWI 169
VLYD+ +NT+R L VQTDTWCSSG+VL NGTLVQTGG+NDGER RMF+PC +++ CDWI
Sbjct: 97 VLYDVVSNTYRPLNVQTDTWCSSGAVLPNGTLVQTGGYNDGERAARMFSPCGYSDTCDWI 156
Query: 170 EFPSYLSERRWYATNQILPDNRIIIIGGRRQFNYEFIPKTTTSSSSSSSSSIHLSFLQET 229
EFP YLS+RRWYATNQILPD RII++GGRRQFNYE P+ + S SS L FL+ET
Sbjct: 157 EFPQYLSQRRWYATNQILPDGRIIVVGGRRQFNYELFPRHDSRSRSS-----RLEFLRET 211
Query: 230 NDPS-ENNLYPFVHLLPNGNLFIFANTRSILFDYKQNVVVKEFPEIPGGDPHNYPSSGSS 288
+D S ENNLYPF+HLLP+GNLF+FANTRSI+FDYK+N +VKEFPEIPGGDP NYPSSGSS
Sbjct: 212 SDGSNENNLYPFIHLLPDGNLFVFANTRSIVFDYKKNRIVKEFPEIPGGDPRNYPSSGSS 271
Query: 289 VLLPLDE-NQISMEATIMICGGAPRGSFEAAKGKNFMPALKTCGFLKVTDSNPSWIIENM 347
+L PLD+ N ++E IM+CGG+P+G F + F A TCG LK++D +PSW +E M
Sbjct: 272 ILFPLDDTNDANVEVEIMVCGGSPKGGF----SRGFTRATSTCGRLKLSDQSPSWEMETM 327
Query: 348 PMARVMGDMLILPNGDVIIINGAGSGTAGWENGRQPVLTPVIFRSSETKSDKRFSVMSPA 407
P+ RVMGDML+LP GDVII+NGAG+GTAGWE R P++ PVI++ D F+VMS
Sbjct: 328 PLPRVMGDMLLLPTGDVIIVNGAGAGTAGWEKARDPIIQPVIYQ----PFDHLFTVMSTP 383
Query: 408 SRPRLYHSSAIVLRDGRVLVGGSNPHVNYNFTGVEFPTDLSLEAFSPPYLSLEFDLVRPT 467
SRPR+YHSSAI+L DGRVLVGGSNPHV YNFT VE+PTDLSLEA+SPPYL D +RP
Sbjct: 384 SRPRMYHSSAILLPDGRVLVGGSNPHVYYNFTNVEYPTDLSLEAYSPPYLFFTSDPIRPK 443
Query: 468 IWHVTNKILGYRVFYYVTFTVAKFASASEVSVRLLAPSFTTHSFGMNQRMVVLKLIGVTM 527
I ++K+L Y+ + V F++A+F + +SVR++APSFTTHSF MNQRMV+LKL+ VT
Sbjct: 444 ILLTSDKVLSYKRLFNVDFSIAQFLTVDLLSVRIVAPSFTTHSFAMNQRMVILKLLSVTR 503
Query: 528 VNL-DIYYATVVGPSTQEIAPPGYYLLFLVHAGVPSSGEWVQL 569
L + Y + +GPST EIAPPGYY++FLVHAG+PSS WVQ+
Sbjct: 504 DQLTNSYRVSALGPSTAEIAPPGYYMIFLVHAGIPSSAAWVQI 546
>At1g14430 hypothetical protein
Length = 849
Score = 598 bits (1543), Expect = e-171
Identities = 304/567 (53%), Positives = 401/567 (70%), Gaps = 35/567 (6%)
Query: 16 LIPHFHILIVSSSSSSPSPSPLPSLLSSPSSNQGEWELIQPTIGISAMHMQLSHNNKIII 75
+I F + S+S SPL L G W+L+QP++GISAMHMQL HNNK++I
Sbjct: 14 VISFFFFFLCSTSDLLLPRSPLAILTG------GRWDLLQPSVGISAMHMQLLHNNKVVI 67
Query: 76 FDRTDFGPSNLPLSNGRCRMDPFDTALKIDCTAHSVLYDIATNTFRSLTVQTDTWCSSGS 135
FDRTD+GPSN+ L + C+ A DC+AHS+LYD+A+NTFR LT++ DTWCSSGS
Sbjct: 68 FDRTDYGPSNVSLPSQTCQ-----NATVFDCSAHSILYDVASNTFRPLTLRYDTWCSSGS 122
Query: 136 VLSNGTLVQTGGFNDGERRIRMFTPCF----NENCDWIEFPSYLSERRWYATNQILPDNR 191
+ ++G+L+QTGG+ +GER +R+FTPC + +CDWIE +YLS RRWY+TNQILPD R
Sbjct: 123 LNASGSLIQTGGYGNGERTVRVFTPCDGGVGSVSCDWIENRAYLSSRRWYSTNQILPDGR 182
Query: 192 IIIIGGRRQFNYEFIPKTTTSSSSSSSSSIHLSFLQETNDPSE-NNLYPFVHLLPNGNLF 250
III+GGRR FNYEF PK S +L FL ET DP+E NNLYPF+HLLP+GNLF
Sbjct: 183 IIIVGGRRAFNYEFYPK------DPGESVFNLRFLAETRDPNEENNLYPFLHLLPDGNLF 236
Query: 251 IFANTRSILFDYKQNVVVKEFPEIPGGDPHNYPSSGSSVLLPL----DENQISMEATIMI 306
IFAN RSILFD+ + ++KEFP+IPGGD NYPS+GSSVLLPL D N+ + A +M+
Sbjct: 237 IFANRRSILFDFVNHRIIKEFPQIPGGDKRNYPSTGSSVLLPLFLTGDINRTKITAEVMV 296
Query: 307 CGGAPRGSF-EAAKG--KNFMPALKTCGFLKVTDSNPSWIIENMPMARVMGDMLILPNGD 363
CGGAP G+F +AA+ K F+ +TCG LKVTD +P W++E MP RVM DML+LPNGD
Sbjct: 297 CGGAPPGAFFKAARTIPKIFVAGSRTCGRLKVTDPDPKWVMEQMPSPRVMSDMLLLPNGD 356
Query: 364 VIIINGAGSGTAGWENGRQPVLTPVIFRSSETKSDKRFSVMSPASRPRLYHSSAIVLRDG 423
V+IINGA +GTAGWE+ VL P+++ E +RF +++P PR+YHS++++L DG
Sbjct: 357 VLIINGAANGTAGWEDATNAVLNPILYLPEEPDQTRRFEILTPTRIPRMYHSASLLLSDG 416
Query: 424 RVLVGGSNPHVNYNFTGVEFPTDLSLEAFSPPYLSLEFDLVRPTIWHVTNKILG---YRV 480
RVLVGGSNPH NYNFT +PT+LSLEA+ P YL ++ VRPTI +T ++ G Y
Sbjct: 417 RVLVGGSNPHRNYNFTARPYPTELSLEAYLPRYLDPQYARVRPTI--ITVELAGNMLYGQ 474
Query: 481 FYYVTFTVAKFAS-ASEVSVRLLAPSFTTHSFGMNQRMVVLKLIGVTMVNLDIYYATVVG 539
+ VTF + F VSVRL+APSF+THS MNQR++VL++ V+ +++ Y A V G
Sbjct: 475 AFAVTFAIPAFGMFDGGVSVRLVAPSFSTHSTAMNQRLLVLRVRRVSQLSVFAYKADVDG 534
Query: 540 PSTQEIAPPGYYLLFLVHAGVPSSGEW 566
P+ +APPGYY++F+VH G+PS W
Sbjct: 535 PTNSYVAPPGYYMMFVVHRGIPSVAVW 561
>At1g19900 unknown protein
Length = 548
Score = 567 bits (1462), Expect = e-162
Identities = 285/532 (53%), Positives = 369/532 (68%), Gaps = 18/532 (3%)
Query: 45 SSNQGEWELIQPTIGISAMHMQLSHNNKIIIFDRTDFGPSNLPLSNGRCRMDPFDTALKI 104
S+ +G W+ I P +GISAMHMQL HN++++++DRT+FGPSN+ L NG CR +P D KI
Sbjct: 27 SAARGLWKYIAPNVGISAMHMQLLHNDRVVMYDRTNFGPSNISLPNGNCRDNPQDAVSKI 86
Query: 105 DCTAHSVLYDIATNTFRSLTVQTDTWCSSGSVLSNGTLVQTGGFNDGERRIRMFTPCFNE 164
DCTAHS+ YD+ATNT R LTVQ++TWCSSGSV +G LVQTGG DGE + R F+PC N
Sbjct: 87 DCTAHSIEYDVATNTIRPLTVQSNTWCSSGSVRPDGVLVQTGGDRDGELKTRTFSPCNNN 146
Query: 165 NCDWIEFPSYLSERRWYATNQILPDNRIIIIGGRRQFNYEFIPKTTTSSSSSSSSSIHLS 224
CDW+E + L +RRWYA+N ILPD + I++GG+ QFNYEF PKTT + + + L
Sbjct: 147 QCDWVEMNNGLKKRRWYASNHILPDGKQIVMGGQGQFNYEFFPKTT------NPNVVALP 200
Query: 225 FLQETNDP-SENNLYPFVHLLPNGNLFIFANTRSILFDYKQNVVVKEFPEIPGGDPHNYP 283
FL ET+D ENNLYPFV + +GNLF+FAN R+IL DY +N VVK FP IPGGDP NYP
Sbjct: 201 FLAETHDQGQENNLYPFVFMNTDGNLFMFANNRAILLDYVKNTVVKTFPAIPGGDPRNYP 260
Query: 284 SSGSSVLLPLDENQI-SMEATIMICGGAPRGSFEAAKGKNFMPALKTCGFLKVTDSNPSW 342
S+GS+VLLPL + ++E +++CGGAP+GS+ A+ K F+ AL TC +K+ D+ P W
Sbjct: 261 STGSAVLLPLKNLEADNVETEVLVCGGAPKGSYNLARKKTFVKALDTCARIKINDAKPEW 320
Query: 343 IIENMPMARVMGDMLILPNGDVIIINGAGSGTAGWENGRQPVLTPVIFRSSETKSDKRFS 402
+E MP ARVMGDM+ LPNGDV++ING GTA WE GR PVL P ++ E RF
Sbjct: 321 AVEKMPHARVMGDMIPLPNGDVLLINGGSFGTAAWELGRTPVLAPDLYH-PENPVGSRFE 379
Query: 403 VMSPASRPRLYHSSAIVLRDGRVLVGGSNPHVNYNFTGVEFPTDLSLEAFSPPYLSLEFD 462
+ P + PR+YHS+AI+LRDGRVLVGGSNPH YN+TGV FPT+LSLEAFSP YL EF
Sbjct: 380 SLRPTTIPRMYHSAAILLRDGRVLVGGSNPHAFYNYTGVLFPTELSLEAFSPVYLQREFS 439
Query: 463 LVRPTIWHVTNKILGYRVFYYVTFTVAKFASASEVS----VRLLAPSFTTHSFGMNQRMV 518
+RP I + + Y T KF+ EV+ V ++ P+FTTHSF MNQR++
Sbjct: 440 NLRPKIISPEPQ----SMIKYGTNLKLKFSVTGEVTTPAKVTMVFPTFTTHSFAMNQRVL 495
Query: 519 VLKLIGVTMVNLD-IYYATVVGPSTQEIAPPGYYLLFLVHAGVPSSGEWVQL 569
VL + T +Y V P + IA PGYY++F+V+ +PS G WV+L
Sbjct: 496 VLDNVKFTRKGKSPMYEVQVRTPRSANIAWPGYYMIFVVNQDIPSEGVWVKL 547
>At1g75620 unknown protein
Length = 547
Score = 565 bits (1455), Expect = e-161
Identities = 283/528 (53%), Positives = 366/528 (68%), Gaps = 15/528 (2%)
Query: 45 SSNQGEWELIQPTIGISAMHMQLSHNNKIIIFDRTDFGPSNLPLSNGRCRMDPFDTALKI 104
S ++G WEL+ P +GISAMH QL HN+++I++DRT+FGPSN+ L NG CR P D K
Sbjct: 31 SGDEGTWELLLPNVGISAMHSQLLHNDRVIMYDRTNFGPSNISLPNGACRSSPGDAVSKT 90
Query: 105 DCTAHSVLYDIATNTFRSLTVQTDTWCSSGSVLSNGTLVQTGGFNDGERRIRMFTPCFNE 164
DCTAHSV YD+A N R LTVQ++TWCSSG V +GTL+QTGG DGER++R+ PC +
Sbjct: 91 DCTAHSVEYDVALNRIRPLTVQSNTWCSSGGVTPDGTLLQTGGDLDGERKVRLMDPCDDN 150
Query: 165 NCDWIEFPSYLSERRWYATNQILPDNRIIIIGGRRQFNYEFIPKTTTSSSSSSSSSIHLS 224
+CDWIE + L+ RRWYATN ILPD R IIIGGR QFNYEF PKT + S +
Sbjct: 151 SCDWIEVDNGLAARRWYATNHILPDGRQIIIGGRGQFNYEFFPKTNAPNFYS------IP 204
Query: 225 FLQETNDP-SENNLYPFVHLLPNGNLFIFANTRSILFDYKQNVVVKEFPEIPGGDPHNYP 283
FL ETNDP ENNLYPFV L +GNLFIFAN R+IL DY N VV+ +PEIPGGDP +YP
Sbjct: 205 FLSETNDPGDENNLYPFVFLNTDGNLFIFANNRAILLDYSTNTVVRTYPEIPGGDPRSYP 264
Query: 284 SSGSSVLLPLDENQISMEATIMICGGAPRGSFEAAKGKNFMPALKTCGFLKVTDSNPSWI 343
S+GS+VLLP+ ++ +++CGGAP+GS+ + F+ AL TC + + D NP WI
Sbjct: 265 STGSAVLLPIK----NLVLEVLVCGGAPKGSYNLSWRNTFVKALDTCARININDVNPQWI 320
Query: 344 IENMPMARVMGDMLILPNGDVIIINGAGSGTAGWENGRQPVLTPVIFRSSETKSDKRFSV 403
+E MP ARVMGDM++LP+G+V++ING SGTA WE GR+PVL P ++ + RF V
Sbjct: 321 VEKMPRARVMGDMMLLPDGNVLLINGGSSGTAAWELGREPVLHPDLYHPDKPVG-SRFEV 379
Query: 404 MSPASRPRLYHSSAIVLRDGRVLVGGSNPHVNYNFTGVEFPTDLSLEAFSPPYLSLEFDL 463
+P++ PR+YHS A +LRDGR+LVGGSNPH YNFTGV FPT+L LEAFSP YL ++
Sbjct: 380 QNPSTIPRMYHSIATLLRDGRILVGGSNPHAFYNFTGVLFPTELRLEAFSPSYLDTKYSS 439
Query: 464 VRPTIWHV-TNKILGYRVFYYVTFTVAKFASASEVSVRLLAPSFTTHSFGMNQRMVVL-K 521
+RP+I + Y + F V+ S V V +L PSFTTHSF M+QR++VL
Sbjct: 440 LRPSIVDPRPQTTVNYGRVLRLRFIVSGRVK-SPVKVTMLFPSFTTHSFSMHQRLLVLDH 498
Query: 522 LIGVTMVNLDIYYATVVGPSTQEIAPPGYYLLFLVHAGVPSSGEWVQL 569
+I + IY V PS+ +APPGYY++F+V+ +PS G WV+L
Sbjct: 499 VISFKLGISKIYEVRVRTPSSAILAPPGYYMVFVVNQDIPSEGLWVRL 546
>At3g53950 unknown protein
Length = 545
Score = 434 bits (1115), Expect = e-122
Identities = 232/527 (44%), Positives = 317/527 (60%), Gaps = 21/527 (3%)
Query: 49 GEWELIQPTIGISAMHMQLSHNNKIIIFDRTDFGPSNLPLSNGRCRMDPFDTALKIDCTA 108
G WELI GI++MH ++ N +I+ DRT+ GPS L RCR DP D ALK DC A
Sbjct: 34 GSWELIVQDAGIASMHTAVTRFNTVILLDRTNIGPSRKALDRHRCRRDPKDAALKRDCYA 93
Query: 109 HSVLYDIATNTFRSLTVQTDTWCSSGSVLSNGTLVQTGGFNDGERRIRMFTPCF-NENCD 167
HSVL+D+ TN R L +QTDTWCSSG LS+G+L+QTGG DG ++IR F PC NE CD
Sbjct: 94 HSVLFDLGTNQIRPLMIQTDTWCSSGQFLSDGSLLQTGGDKDGFKKIRKFEPCDPNETCD 153
Query: 168 WIEF-PSYLSERRWYATNQILPDNRIIIIGGRRQFNYEFIPKTTTSSSSSSSSSIHLSFL 226
W+E + L RWYA+NQILPD +II+GGR E+ P + ++ FL
Sbjct: 154 WVELQDTELITGRWYASNQILPDGSVIIVGGRGTNTVEYYP-------PRENGAVPFQFL 206
Query: 227 QETNDPSENNLYPFVHLLPN---GNLFIFANTRSILFDYKQNVVVKEFPEIPGGDPHNYP 283
+ D +NLYP+VHLLP+ GNLFIFAN+R++ +D++ N VVKE+P + GG P NYP
Sbjct: 207 ADVEDKQMDNLYPYVHLLPDDDGGNLFIFANSRAVKYDHRINAVVKEYPPLDGG-PRNYP 265
Query: 284 SSGSSVLLPLDENQISMEATIMICGGAPRGSFEAAKGKNFMPALKTCGFLKVTDSNPSWI 343
S GSS +L + + + E I+ICGGA G+F A PA TCG + T ++P W+
Sbjct: 266 SGGSSAMLAIQGDFTTAE--ILICGGAQSGAFTARAID--APAHGTCGRIVATAADPVWV 321
Query: 344 IENMPMARVMGDMLILPNGDVIIINGAGSGTAGWENGRQPVLTPVIFRSSETKSDKRFSV 403
E MP R+MGDM+ LP G+++IINGA +G+ G+E G P L P+++R + RF
Sbjct: 322 TEEMPFGRIMGDMVNLPTGEILIINGAQAGSQGFEMGSDPCLYPLLYRPDQ-PIGLRFMT 380
Query: 404 MSPASRPRLYHSSAIVLRDGRVLVGGSNPHVNYNFTGVEFPTDLSLEAFSPPYLSLEFDL 463
++P + PR+YHS+A +L DGR+L+ GSNPH Y F EFPT+L +EAFSP YLS +
Sbjct: 381 LNPGTVPRMYHSTANLLPDGRILLAGSNPHYFYKF-NAEFPTELRIEAFSPEYLSPDRAN 439
Query: 464 VRPTIWHVTNKILGYRVFYYVTFTVAKFASASEVSVRLLAPSFTTHSFGMNQRMVVLKLI 523
+RP I + I VF F + + + F THSF QR+V L +
Sbjct: 440 LRPEIQEIPQIIRYGEVF--DVFVTVPLPVVGIIQMNWGSAPFATHSFSQGQRLVKLTVA 497
Query: 524 GVTMVNLDIYYATVVGPSTQEIAPPGYYLLFLVHAGVPSSGEWVQLM 570
+ Y P ++PPGYY+ F V+ GVPS W++++
Sbjct: 498 PSVPDGVGRYRIQCTAPPNGAVSPPGYYMAFAVNQGVPSIARWIRIV 544
>At5g19580 putative protein
Length = 594
Score = 431 bits (1107), Expect = e-121
Identities = 231/529 (43%), Positives = 320/529 (59%), Gaps = 18/529 (3%)
Query: 49 GEWELIQPTIGISAMHMQLSHN-NKIIIFDRTDFGPSNLPLSNG-RCRMDPFDTALKIDC 106
G+WEL G+S MH L NK+ +D T + S + L G C + T K+DC
Sbjct: 77 GKWELFLENSGVSGMHAILMPVINKVQYYDATIWRISKIKLPPGVPCHVVDAKTN-KVDC 135
Query: 107 TAHSVLYDIATNTFRSLTVQTDTWCSSGSVLSNGTLVQTGGFNDGERRIRMFTPCFNENC 166
AHS+L D+ T + L + TDTWCSSG + NGTLV TGG+ G R + C ENC
Sbjct: 136 WAHSILMDVNTGALKPLGLSTDTWCSSGGLTVNGTLVSTGGYGGGANTARYLSSC--ENC 193
Query: 167 DWIEFPSYLSERRWYATNQILPDNRIIIIGGRRQFNYEFIPKTTTSSSSSSSSSIHLSFL 226
W E+P L+ +RWY+T LPD + +IGGR NYE+IP+ ++ S + L
Sbjct: 194 KWEEYPQALAAKRWYSTQATLPDGKFFVIGGRDALNYEYIPEEGQNNRKLFDSLL----L 249
Query: 227 QETNDPSENNLYPFVHLLPNGNLFIFANTRSILFDYKQNVVVKEFPEIPGGDPHNYPSSG 286
++T+DP ENNLYPFV L +GNLFIFAN RSIL K N V+KEFP++PGG NYP SG
Sbjct: 250 RQTDDPEENNLYPFVWLNTDGNLFIFANNRSILLSPKTNQVIKEFPQLPGG-ARNYPGSG 308
Query: 287 SSVLLPLD---ENQISMEATIMICGGAPRGSFEAAKGKNFMPALKTCGFLKVTDSNPSWI 343
SS LLP+ +N + A +++CGG+ + ++ A K + PAL+ C +++ + P W
Sbjct: 309 SSALLPIQLYVKNPKVIPAEVLVCGGSKQDAYYKAGKKIYEPALQDCARIRINSAKPRWK 368
Query: 344 IENMPMARVMGDMLILPNGDVIIINGAGSGTAGWENGRQPVLTPVIFRSSETKSDKRFSV 403
E MP R+M D +ILPNGD++++NGA G +GW G+ P P++++ + KRF
Sbjct: 369 TEMMPTPRIMSDTVILPNGDILLVNGAKRGCSGWGYGKDPAFAPLLYKPHAARG-KRFRQ 427
Query: 404 MSPASRPRLYHSSAIVLRDGRVLVGGSNPHVNYNFTGVEFPTDLSLEAFSPPYLSLEFDL 463
+ P + PR+YHSSAI+L DG+VLVGGSN + Y + VEFPT+L +E FSPPYL
Sbjct: 428 LKPTTIPRMYHSSAIILPDGKVLVGGSNTNDGYKY-NVEFPTELRVEKFSPPYLDPALAN 486
Query: 464 VRPTIWHV-TNKILGYRVFYYVTFTV-AKFASASEVSVRLLAPSFTTHSFGMNQRMVVLK 521
+RP I T K + Y F+ V + K A+ + V +LAP+FTTHS MN RM++L
Sbjct: 487 IRPKIVTTGTPKQVKYGQFFNVKVDLKEKGATKGNLKVTMLAPAFTTHSISMNMRMLILG 546
Query: 522 LIGVTMVNLDIYYATVVGPSTQEIAPPGYYLLFLVHAGVPSSGEWVQLM 570
+ V Y V P IAPPGYYL+F ++ GVPS+GEW+Q++
Sbjct: 547 VNNVKPAGAG-YDIQAVAPPNGNIAPPGYYLIFAIYKGVPSTGEWIQVV 594
>At1g67290 unknown protein
Length = 615
Score = 427 bits (1098), Expect = e-120
Identities = 242/542 (44%), Positives = 330/542 (60%), Gaps = 21/542 (3%)
Query: 39 SLLSSPSSN-QGEWELIQPTIGISAMHMQLSHN-NKIIIFDRTDFGPSNLPLSNG-RCRM 95
++ + P N G+WEL G+SAMH L NK+ +D T + S + L G C +
Sbjct: 85 TVAAGPEMNWPGQWELFMKNSGVSAMHAILMPLINKVQFYDATIWRISQIKLPPGVPCHV 144
Query: 96 DPFDTAL-KIDCTAHSVLYDIATNTFRSLTVQTDTWCSSGSVLSNGTLVQTGGFNDGERR 154
FD K+DC AHSVL DI T + L + TDTWCSSG + NGTLV TGGF G
Sbjct: 145 --FDAKKNKVDCWAHSVLVDINTGDIKPLALTTDTWCSSGGLTVNGTLVSTGGFQGGANT 202
Query: 155 IRMFTPCFNENCDWIEFPSYLSERRWYATNQILPDNRIIIIGGRRQFNYEFIPKTTTSSS 214
R + C ENC WIE+P L+ RRWY+T LPD I++GGR NYE+I ++
Sbjct: 203 ARYLSTC--ENCVWIEYPKALAARRWYSTQATLPDGTFIVVGGRDALNYEYILPEGQNNK 260
Query: 215 SSSSSSIHLSFLQETNDPSENNLYPFVHLLPNGNLFIFANTRSILFDYKQNVVVKEFPEI 274
S + L++T+DP ENNLYPFV L +GNLFIFAN RSIL K N V+KEFP++
Sbjct: 261 KLYDSQL----LRQTDDPEENNLYPFVWLNTDGNLFIFANNRSILLSPKTNKVLKEFPQL 316
Query: 275 PGGDPHNYPSSGSSVLLPLD---ENQISMEATIMICGGAPRGS-FEAAKGKNFMPALKTC 330
PGG NYP S SS LLP+ +N + A +++CGGA + + F A + K + ALK C
Sbjct: 317 PGG-ARNYPGSASSALLPIRLYVQNPAIIPADVLVCGGAKQDAYFRAERLKIYDWALKDC 375
Query: 331 GFLKVTDSNPSWIIENMPMARVMGDMLILPNGDVIIINGAGSGTAGWENGRQPVLTPVIF 390
L + + P W E MP +RVM D +ILPNG+++IINGA G++GW ++P P+++
Sbjct: 376 ARLNINSAKPVWKTETMPTSRVMSDTVILPNGEILIINGAKRGSSGWHLAKEPNFAPLLY 435
Query: 391 RSSETKSDKRFSVMSPASRPRLYHSSAIVLRDGRVLVGGSNPHVNYNFTGVEFPTDLSLE 450
+ ++ +RF ++P++ PR+YHS AI L DG+VLVGGSN + Y F VE+PT+L +E
Sbjct: 436 KPNKPLG-QRFKELAPSTIPRVYHSIAIALPDGKVLVGGSNTNNGYQFN-VEYPTELRIE 493
Query: 451 AFSPPYLSLEFDLVRPTIWHV-TNKILGYRVFYYVTFTVAKFASASE-VSVRLLAPSFTT 508
FSPPYL +RP I + T K + Y + V + + A E V V +LAPSFTT
Sbjct: 494 KFSPPYLDPALANMRPRIVNTATPKQIKYGQMFDVKIELKQQNVAKENVMVTMLAPSFTT 553
Query: 509 HSFGMNQRMVVLKLIGVTMVNLDIYYATVVGPSTQEIAPPGYYLLFLVHAGVPSSGEWVQ 568
HS MN R+++L + V V D + V P + ++APPGYYLLF V+ GVPS GEW+Q
Sbjct: 554 HSVSMNMRLLMLGINNVKNVGGDNHQIQAVAPPSGKLAPPGYYLLFAVYNGVPSVGEWIQ 613
Query: 569 LM 570
++
Sbjct: 614 IV 615
>At1g70280 NHL repeat-containing protein
Length = 509
Score = 34.7 bits (78), Expect = 0.15
Identities = 20/80 (25%), Positives = 39/80 (48%), Gaps = 4/80 (5%)
Query: 208 KTTTSSSSSSSSSIHLS---FLQETNDPSENNLYPF-VHLLPNGNLFIFANTRSILFDYK 263
KTTT ++ ++ S + ++ D S+ + P+ + +LPNG L I + S ++
Sbjct: 51 KTTTKTTIATRSMVKFENGYSVETVFDGSKLGIEPYSIEVLPNGELLILDSENSNIYKIS 110
Query: 264 QNVVVKEFPEIPGGDPHNYP 283
++ + P + G P YP
Sbjct: 111 SSLSLYSRPRLVTGSPEGYP 130
>At5g32640 putative protein
Length = 402
Score = 34.3 bits (77), Expect = 0.19
Identities = 29/87 (33%), Positives = 42/87 (47%), Gaps = 12/87 (13%)
Query: 7 ILIFFILSSLIPHFHILIVSSSSS---SPSPSPLPSLLSSPSS-NQGEWELIQPTIGISA 62
I IFF +S PH ++S SS SPS PLP+ L P + + + ++ PTI A
Sbjct: 123 IYIFFDVSHSQPHLSTFLISFSSDKAFSPSQPPLPNRLREPDAILKLFFTMVAPTIATQA 182
Query: 63 MHMQLSHNNKIIIFDRTDFGPSNLPLS 89
+ N ++ F S+LPLS
Sbjct: 183 VEC----NGTCLVV----FTSSSLPLS 201
>At1g23880 unknown protein
Length = 485
Score = 31.6 bits (70), Expect = 1.2
Identities = 19/80 (23%), Positives = 37/80 (45%), Gaps = 4/80 (5%)
Query: 208 KTTTSSSSSSSSSIHLS---FLQETNDPSENNLYPF-VHLLPNGNLFIFANTRSILFDYK 263
KTTT ++ + S + ++ D S+ + P+ + +L NG L I + S ++
Sbjct: 53 KTTTKTAVPTKSMVKFENGYSVETVLDGSKLGIEPYSIQVLSNGELLILDSQNSNIYQIS 112
Query: 264 QNVVVKEFPEIPGGDPHNYP 283
++ + P + G P YP
Sbjct: 113 SSLSLYSRPRLVTGSPEGYP 132
>At5g41960 unknown protein
Length = 217
Score = 30.8 bits (68), Expect = 2.1
Identities = 18/44 (40%), Positives = 27/44 (60%)
Query: 1 MSSITLILIFFILSSLIPHFHILIVSSSSSSPSPSPLPSLLSSP 44
M+S++ + I + SS + ++SSSSSSPSP PL L +P
Sbjct: 1 MASLSCLPICGLSSSASLYTKARLLSSSSSSPSPLPLAYPLQNP 44
>At2g05220 40S ribosomal protein S17
Length = 179
Score = 30.8 bits (68), Expect = 2.1
Identities = 16/31 (51%), Positives = 17/31 (54%)
Query: 10 FFILSSLIPHFHILIVSSSSSSPSPSPLPSL 40
F S + H I I SSSPSPSP PSL
Sbjct: 7 FLYFSPPLSHLQIEIALCLSSSPSPSPSPSL 37
>rpoC2 -chloroplast genome- RNA polymerase beta' subunit-2
Length = 1376
Score = 30.4 bits (67), Expect = 2.8
Identities = 29/108 (26%), Positives = 45/108 (40%), Gaps = 15/108 (13%)
Query: 200 QFNYEFIPKTTTSSSSSSSSSIHLS---FLQETNDPSENNLYPFVHLLPNGNL------- 249
Q N F+ S SS S ++ +S F + DP + +Y + L NGNL
Sbjct: 499 QMNIPFLSAERKSISSLSVNNDQVSQKFFSSDFADPKKLGIYDYSEL--NGNLGTSHYNL 556
Query: 250 ---FIFANTRSILFDYKQNVVVKEFPEIPGGDPHNYPSSGSSVLLPLD 294
IF +L ++N + F I + P SG SV +P++
Sbjct: 557 IYSAIFHENSDLLAKRRRNRFLIPFQSIQEQEKEFIPQSGISVEIPIN 604
>At5g55020 putative transcription factor MYB120 (MYB120)
Length = 523
Score = 30.4 bits (67), Expect = 2.8
Identities = 19/47 (40%), Positives = 27/47 (57%), Gaps = 7/47 (14%)
Query: 26 SSSSSSPSPSPLPSLLSSPSSNQGEWELIQPTIGISAM-HMQLSHNN 71
S SS+P P PL S L SP +NQ PT+ + A+ Q+++NN
Sbjct: 228 SQLSSTPPPPPLSSPLCSPRNNQ------YPTLPLFALPRSQINNNN 268
>At4g02070 G/T DNA mismatch repair enzyme
Length = 1324
Score = 30.4 bits (67), Expect = 2.8
Identities = 16/24 (66%), Positives = 19/24 (78%), Gaps = 2/24 (8%)
Query: 26 SSSSSSPSPSPLPSLLS--SPSSN 47
+SSSSSPSPSP PSL + +P SN
Sbjct: 29 ASSSSSPSPSPSPSLSNKKTPKSN 52
>At3g19120 unknown protein
Length = 446
Score = 30.0 bits (66), Expect = 3.6
Identities = 18/37 (48%), Positives = 22/37 (58%), Gaps = 4/37 (10%)
Query: 8 LIFFILSSLIPHFHI----LIVSSSSSSPSPSPLPSL 40
L+FF L+SL+ + SSSS SPSPSP P L
Sbjct: 51 LLFFTLASLLSFLAVNRSSTESSSSSESPSPSPPPPL 87
>At3g02150 unknown protein
Length = 355
Score = 30.0 bits (66), Expect = 3.6
Identities = 19/60 (31%), Positives = 31/60 (51%), Gaps = 1/60 (1%)
Query: 13 LSSLIPHFHILIVSSSSSSPSPSPLPSLL-SSPSSNQGEWELIQPTIGISAMHMQLSHNN 71
L SL+P ++V ++SP+ + + SL SS S+ G E + P +S M L N+
Sbjct: 231 LHSLVPFPSQILVCPMTTSPTTTTIQSLFPSSSSAGSGTMETLDPRQMVSHFQMPLMGNS 290
>At5g27820 putative protein
Length = 114
Score = 29.6 bits (65), Expect = 4.7
Identities = 20/52 (38%), Positives = 29/52 (55%), Gaps = 5/52 (9%)
Query: 451 AFSPPYLSLEFDLVRPTI--WHVTNKILGYRVFYYVTFTVAKFASASEVSVR 500
A PP L FD ++P + H TNK + +V + T TVA AS+ E ++R
Sbjct: 6 AVRPPRL---FDYLKPYVLKMHFTNKFVHAQVIHSPTATVACSASSQEKALR 54
>At4g29880 putative protein
Length = 404
Score = 29.6 bits (65), Expect = 4.7
Identities = 28/107 (26%), Positives = 48/107 (44%), Gaps = 11/107 (10%)
Query: 19 HFHILIVSSSSSSPSPSPLPSLLS--SPSSNQGEWELIQPTIGISAMHMQLSHNNKIIIF 76
H +++ S+SP SPL LS SPS N E E ++ + +S M +Q N + +
Sbjct: 18 HMMTMMMMDLSTSPPSSPLSPSLSPKSPSYNNNEEERLE-VVNLSGMALQSLPNPSLNLA 76
Query: 77 DRTDFGPSNLPLSNGRCRMDPFDTALKIDCTAHSVLYDIATNTFRSL 123
+ L LSN + P ++ + + DI +N ++L
Sbjct: 77 N-----ICKLDLSNNHIKKIPESLTARL---LNLIALDIHSNQIKAL 115
>At5g26780 glycine hydroxymethyltransferase - like protein
Length = 533
Score = 29.3 bits (64), Expect = 6.2
Identities = 12/36 (33%), Positives = 22/36 (60%), Gaps = 1/36 (2%)
Query: 256 RSILFDYKQNVVVKEFPEIPGGDPHNYPSSGSSVLL 291
+ +++DY+ + FP + GG PHN+ +G +V L
Sbjct: 310 KEVMYDYEDRINQAVFPGLQGG-PHNHTITGLAVAL 344
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.136 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,819,421
Number of Sequences: 26719
Number of extensions: 645140
Number of successful extensions: 2584
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 14
Number of HSP's successfully gapped in prelim test: 14
Number of HSP's that attempted gapping in prelim test: 2484
Number of HSP's gapped (non-prelim): 42
length of query: 570
length of database: 11,318,596
effective HSP length: 105
effective length of query: 465
effective length of database: 8,513,101
effective search space: 3958591965
effective search space used: 3958591965
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 63 (28.9 bits)
Medicago: description of AC148345.6


