

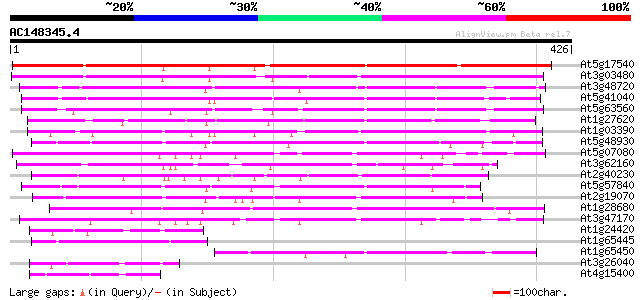
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148345.4 - phase: 0
(426 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g17540 unknown protein 352 2e-97
At3g03480 putative hypersensitivity-related gene 320 7e-88
At3g48720 unknown protein 173 2e-43
At5g41040 N-hydroxycinnamoyl/benzoyltransferase-like protein 167 1e-41
At5g63560 acyltransferase-like protein 166 2e-41
At1g27620 putative hypersensitivity-related protein 141 6e-34
At1g03390 hypothetical protein 127 9e-30
At5g48930 anthranilate N-benzoyltransferase 124 1e-28
At5g07080 unknown protein 112 3e-25
At3g62160 unknown protein 112 4e-25
At2g40230 putative anthranilate N-hydroxycinnamoyl/benzoyltransf... 108 4e-24
At5g57840 N-hydroxycinnamoyl/benzoyltransferase 105 5e-23
At2g19070 putative anthranilate N-hydroxycinnamoyl/benzoyltransf... 105 6e-23
At1g28680 anthranilate N-hydroxycinnamoyl/benzoyltransferase lik... 103 2e-22
At3g47170 hypersensitivity-related protein-like protein 92 5e-19
At1g24420 unknown protein 62 5e-10
At1g65445 unknown protein 60 3e-09
At1g65450 unknown protein 59 4e-09
At3g26040 hypothetical protein 59 5e-09
At4g15400 HSR201 like protein 57 2e-08
>At5g17540 unknown protein
Length = 461
Score = 352 bits (904), Expect = 2e-97
Identities = 199/449 (44%), Positives = 280/449 (62%), Gaps = 47/449 (10%)
Query: 3 SSPLLFTVRRCQPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEKDPA 62
S L F + R +PELV PA PTPRE+K LSDIDDQE LRF++P IF YRH P+ DP
Sbjct: 2 SGSLTFKIYRQKPELVSPAKPTPRELKPLSDIDDQEGLRFHIPTIFFYRHNPTT-NSDPV 60
Query: 63 KVLKRALSQTLVYYYPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDEFG--DALHP 120
V++RAL++TLVYYYP AGR+REG KL VDCTGEGV+FIEA+ DVTL EF DAL P
Sbjct: 61 AVIRRALAETLVYYYPFAGRLREGPNRKLAVDCTGEGVLFIEADADVTLVEFEEKDALKP 120
Query: 121 PFPCFQELIYDVPGTNQIIDHPILLIQMAR------------------------------ 150
PFPCF+EL+++V G+ ++++ P++L+Q+ R
Sbjct: 121 PFPCFEELLFNVEGSCEMLNTPLMLMQVTRLKCGGFIFAVRINHAMSDAGGLTLFLKTMC 180
Query: 151 ----GAHQPSIQPVWRREILMARDPPRVTCNHHEYEQI--LSSNINKEEDVAATIVHHSF 204
G H P++ PVW R +L AR RVT H EY+++ + + + D +V S
Sbjct: 181 EFVRGYHAPTVAPVWERHLLSARVLLRVTHAHREYDEMPAIGTELGSRRD---NLVGRSL 237
Query: 205 FFTLTRIAEIRRLIPFHLRQCST-FDLITACFWCCRTKALQLEPDEEVRMMCIVNARSRF 263
FF ++ IRRL+P +L ST +++T+ W RT AL+ + D+E+R++ IVNARS+
Sbjct: 238 FFGPCEMSAIRRLLPPNLVNSSTNMEMLTSFLWRYRTIALRPDQDKEMRLILIVNARSKL 297
Query: 264 RADHSPLVGYYGNCFAFPAAVTTAGKLCGNPLGYGVELIRKAKAQVTEEYMHSLADLMVI 323
+ P GYYGN FAFP A+ TA +L PL + + LI++AK+ VTEEYM SLADLMVI
Sbjct: 298 KNPPLPR-GYYGNAFAFPVAIATANELTKKPLEFALRLIKEAKSSVTEEYMRSLADLMVI 356
Query: 324 KERCLFTTIRSCVVSDLTRAKYAEVNFG-WGEAVYGGVVNCVAGPLPRATFIIPHKNAEG 382
K R F++ + +VSD+ +A+++FG WG+ VYGG+ LP A+F + + G
Sbjct: 357 KGRPSFSSDGAYLVSDV--RIFADIDFGIWGKPVYGGIGTAGVEDLPGASFYVSFEKRNG 414
Query: 383 EEGLILLIFLTSEVMKRFAEELDKMFGNQ 411
E G+++ + L + M+RF EEL+ +F Q
Sbjct: 415 EIGIVVPVCLPEKAMQRFVEELEGVFNGQ 443
>At3g03480 putative hypersensitivity-related gene
Length = 454
Score = 320 bits (821), Expect = 7e-88
Identities = 186/446 (41%), Positives = 258/446 (57%), Gaps = 52/446 (11%)
Query: 2 TSSPLLFTVRRCQPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEKDP 61
T++ L F V R Q ELV PA PTPRE+K LSDIDDQ+ LRF +P+IF YR S + DP
Sbjct: 11 TTTGLSFKVHRQQRELVTPAKPTPRELKPLSDIDDQQGLRFQIPVIFFYRPNLSS-DLDP 69
Query: 62 AKVLKRALSQTLVYYYPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDEF--GDALH 119
+V+K+AL+ LVYYYP AGR+RE + KL VDCTGEGV+FIEAE DV L E DAL
Sbjct: 70 VQVIKKALADALVYYYPFAGRLRELSNRKLAVDCTGEGVLFIEAEADVALAELEEADALL 129
Query: 120 PPFPCFQELIYDVPGTNQIIDHPILLIQMAR----------------------------- 150
PPFP +EL++DV G++ +++ P+LL+Q+ R
Sbjct: 130 PPFPFLEELLFDVEGSSDVLNTPLLLVQVTRLKCCGFIFALRFNHTMTDGAGLSLFLKSL 189
Query: 151 -----GAHQPSIQPVWRREIL-MARDPPRVTCNHHEYEQILSSNINKEEDVAAT---IVH 201
G H PS+ PVW R +L ++ RVT H EY+ + DV AT +V
Sbjct: 190 CELACGLHAPSVPPVWNRHLLTVSASEARVTHTHREYDDQVGI------DVVATGHPLVS 243
Query: 202 HSFFFTLTRIAEIRRLIPFHLRQCSTFDLITACFWCCRTKALQLEPDEEVRMMCIVNARS 261
SFFF I+ IR+L+P L S F+ +++ W CRT AL +P+ E+R+ CI+N+RS
Sbjct: 244 RSFFFRAEEISAIRKLLPPDLHNTS-FEALSSFLWRCRTIALNPDPNTEMRLTCIINSRS 302
Query: 262 RFRADHSPLV-GYYGNCFAFPAAVTTAGKLCGNPLGYGVELIRKAKAQVTEEYMHSLADL 320
+ R + PL GYYGN F PAA+ TA L PL + + LI++ K+ VTE+Y+ S+ L
Sbjct: 303 KLR--NPPLEPGYYGNVFVIPAAIATARDLIEKPLEFALRLIQETKSSVTEDYVRSVTAL 360
Query: 321 MVIKERCLFTTIRSCVVSDLTRAKYAEVNFG-WGEAVYGGVVNCVAGPLPRATFIIPHKN 379
M + R +F + ++SDL +++FG WG+ VYGG P +F +P KN
Sbjct: 361 MATRGRPMFVASGNYIISDLRHFDLGKIDFGPWGKPVYGGTAKAGIALFPGVSFYVPFKN 420
Query: 380 AEGEEGLILLIFLTSEVMKRFAEELD 405
+GE G ++ I L M+ F EL+
Sbjct: 421 KKGETGTVVAISLPVRAMETFVAELN 446
>At3g48720 unknown protein
Length = 430
Score = 173 bits (438), Expect = 2e-43
Identities = 121/437 (27%), Positives = 218/437 (49%), Gaps = 50/437 (11%)
Query: 8 FTVRRCQPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEKDPAKVLKR 67
F V R PEL+PP + TP LS++D + + ++ Y+ E S ++ V+K+
Sbjct: 7 FIVTRKNPELIPPVSETPNGHYYLSNLDQN--IAIIVKTLYYYKSE-SRTNQESYNVIKK 63
Query: 68 ALSQTLVYYYPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDEFGDALHPP-FPCFQ 126
+LS+ LV+YYP+AGR+ K+ V+CTGEGV+ +EAE + +D +A+ +
Sbjct: 64 SLSEVLVHYYPVAGRLTISPEGKIAVNCTGEGVVVVEAEANCGIDTIKEAISENRMETLE 123
Query: 127 ELIYDVPGTNQIIDHPILLIQ----------------------------------MARGA 152
+L+YDVPG I++ P +++Q MA+G
Sbjct: 124 KLVYDVPGARNILEIPPVVVQVTNFKCGGFVLGLGMSHNMFDGVAAAEFLNSWCEMAKGL 183
Query: 153 HQPSIQPVWRREILMARDPPRVTCNHHEYEQILSSNINKEEDVAATIVHHSFFFTLTRIA 212
S+ P R IL +R+PP++ H+E+++I + + +++ SF F ++
Sbjct: 184 PL-SVPPFLDRTILRSRNPPKIEFPHNEFDEIEDISDTGKIYDEEKLIYKSFLFEPEKLE 242
Query: 213 EIRRLI--PFHLRQCSTFDLITACFWCCRTKALQLEPDEEVRMMCIVNARSRFRADHSPL 270
+++ + + + STF +T W R +AL+ +PD+ V+++ + RSRF P
Sbjct: 243 KLKIMAIEENNNNKVSTFQALTGFLWKSRCEALRFKPDQRVKLLFAADGRSRF-IPRLPQ 301
Query: 271 VGYYGNCFAFPAAVTTAGKLCGNPLGYGVELIRKAKAQVTEEYMHSLADLMVIKERCLFT 330
GY GN VT++G+L GNPL + V L+++ VT+ +M S D + R +
Sbjct: 302 -GYCGNGIVLTGLVTSSGELVGNPLSHSVGLVKRLVELVTDGFMRSAMDYFEV-NRTRPS 359
Query: 331 TIRSCVVSDLTRAKYAEVNFGWGEAVYGGVVNCVAGPLPRATFIIPHKNAEGEEGLILLI 390
+ +++ ++ +++FGWGE V+ G V LP I+ + + + + + +
Sbjct: 360 MNATLLITSWSKLTLHKLDFGWGEPVFSGPVG-----LPGREVILFLPSGDDMKSINVFL 414
Query: 391 FLTSEVMKRFAEELDKM 407
L + M+ F EEL K+
Sbjct: 415 GLPTSAMEVF-EELMKI 430
>At5g41040 N-hydroxycinnamoyl/benzoyltransferase-like protein
Length = 457
Score = 167 bits (423), Expect = 1e-41
Identities = 125/433 (28%), Positives = 208/433 (47%), Gaps = 51/433 (11%)
Query: 10 VRRCQPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEKDPAKVLKRAL 69
V + +P LV P + T + + LS++D + + I+ ++ E E + +V+K+AL
Sbjct: 33 VHQKEPALVKPESETRKGLYFLSNLDQN--IAVIVRTIYCFKSEERGNE-EAVQVIKKAL 89
Query: 70 SQTLVYYYPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDEFGDALHPPFPCFQELI 129
SQ LV+YYPLAGR+ KL VDCT EGV+F+EAE + +DE GD P +L+
Sbjct: 90 SQVLVHYYPLAGRLTISPEGKLTVDCTEEGVVFVEAEANCKMDEIGDITKPDPETLGKLV 149
Query: 130 YDVPGTNQIIDHPILLIQMAR--------------------GAHQ-------------PS 156
YDV I++ P + Q+ + GA + +
Sbjct: 150 YDVVDAKNILEIPPVTAQVTKFKCGGFVLGLCMNHCMFDGIGAMEFVNSWGQVARGLPLT 209
Query: 157 IQPVWRREILMARDPPRVTCNHHEYEQILS-SNINKEEDVAATIVHHSFFFTLTRIAEIR 215
P R IL AR+PP++ H E+E+I SNIN T+ + SF F +I +++
Sbjct: 210 TPPFSDRTILNARNPPKIENLHQEFEEIEDKSNINSLYTKEPTL-YRSFCFDPEKIKKLK 268
Query: 216 RLIPFHLRQ-----CSTFDLITACFWCCRTKALQLEPDEEVRMMCIVNARSRFRADHSPL 270
+ C++F+ ++A W RTK+L++ D++ +++ V+ R++F
Sbjct: 269 LQATENSESLLGNSCTSFEALSAFVWRARTKSLKMLSDQKTKLLFAVDGRAKFEPQLPK- 327
Query: 271 VGYYGNCFAFPAAVTTAGKLCGNPLGYGVELIRKAKAQVTEEYMHSLADLMVIKERCLFT 330
GY+GN ++ AG+L PL + V L+R+A VT+ YM S D + R +
Sbjct: 328 -GYFGNGIVLTNSICEAGELIEKPLSFAVGLVREAIKMVTDGYMRSAIDYFEV-TRARPS 385
Query: 331 TIRSCVVSDLTRAKYAEVNFGWGEAVYGGVVNCVAGPLPRATFIIPHKNAEGEEGLILLI 390
+ +++ +R + +FGWGE + G VA P T + H E + +L+
Sbjct: 386 LSSTLLITTWSRLGFHTTDFGWGEPILSGP---VALPEKEVTLFLSH--GEQRRSINVLL 440
Query: 391 FLTSEVMKRFAEE 403
L + M F E+
Sbjct: 441 GLPATAMDVFQEQ 453
>At5g63560 acyltransferase-like protein
Length = 426
Score = 166 bits (420), Expect = 2e-41
Identities = 124/443 (27%), Positives = 208/443 (45%), Gaps = 73/443 (16%)
Query: 10 VRRCQPELVPPAAPTPREVKLLSDIDDQECLRFNMPII---FIYRHEPSMIEKDPAKVLK 66
V R +P LV PA+ TP+ + LS++D N+ II F Y S ++ +V+K
Sbjct: 9 VTRKEPVLVSPASETPKGLHYLSNLDQ------NIAIIVKTFYYFKSNSRSNEESYEVIK 62
Query: 67 RALSQTLVYYYPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDEFGDALHP--PFPC 124
++LS+ LV+YYP AGR+ K+ VDCTGEGV+ +EAE + +++ A+
Sbjct: 63 KSLSEVLVHYYPAAGRLTISPEGKIAVDCTGEGVVVVEAEANCGIEKIKKAISEIDQPET 122
Query: 125 FQELIYDVPGTNQIIDHPILLIQM----------------------------------AR 150
++L+YDVPG I++ P +++Q+ AR
Sbjct: 123 LEKLVYDVPGARNILEIPPVVVQVTNFKCGGFVLGLGMNHNMFDGIAAMEFLNSWAETAR 182
Query: 151 GAHQPSIQPVWRREILMARDPPRVTCNHHEYEQILSSNINKEEDVAAT--------IVHH 202
G S+ P R +L R PP++ H+E+E + ED++ T +V+
Sbjct: 183 GLPL-SVPPFLDRTLLRPRTPPKIEFPHNEFEDL--------EDISGTGKLYSDEKLVYK 233
Query: 203 SFFFTLTRIAEIRRLIPFHLRQCSTFDLITACFWCCRTKALQLEPDEEVRMMCIVNARSR 262
SF F ++ ++ + + +TF +T W R +AL L+PD+ ++++ + RSR
Sbjct: 234 SFLFGPEKLERLKIMAE---TRSTTFQTLTGFLWRARCQALGLKPDQRIKLLFAADGRSR 290
Query: 263 FRADHSPLVGYYGNCFAFPAAVTTAGKLCGNPLGYGVELIRKAKAQVTEEYMHSLADLMV 322
F + GY GN F VTTAG++ NPL + V L+++A V + +M S D
Sbjct: 291 FVPELPK--GYSGNGIVFTYCVTTAGEVTLNPLSHSVCLVKRAVEMVNDGFMRSAIDYFE 348
Query: 323 IKERCLFTTIRSCVVSDLTRAKYAEVNFGWGEAVYGGVVNCVAGPLPRATFIIPHKNAEG 382
+ R + + +++ + + +FGWGE V G V LP I+
Sbjct: 349 V-TRARPSLTATLLITSWAKLSFHTKDFGWGEPVVSGPVG-----LPEKEVILFLPCGSD 402
Query: 383 EEGLILLIFLTSEVMKRFAEELD 405
+ + +L+ L MK F +D
Sbjct: 403 TKSINVLLGLPGSAMKVFQGIMD 425
>At1g27620 putative hypersensitivity-related protein
Length = 442
Score = 141 bits (356), Expect = 6e-34
Identities = 116/430 (26%), Positives = 201/430 (45%), Gaps = 66/430 (15%)
Query: 14 QPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEKDPAKVLKRALSQTL 73
QP L+ P +PTP LS++DD LRF++ +++++ S + LK +LS+ L
Sbjct: 13 QPTLITPLSPTPNHSLYLSNLDDHHFLRFSIKYLYLFQKSISPL------TLKDSLSRVL 66
Query: 74 VYYYPLAGRIR---EGARDKLMVDCTGEGVMFIEAETDVTLDEFGDALHPPFPCFQELIY 130
V YYP AGRIR EG+ KL VDC GEG +F EA D+T +F P +++L++
Sbjct: 67 VDYYPFAGRIRVSDEGS--KLEVDCNGEGAVFAEAFMDITCQDFVQLSPKPNKSWRKLLF 124
Query: 131 DVPGTNQIIDHPILLIQM-----------------------------------ARGAHQP 155
V +D P L+IQ+ AH P
Sbjct: 125 KVQ-AQSFLDIPPLVIQVTYLRCGGMILCTAINHCLCDGIGTSQFLHAWAHATTSQAHLP 183
Query: 156 SIQPVWRREILMARDPPRVTCNHHEYEQILSSNINKEEDV-----AATIVHHSFFFTLTR 210
+ +P R +L R+PPRVT +H + + + + + D+ + + + F +
Sbjct: 184 T-RPFHSRHVLDPRNPPRVTHSHPGFTRTTTVDKSSTFDISKYLQSQPLAPATLTFNQSH 242
Query: 211 IAEIRRLIPFHLRQCSTFDLITACFWCCRTKALQLEPDEEVRMMCIVNARSRFRADHSPL 270
+ +++ L+ C+TF+ + A W ++L L V+++ VN R R +
Sbjct: 243 LLRLKKTCAPSLK-CTTFEALAANTWRSWAQSLDLPMTMLVKLLFSVNMRKRLTPELPQ- 300
Query: 271 VGYYGNCFAFPAAVTTAGKLCGNPLGYGVELIRKAKAQVTEEYMHSLADLMVIKERCLFT 330
GYYGN F A + L + + V+ I++AK+++T+EY+ S DL ++++ + T
Sbjct: 301 -GYYGNGFVLACAESKVQDLVNGNIYHAVKSIQEAKSRITDEYVRSTIDL--LEDKTVKT 357
Query: 331 TIR-SCVVSDLTRAKYAEVNFGWGEAVYGGVVNCVAGPLPRATFIIPHKNAEGEEGLILL 389
+ S V+S + E++ G G+ +Y GPL + + A + + +
Sbjct: 358 DVSCSLVISQWAKLGLEELDLGGGKPMY-------MGPLTSDIYCLFLPVASDNDAIRVQ 410
Query: 390 IFLTSEVMKR 399
+ L EV+KR
Sbjct: 411 MSLPEEVVKR 420
>At1g03390 hypothetical protein
Length = 461
Score = 127 bits (320), Expect = 9e-30
Identities = 124/448 (27%), Positives = 198/448 (43%), Gaps = 70/448 (15%)
Query: 14 QPELVPPAAPTPREVK-----LLSDIDDQECLRFNMPIIFIYRHEPSMIEKDP--AKVLK 66
Q LV P++PTP LS++DD R P ++ Y PS ++ K L+
Sbjct: 21 QQFLVHPSSPTPANQSPHHSLYLSNLDDIIGARVFTPSVYFY---PSTNNRESFVLKRLQ 77
Query: 67 RALSQTLVYYYPLAGRIREGARDKLMVDCTGE-GVMFIEAETDVTLDEFGDALHPPFPCF 125
ALS+ LV YYPL+GR+RE KL V E GV+ + A + + L + GD L P P +
Sbjct: 78 DALSEVLVPYYPLSGRLREVENGKLEVFFGEEQGVLMVSANSSMDLADLGD-LTVPNPAW 136
Query: 126 QELIYDVPGTN--QIIDHPILLIQMAR--------------------GAHQ--------- 154
LI+ PG +I++ P+L+ Q+ GA Q
Sbjct: 137 LPLIFRNPGEEAYKILEMPLLIAQVTFFTCGGFSLGIRLCHCICDGFGAMQFLGSWAATA 196
Query: 155 ------PSIQPVWRREILMARDPPRVTCNHHEYEQI-----LSSNINKEEDVAATI-VHH 202
+PVW RE R+PP V HHEY I L++++ + + +
Sbjct: 197 KTGKLIADPEPVWDRETFKPRNPPMVKYPHHEYLPIEERSNLTNSLWDTKPLQKCYRISK 256
Query: 203 SFFFTLTRIA--EIRRLIPFHLRQCSTFDLITACFWCCRTKALQLEP-DEEVRMMCIVNA 259
F + IA E L+ CSTFD + A W KAL ++P D +R+ VN
Sbjct: 257 EFQCRVKSIAQGEDPTLV------CSTFDAMAAHIWRSWVKALDVKPLDYNLRLTFSVNV 310
Query: 260 RSRFRADHSPLVGYYGNCFAFPAAVTTAGKLCGNPLGYGVELIRKAKAQVTEEYMHSLAD 319
R+R G+YGN A+++ L + L L++ A+ +V+E+Y+ S+ D
Sbjct: 311 RTRLET-LKLRKGFYGNVVCLACAMSSVESLINDSLSKTTRLVQDARLRVSEDYLRSMVD 369
Query: 320 LMVIKERCLFTTIRSCVVSDLTRAK-YAEVNFGWGEAVYGGVVNCVAGPLPRATFIIPHK 378
+ +K ++ TR + Y +FGWG+ VY G ++ P P+ ++P
Sbjct: 370 YVDVKRPKRLEFGGKLTITQWTRFEMYETADFGWGKPVYAGPID--LRPTPQVCVLLPQG 427
Query: 379 NAE--GEEGLILLIFLTSEVMKRFAEEL 404
E ++ +++ + L + F L
Sbjct: 428 GVESGNDQSMVVCLCLPPTAVHTFTRLL 455
>At5g48930 anthranilate N-benzoyltransferase
Length = 433
Score = 124 bits (310), Expect = 1e-28
Identities = 115/437 (26%), Positives = 187/437 (42%), Gaps = 65/437 (14%)
Query: 17 LVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEKDPAKVLKRALSQTLVYY 76
+V PA TP S++D RF+ P ++ YR + DP +V+K ALS+ LV +
Sbjct: 10 MVRPATETPITNLWNSNVD-LVIPRFHTPSVYFYRPTGASNFFDP-QVMKEALSKALVPF 67
Query: 77 YPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDEFGDALHPPFPCFQELIYDVPGTN 136
YP+AGR++ ++ +DC G GV+F+ A+T +D+FGD P ++LI +V +
Sbjct: 68 YPMAGRLKRDDDGRIEIDCNGAGVLFVVADTPSVIDDFGD--FAPTLNLRQLIPEVDHSA 125
Query: 137 QIIDHPILLIQ----------------------------------MARGAHQPSIQPVWR 162
I P+L++Q MARG +I P
Sbjct: 126 GIHSFPLLVLQVTFFKCGGASLGVGMQHHAADGFSGLHFINTWSDMARGL-DLTIPPFID 184
Query: 163 REILMARDPPRVTCNHHEYEQILSSNINKEEDVAATIVHHSFFFTLTR----IAEIRRLI 218
R +L ARDPP+ +H EY+ S I + + F LTR + +
Sbjct: 185 RTLLRARDPPQPAFHHVEYQPAPSMKIPLDPSKSGPENTTVSIFKLTRDQLVALKAKSKE 244
Query: 219 PFHLRQCSTFDLITACFWCCRTKALQLEPDEEVRMMCIVNARSRFRADHSPLVGYYGNCF 278
+ S+++++ W KA L D+E ++ + RSR R P GY+GN
Sbjct: 245 DGNTVSYSSYEMLAGHVWRSVGKARGLPNDQETKLYIATDGRSRLRPQLPP--GYFGNVI 302
Query: 279 AFPAAVTTAGKLCGNPLGYGVELIRKAKAQVTEEYMHSLADLMVIKERCLFTTIR----- 333
+ AG L P Y I ++ + Y+ S D + ++ L +R
Sbjct: 303 FTATPLAVAGDLLSKPTWYAAGQIHDFLVRMDDNYLRSALDYLEMQPD-LSALVRGAHTY 361
Query: 334 ---SCVVSDLTRAKYAEVNFGWGEAVY---GGVVNCVAGPLPRATFIIPHKNAEGEEGLI 387
+ ++ R + +FGWG ++ GG+ P +F++P +G L
Sbjct: 362 KCPNLGITSWVRLPIYDADFGWGRPIFMGPGGI------PYEGLSFVLPSPTNDG--SLS 413
Query: 388 LLIFLTSEVMKRFAEEL 404
+ I L SE MK F + L
Sbjct: 414 VAIALQSEHMKLFEKFL 430
>At5g07080 unknown protein
Length = 450
Score = 112 bits (281), Expect = 3e-25
Identities = 121/465 (26%), Positives = 190/465 (40%), Gaps = 83/465 (17%)
Query: 3 SSPLLFTVRRCQPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRH-EPSMIEKDP 61
SSP V++ Q +V P+ TP LS +D+ L I++Y P+ DP
Sbjct: 7 SSPSPLVVKKSQVVIVKPSKATPDVSLSLSTLDNDPYLETLAKTIYVYAPPSPNDHVHDP 66
Query: 62 AKVLKRALSQTLVYYYPLAGRIREGARD-KLMVDCT-GEGVMFIEAETDVTLD------E 113
A + ++ALS LVYYYPLAG++ G+ D +L + C+ EGV F+ A D TL +
Sbjct: 67 ASLFQQALSDALVYYYPLAGKLHRGSHDHRLELRCSPAEGVPFVRATADCTLSSLNYLKD 126
Query: 114 FGDALHPPFPC-------------FQELIYDVPGTN--QIIDHPI-----------LLIQ 147
L+ PC Q ++ G + H + L +
Sbjct: 127 MDTDLYQLVPCDVAAPSGDYNPLALQITLFACGGLTLATALSHSLCDGFGASQFFKTLTE 186
Query: 148 MARGAHQPSIQPVWRREILMARD-----------PPRVTCNHHEYEQILSSNINKEEDVA 196
+A G QPSI PVW R L + + P++ +S D+
Sbjct: 187 LAAGKTQPSIIPVWDRHRLTSNNFSLNDQVEEGQAPKLVDFGEACSSAATSPYTPSNDMV 246
Query: 197 ATIVHHSFFFTLTRIAEIRRLIPFHLRQCSTFDLITACFWCCRTKALQLEPDEEVRMMCI 256
I++ T I +++ + +T +++ A W R +AL+L PD
Sbjct: 247 CEILN----VTSEDITQLKEKV---AGVVTTLEILAAHVWRARCRALKLSPDGTSLFGMA 299
Query: 257 VNARSRFRADHSPL-VGYYGNCFAFPAAVTTAGKLCGNPLGYGVELIRKAKAQVTE---- 311
V R R PL GYYGN F AG+L +PL + V+LI++AK E
Sbjct: 300 VGIR---RIVEPPLPEGYYGNAFVKANVAMKAGELSNSPLSHVVQLIKEAKKAAQEKRYV 356
Query: 312 -EYMHSLADLMVIKERC--------LFTTIRSCVVSDLTRAKYAEVNFGWGEAVYGGVVN 362
E + + + C L T R + D E++FG+G +V ++
Sbjct: 357 LEQLRETEKTLKMNVACEGGKGAFMLLTDWRQLGLLD-------EIDFGYGGSV--NIIP 407
Query: 363 CVAGPLPRATFIIPHKNAEGEEGLILLIFLTSEVMKRFAEELDKM 407
V LP +P K + G+ +L+ L VM F E ++ +
Sbjct: 408 LVPKYLPDICIFLPRK----QGGVRVLVTLPKSVMVNFKEHINPL 448
>At3g62160 unknown protein
Length = 428
Score = 112 bits (280), Expect = 4e-25
Identities = 114/426 (26%), Positives = 180/426 (41%), Gaps = 91/426 (21%)
Query: 6 LLFTVRRCQPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEKDPAKVL 65
+ F++ R L+ TP V LS ID+ LR I ++ H P D A+ +
Sbjct: 1 MAFSLTRVNRVLIQACETTPSVVLDLSLIDNIPVLRCFARTIHVFTHGP-----DAARAI 55
Query: 66 KRALSQTLVYYYPLAGRIREGARDKLMVDCTGE-GVMFIEAETDVTLDEFG--------- 115
+ AL++ LV YYPL+GR++E + KL +DCTG+ G+ F++A TDV L+ G
Sbjct: 56 QEALAKALVSYYPLSGRLKELNQGKLQIDCTGKTGIWFVDAVTDVKLESVGYFDNLMEIP 115
Query: 116 -DALHP------------------PFPC--------FQELIYDVPGTNQIIDHPILLIQM 148
D L P F C F I D G Q + + ++
Sbjct: 116 YDDLLPDQIPKDDDAEPLVQMQVTQFSCGGFVMGLRFCHAICDGLGAAQFL---TAVGEI 172
Query: 149 ARGAHQPSIQPVWRREILMARDPPRVTCNHHEYEQILSSNINKEEDVA---ATIVHHSFF 205
A G + PVW R+ + H ++S I A + H SF
Sbjct: 173 ACGQTNLGVSPVWHRDFIP---------QQHRANDVISPAIPPPFPPAFPEYKLEHLSFD 223
Query: 206 FTLTRIAEIRRLIPFHLRQ-CSTFDLITACFWCCRTKALQLEPDEEVRMMCIVNARSRFR 264
I +R + CS F++I ACFW RT+A+ L D EV+++ N R
Sbjct: 224 IPSDLIERFKREFRASTGEICSAFEVIAACFWKLRTEAINLREDTEVKLVFFANCR---H 280
Query: 265 ADHSPL-VGYYGNCFAFPAAVTTAGKLCGNPLGY--GVELIRKAKAQVTEEYMHSLAD-- 319
+ PL +G+YGNCF FP ++ V+LI++AK ++ EE+ ++
Sbjct: 281 LVNPPLPIGFYGNCF-FPVKISAESHEISKEKSVFDVVKLIKQAKTKLPEEFEKFVSGDG 339
Query: 320 ----LMVIKERCLFTTIRSCVVSDLTRAKYAEVNFGWGEAVY-----------GGVVNCV 364
+ LF +S+ + + +V++GWG ++ G+V V
Sbjct: 340 DDPFAPAVGYNTLF-------LSEWGKLGFNQVDYGWGPPLHVAPIQGLSIVPAGIVGSV 392
Query: 365 AGPLPR 370
PLP+
Sbjct: 393 --PLPK 396
>At2g40230 putative anthranilate
N-hydroxycinnamoyl/benzoyltransferase
Length = 433
Score = 108 bits (271), Expect = 4e-24
Identities = 106/391 (27%), Positives = 185/391 (47%), Gaps = 52/391 (13%)
Query: 17 LVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEKDPAKVLKRALSQTLVYY 76
++ P+ TP V LS +D Q LRF + + +Y S E + LK ALS+ LV Y
Sbjct: 12 VITPSDQTPSSVLSLSALDSQLFLRFTIEYLLVYP-PVSDPEYSLSGRLKSALSRALVPY 70
Query: 77 YPLAGRIRE--GARDKLMVDCTGEGVMFIEAETDV-TLDEFGD------------ALH-- 119
+P +GR+RE L V+C G+G +F+EA +D+ T +F +LH
Sbjct: 71 FPFSGRVREKPDGGGGLEVNCRGQGALFLEAVSDILTCLDFQKPPRHVTSWRKLLSLHVI 130
Query: 120 ------PPFPCFQELIYD-----VPGTNQIIDHPI-------LLIQMARGAHQPSI---Q 158
PP + D G N + I L ++++ + + +
Sbjct: 131 DVLAGAPPLVVQLTWLRDGGAALAVGVNHCVSDGIGSAEFLTLFAELSKDSLSQTELKRK 190
Query: 159 PVWRREILMARDPPRVTCNHHEYEQI--LSSNINKEEDVAATIVHHSFFFTLTRIAEIRR 216
+W R++LM P R + +H E+ ++ L +N+ A +V S F ++ E+++
Sbjct: 191 HLWDRQLLMP-SPIRDSLSHPEFNRVPDLCGFVNRFN--AERLVPTSVVFERQKLNELKK 247
Query: 217 LIP----FHLRQCSTFDLITACFWCCRTKALQLEPDEEVRMMCIVNARSRFRADHSPLVG 272
L F+ + S F++++A W ++L L ++ ++++ VN R R + S G
Sbjct: 248 LASRLGEFNSKHTS-FEVLSAHVWRSWARSLNLPSNQVLKLLFSVNIRDRVKP--SLPSG 304
Query: 273 YYGNCFAFPAAVTTAGKLCGNPLGYGVELIRKAKAQVTEEYMHSLADLMVIKERCLFTTI 332
+YGN F A TT L L Y L+++AK +V +EY+ S+ + V KER ++
Sbjct: 305 FYGNAFVVGCAQTTVKDLTEKGLSYATMLVKQAKERVGDEYVRSVVE-AVSKERASPDSV 363
Query: 333 RSCVVSDLTRAKYAEVNFGWGEAVYGGVVNC 363
++S +R +++FG G+ V+ G V C
Sbjct: 364 GVLILSQWSRLGLEKLDFGLGKPVHVGSVCC 394
>At5g57840 N-hydroxycinnamoyl/benzoyltransferase
Length = 443
Score = 105 bits (262), Expect = 5e-23
Identities = 102/403 (25%), Positives = 172/403 (42%), Gaps = 67/403 (16%)
Query: 10 VRRCQPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEKDPAKVLKRAL 69
+R Q +V PA TP LS++D + +R +M ++ Y+ S ++ + L AL
Sbjct: 3 IRVKQATIVKPAEETPTHNLWLSNLDLIQ-VRLHMGTLYFYK-PCSSSDRPNTQSLIEAL 60
Query: 70 SQTLVYYYPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDEFGDALHPPFPCFQELI 129
S+ LV++YP AGR+++ +L V C GEGV+F+EAE+D T+ + G L +L+
Sbjct: 61 SKVLVFFYPAAGRLQKNTNGRLEVQCNGEGVLFVEAESDSTVQDIG--LLTQSLDLSQLV 118
Query: 130 YDVPGTNQIIDHPILLIQM----------------------------------ARGAHQP 155
V I +P+LL Q+ ARG
Sbjct: 119 PTVDYAGDISSYPLLLFQVTYFKCGTICVGSSIHHTFGEATSLGYIMEAWSLTARGL-LV 177
Query: 156 SIQPVWRREILMARDPPRVTCNHHEYE---------QILSSNINKEEDVAATIVHHSFFF 206
+ P R +L AR+PP H EY+ + L+ N E D A +
Sbjct: 178 KLTPFLDRTVLHARNPPSSVFPHTEYQSPPFHNHPMKSLAYRSNPESDSAIATLK----L 233
Query: 207 TLTRIAEIRRLIPFHLRQCSTFDLITACFWCCRTKALQ-LEPDEEVRMMCIVNARSRFRA 265
T ++ ++ + ST++++ A W C + A + L + R+ I++ R R +
Sbjct: 234 TRLQLKALKARAEIADSKFSTYEVLVAHTWRCASFANEDLSEEHSTRLHIIIDGRPRLQP 293
Query: 266 DHSPLVGYYGNCFAFPAAVTTAGKLCGNPLGYGVELIRKAKAQVTEEYMHSLAD------ 319
GY GN V+ G VE + + ++ EY+ S D
Sbjct: 294 KLPQ--GYIGNTLFHARPVSPLGAFHRESFSETVERVHREIRKMDNEYLRSAIDYLERHP 351
Query: 320 ----LMVIKERCLFTTIRS-CVVSDLTRAKYAEVNFGWGEAVY 357
L+ + +F+ + C+VS + + Y E++FGWG+A Y
Sbjct: 352 DLDQLVPGEGNTIFSCAANFCIVSLIKQTAY-EMDFGWGKAYY 393
>At2g19070 putative anthranilate
N-hydroxycinnamoyl/benzoyltransferase
Length = 451
Score = 105 bits (261), Expect = 6e-23
Identities = 110/405 (27%), Positives = 176/405 (43%), Gaps = 72/405 (17%)
Query: 18 VPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEKDPAKVLKRALSQTLVYYY 77
+ PA PT L++ D + ++P ++ Y + + ++LK +LS+ LV++Y
Sbjct: 12 IVPAEPTWSGRFPLAEWDQVGTIT-HIPTLYFYDKPSESFQGNVVEILKTSLSRVLVHFY 70
Query: 78 PLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDEFGDALHPPFPCFQELIYDVPGTNQ 137
P+AGR+R R + ++C EGV FIEAE++ L +F D P P F+ L+ V N
Sbjct: 71 PMAGRLRWLPRGRFELNCNAEGVEFIEAESEGKLSDFKD--FSPTPEFENLMPQVNYKNP 128
Query: 138 IIDHPILLIQ----------------------------------MARGAHQPSIQPVWRR 163
I P+ L Q +ARG ++ P R
Sbjct: 129 IETIPLFLAQVTKFKCGGISLSVNVSHAIVDGQSALHLISEWGRLARGEPLETV-PFLDR 187
Query: 164 EILMARD--PPRVT---CNHHEYEQ---ILSSNINKEEDVAATIVHHSFFFTLTRIAEIR 215
+IL A + PP V+ +H E++Q ++ N EE TIV + +++ ++R
Sbjct: 188 KILWAGEPLPPFVSPPKFDHKEFDQPPFLIGETDNVEERKKKTIV-VMLPLSTSQLQKLR 246
Query: 216 RLI-----PFHLRQCSTFDLITACFWCCRTKALQLEPDEEVRMMCIVNARSRFRADHSPL 270
+ + ++ +T W C KA P++ + ++ RSR PL
Sbjct: 247 SKANGSKHSDPAKGFTRYETVTGHVWRCACKARGHSPEQPTALGICIDTRSRM---EPPL 303
Query: 271 V-GYYGNCFAFPAAVTTAGKLCGNPLGYGVELIRKAKAQVTEEY-MHSLADLMVIKERCL 328
GY+GN A +T+G+L N LG+ LI KA VT EY M + L K+
Sbjct: 304 PRGYFGNATLDVVAASTSGELISNELGFAASLISKAIKNVTNEYVMIGIEYLKNQKDLKK 363
Query: 329 FTTIRSC--------------VVSDLTRAKYAEVNFGWGEAVYGG 359
F + + VVS LT Y ++FGWG+ Y G
Sbjct: 364 FQDLHALGSTEGPFYGNPNLGVVSWLTLPMYG-LDFGWGKEFYTG 407
>At1g28680 anthranilate N-hydroxycinnamoyl/benzoyltransferase like
protein
Length = 451
Score = 103 bits (257), Expect = 2e-22
Identities = 102/422 (24%), Positives = 181/422 (42%), Gaps = 52/422 (12%)
Query: 31 LSDIDDQECLRFNMPIIFIYRHEPSMIE-KDPAKVLKRALSQTLVYYYPLAGRIREGARD 89
LS +D+ L + + +Y S + + P+ V+ +L+ LV+YYPLAG +R A D
Sbjct: 27 LSHLDNDNNLHVSFRYLRVYSSSSSTVAGESPSAVVSASLATALVHYYPLAGSLRRSASD 86
Query: 90 KL--MVDCTGEGVMFIEAETDVTLDEFGDALHPPFPCFQELIYDVPGTNQIIDHPILL-- 145
++ G+ V + A + TL+ G L P P F E + P + + +P +L
Sbjct: 87 NRFELLCSAGQSVPLVNATVNCTLESVG-YLDGPDPGFVERLVPDPTREEGMVNPCILQV 145
Query: 146 -------------------------------IQMARGAHQPSIQPVWRREILMA-RDPPR 173
++ARGA + SI+PVW RE L+ R+ P
Sbjct: 146 TMFQCGGWVLGASIHHAICDGLGASLFFNAMAELARGATKISIEPVWDRERLLGPREKPW 205
Query: 174 VTCNHHEYEQILSSNINKEEDVAATIVHHSFFFTLTRIAEIR-RLIPFHLRQCSTFDLIT 232
V ++ L + + + FF T + +++ +L+ +TF+ +
Sbjct: 206 VGAPVRDFLS-LDKDFDPYGQAIGDVKRDCFFVTDDSLDQLKAQLLEKSGLNFTTFEALG 264
Query: 233 ACFWCCRTKALQLEPDEEVRMMCIVNARSRFRADHSPLV-GYYGNCFAFPAAVTTAGKLC 291
A W + +A + E E V+ + +N R R + PL GY+GN A AG+L
Sbjct: 265 AYIWRAKVRAAKTEEKENVKFVYSINIR---RLMNPPLPKGYWGNGCVPMYAQIKAGELI 321
Query: 292 GNPLGYGVELIRKAKAQVTEEYMHSLADLMVIKERCLFTTIRSCV-VSDLTRAKYAEVNF 350
P+ ELI+++K+ ++EY+ S D + + +D ++ ++F
Sbjct: 322 EQPIWKTAELIKQSKSNTSDEYVRSFIDFQELHHKDGINAGTGVTGFTDWRYLGHSTIDF 381
Query: 351 GWGEAV-YGGVVNCVAGPLPRATFIIPHK-----NAEGEEGLILLIFLTSEVMKRFAEEL 404
GWG V + N + G + F +P+ ++ + G +L+ L M F E +
Sbjct: 382 GWGGPVTVLPLSNKLLGSM-EPCFFLPYSTDAAAGSKKDSGFKVLVNLRESAMPEFKEAM 440
Query: 405 DK 406
DK
Sbjct: 441 DK 442
>At3g47170 hypersensitivity-related protein-like protein
Length = 447
Score = 92.0 bits (227), Expect = 5e-19
Identities = 112/466 (24%), Positives = 191/466 (40%), Gaps = 101/466 (21%)
Query: 8 FTVRRCQPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEKD---PAKV 64
F V + + LV P+ PTP LS ID+ + I+ ++ P + + PA
Sbjct: 10 FVVEKSEVVLVKPSKPTPDVTLSLSTIDNDPNNEVILDILCVFSPNPYVRDHTNYHPASF 69
Query: 65 LKRALSQTLVYYYPLAGRIREGARDK-LMVDCT-GEGVMFIEAETDVTLD-----EFGDA 117
++ ALS LVYYYPLAG++ D+ L +DCT G+GV I+A TL E G+
Sbjct: 70 IQLALSNALVYYYPLAGKLHRRTSDQTLQLDCTQGDGVPLIKATASCTLSSLNYLESGNH 129
Query: 118 LHPPFPC-------------FQELIYDVP-------GTNQIIDHPI-----------LLI 146
L + ++ L + + H + ++
Sbjct: 130 LEATYQLVPSHDTLKGCNLGYRPLALQITKFTCGGMTIGSVHSHTVCDGIGMAQFSQAIL 189
Query: 147 QMARGAHQPSIQPVWRREILMA------------RDPPRVTCNHHEYEQILSSNINKEED 194
++A G QP++ PVW R ++ + ++ P++ + E+ SS ED
Sbjct: 190 ELAAGRAQPTVIPVWDRHMITSNQISTLCKLGNDKNNPKLV----DVEKDCSSPDTPTED 245
Query: 195 VAATIVHHSFFFTLTRIAEIRRLI---------PFHLRQCSTFDLITACFWCCRTKALQL 245
+ I++ T I +++ +I + +T +++ A W R +A++L
Sbjct: 246 MVREILN----ITSEDITKLKNIIIEDENLTNEKEKNMKITTVEVLAAYVWRARCRAMKL 301
Query: 246 EPDEEVRMMCIVNARSRFRADHSPL-VGYYGNCFAFPAAVTTAGKLCGNPLGYGVELIRK 304
PD ++ V+ RS PL GYYGN F + TA +L P+ V LI+
Sbjct: 302 NPDTITDLVISVSIRSSI---EPPLPEGYYGNAFTHASVALTAKELSKTPISRLVRLIKD 358
Query: 305 AKAQVTE-----EYMHSLADLMVIKERCLFTTIRSCVVSDLTRAKYAEVNFGWGEAVYGG 359
AK + E + + + M +K I V LT ++ G + V+G
Sbjct: 359 AKRAALDNGYVCEQLREMENTMKLK--LASKEIHGGVFMMLTDWRH----LGLDQDVWG- 411
Query: 360 VVNCVAGPLPRATFIIPHKNAEGEEGLILLIFLTSEVMKRFAEELD 405
A+ +P K+ G+ +L L + M +F EE+D
Sbjct: 412 -----------ASKAVPGKSG----GVRVLTTLPRDAMAKFKEEID 442
>At1g24420 unknown protein
Length = 436
Score = 62.4 bits (150), Expect = 5e-10
Identities = 48/139 (34%), Positives = 74/139 (52%), Gaps = 16/139 (11%)
Query: 16 ELVPPAAPTPREVKLL--SDIDDQECLRFNMPIIFIYRHEPSMI----EKDPAKVLKRAL 69
E+V P++PTP + ++L S +D + ++F Y +P + ++ + LK++L
Sbjct: 11 EIVKPSSPTPDDKRILNLSLLDILSSPMYTGALLF-YAADPQNLLGFSTEETSLKLKKSL 69
Query: 70 SQTLVYYYPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDEFGDALHPPFPCFQELI 129
S+TL +YPLAGRI V+C EG +FIEA D L EF L P P EL+
Sbjct: 70 SKTLPIFYPLAGRIIGS-----FVECNDEGAVFIEARVDHLLSEF---LKCPVPESLELL 121
Query: 130 YDVPG-TNQIIDHPILLIQ 147
V + + + P+LLIQ
Sbjct: 122 IPVEAKSREAVTWPVLLIQ 140
>At1g65445 unknown protein
Length = 166
Score = 59.7 bits (143), Expect = 3e-09
Identities = 39/135 (28%), Positives = 70/135 (50%), Gaps = 4/135 (2%)
Query: 17 LVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEKDPAKVLKRALSQTLVYY 76
+ P + R+ LS++D + L F++ + +R + ++ L++AL + + Y
Sbjct: 26 IFPSEETSERKSMFLSNVD--QILNFDVQTVHFFRPNKEFPPEMVSEKLRKALVKLMDAY 83
Query: 77 YPLAGRIR-EGARDKLMVDCTGEGVMFIEAETDVTLDEFGDALHPPFPCFQELIYDVPGT 135
LAGR+R + + +L VDC G G F+ A +D TL+E GD ++P P F +L+ +
Sbjct: 84 EFLAGRLRVDPSSGRLDVDCNGAGAGFVTAASDYTLEELGDLVYPN-PAFAQLVTSQLQS 142
Query: 136 NQIIDHPILLIQMAR 150
D P+ + Q R
Sbjct: 143 LPKDDQPLFVFQEER 157
>At1g65450 unknown protein
Length = 286
Score = 59.3 bits (142), Expect = 4e-09
Identities = 62/258 (24%), Positives = 104/258 (40%), Gaps = 25/258 (9%)
Query: 156 SIQPVWRREILMARDPPRVTCNHHEYEQILSSNINKEEDVAATIVHHSFFFTLTRIAEIR 215
S P R +L ARDPP V HHE + + AT H F +I+
Sbjct: 32 STPPCNDRTLLKARDPPSVAFPHHEL--VKFQDCETTTVFEATSEHLDFKIFKLSSEQIK 89
Query: 216 RLIPFHLR------QCSTFDLITACFWCCRTKALQLEPDEEVRM------MCIVNARSRF 263
+L + + F+++TA W C+ ++ E EE + + V+ R R
Sbjct: 90 KLKERASETSNGNVRVTGFNVVTALVWRCKALSVAAEEGEETNLERESTILYAVDIRGRL 149
Query: 264 RADHSPLVGYYGNCFAFPAAVTTAGKLCGNPLGYGVELIRKAKAQVTEEYMHSLADLMVI 323
+ P Y GN A L P G VE++ + ++T+EY S D +
Sbjct: 150 NPELPP--SYTGNAVLTAYAKEKCKALLEEPFGRIVEMVGEGSKRITDEYARSAIDWGEL 207
Query: 324 KERCLFTTIRSCVVSDLTRAKYAEVNFGWGEAVYGGVVNCVAGPLPRATFIIPHKNAEGE 383
+ + +VS + +AEV + WG+ Y V R ++ + +G+
Sbjct: 208 YKGFPHGEV---LVSSWWKLGFAEVEYPWGKPKYSCPV-----VYHRKDIVLLFPDIDGD 259
Query: 384 -EGLILLIFLTSEVMKRF 400
+G+ +L L S+ M +F
Sbjct: 260 SKGVYVLAALPSKEMSKF 277
>At3g26040 hypothetical protein
Length = 442
Score = 58.9 bits (141), Expect = 5e-09
Identities = 36/116 (31%), Positives = 62/116 (53%), Gaps = 9/116 (7%)
Query: 16 ELVPPAAPTPREVKL--LSDIDDQECLRFNMPIIFIYRHEPSMIEKDPAKVLKRALSQTL 73
+++ P++PTP +K LS ++ F P++F Y S+ + ++LK++LS+TL
Sbjct: 9 DIIKPSSPTPNHLKKFKLSLLEQLGPTIFG-PMVFFYSANNSIKPTEQLQMLKKSLSETL 67
Query: 74 VYYYPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDEFGDALHPPFPCFQELI 129
++YPLAGR+ + + +DC G F+EA + L L P Q+LI
Sbjct: 68 THFYPLAGRL----KGNISIDCNDSGADFLEARVNSPLSNL--LLEPSSDSLQQLI 117
>At4g15400 HSR201 like protein
Length = 435
Score = 57.0 bits (136), Expect = 2e-08
Identities = 38/99 (38%), Positives = 55/99 (55%), Gaps = 7/99 (7%)
Query: 16 ELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEKDPAKVLKRALSQTLVY 75
E++ PA+P+PR+ +L I D C + IF Y E ++ LK +LS+TL
Sbjct: 11 EVIKPASPSPRD-RLQLSILDLYCPGIYVSTIFFYDLITESSEVF-SENLKLSLSETLSR 68
Query: 76 YYPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDEF 114
+YPLAGRI + L + C EG +F EA TD+ L +F
Sbjct: 69 FYPLAGRI-----EGLSISCNDEGAVFTEARTDLLLPDF 102
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.325 0.140 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,670,928
Number of Sequences: 26719
Number of extensions: 402547
Number of successful extensions: 1179
Number of sequences better than 10.0: 59
Number of HSP's better than 10.0 without gapping: 40
Number of HSP's successfully gapped in prelim test: 19
Number of HSP's that attempted gapping in prelim test: 1018
Number of HSP's gapped (non-prelim): 106
length of query: 426
length of database: 11,318,596
effective HSP length: 102
effective length of query: 324
effective length of database: 8,593,258
effective search space: 2784215592
effective search space used: 2784215592
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC148345.4


