

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148345.12 + phase: 0
(462 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
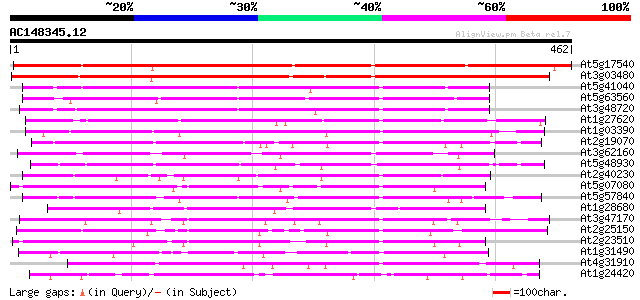
At5g17540 unknown protein 403 e-113
At3g03480 putative hypersensitivity-related gene 373 e-103
At5g41040 N-hydroxycinnamoyl/benzoyltransferase-like protein 220 1e-57
At5g63560 acyltransferase-like protein 219 3e-57
At3g48720 unknown protein 219 3e-57
At1g27620 putative hypersensitivity-related protein 188 6e-48
At1g03390 hypothetical protein 179 2e-45
At2g19070 putative anthranilate N-hydroxycinnamoyl/benzoyltransf... 157 1e-38
At3g62160 unknown protein 156 2e-38
At5g48930 anthranilate N-benzoyltransferase 151 8e-37
At2g40230 putative anthranilate N-hydroxycinnamoyl/benzoyltransf... 150 2e-36
At5g07080 unknown protein 146 3e-35
At5g57840 N-hydroxycinnamoyl/benzoyltransferase 142 5e-34
At1g28680 anthranilate N-hydroxycinnamoyl/benzoyltransferase lik... 137 1e-32
At3g47170 hypersensitivity-related protein-like protein 134 1e-31
At2g25150 unknown protein 121 7e-28
At2g23510 putative anthranilate N-hydroxycinnamoyl/benzoyltransf... 121 7e-28
At1g31490 putative protein 105 7e-23
At4g31910 unknown protein 103 2e-22
At1g24420 unknown protein 101 8e-22
>At5g17540 unknown protein
Length = 461
Score = 403 bits (1036), Expect = e-113
Identities = 219/466 (46%), Positives = 304/466 (64%), Gaps = 13/466 (2%)
Query: 4 SSPLLFTVRRSQPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSMKEKDPV 63
S L F + R +PELV PA PTP E+K LSDIDDQEGLRF++P IF +RH P+ DPV
Sbjct: 2 SGSLTFKIYRQKPELVSPAKPTPRELKPLSDIDDQEGLRFHIPTIFFYRHNPTTNS-DPV 60
Query: 64 KVLKHALSRTLVYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQFG--DALQP 121
V++ AL+ TLVYYYP AGR+REG RKL VDCTGEGV+FIEA+AD+TL +F DAL+P
Sbjct: 61 AVIRRALAETLVYYYPFAGRLREGPNRKLAVDCTGEGVLFIEADADVTLVEFEEKDALKP 120
Query: 122 PFPCFQEILCDVPGSEYIIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSAWA 181
PFPCF+E+L +V GS +++ P+ L+QVTRLKCGGFI A+ +NH M D GL F+
Sbjct: 121 PFPCFEELLFNVEGSCEMLNTPLMLMQVTRLKCGGFIFAVRINHAMSDAGGLTLFLKTMC 180
Query: 182 EIARGANQPSIQPVWNREILMARDPPYITCNHREYEQILPPNTYIKEEDTTIVVHQSFFF 241
E RG + P++ PVW R +L AR +T HREY+++ T + + V +S FF
Sbjct: 181 EFVRGYHAPTVAPVWERHLLSARVLLRVTHAHREYDEMPAIGTELGSRRDNL-VGRSLFF 239
Query: 242 TSAHIAAIRRLVPLHL-SRCTAYDLITACYWCCRTKALQVEPDEDVRMMCIVNARSRFSA 300
++AIRRL+P +L + T +++T+ W RT AL+ + D+++R++ IVNARS+
Sbjct: 240 GPCEMSAIRRLLPPNLVNSSTNMEMLTSFLWRYRTIALRPDQDKEMRLILIVNARSKL-- 297
Query: 301 NNRSSLIGYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAKAQVTEKYMHSLADFMVIK 360
N GYYGN AF A+ TA EL L +A+ LI++AK+ VTE+YM SLAD MVIK
Sbjct: 298 KNPPLPRGYYGNAFAFPVAIATANELTKKPLEFALRLIKEAKSSVTEEYMRSLADLMVIK 357
Query: 361 ERCLFTKGRTCMVSDWTRAKFSEVNFG-WGEAVYGGAAKGGIGSFLGGTFLVTHKNAKGE 419
R F+ +VSD F++++FG WG+ VYGG G+ G +F V+ + GE
Sbjct: 358 GRPSFSSDGAYLVSD--VRIFADIDFGIWGKPVYGGIGTAGVEDLPGASFYVSFEKRNGE 415
Query: 420 EGLILPICLPPEDMKRFVKELDDMLGNQ---NYPTMSGPSFILSTL 462
G+++P+CLP + M+RFV+EL+ + Q N + S I+S+L
Sbjct: 416 IGIVVPVCLPEKAMQRFVEELEGVFNGQVVFNRGSKSSNKLIMSSL 461
>At3g03480 putative hypersensitivity-related gene
Length = 454
Score = 373 bits (957), Expect = e-103
Identities = 204/447 (45%), Positives = 285/447 (63%), Gaps = 11/447 (2%)
Query: 2 TSSSPLLFTVRRSQPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSMKEKD 61
++++ L F V R Q ELV PA PTP E+K LSDIDDQ+GLRF +PVIF +R S + D
Sbjct: 10 STTTGLSFKVHRQQRELVTPAKPTPRELKPLSDIDDQQGLRFQIPVIFFYRPNLS-SDLD 68
Query: 62 PVKVLKHALSRTLVYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQF--GDAL 119
PV+V+K AL+ LVYYYP AGR+RE + RKL VDCTGEGV+FIEAEAD+ L + DAL
Sbjct: 69 PVQVIKKALADALVYYYPFAGRLRELSNRKLAVDCTGEGVLFIEAEADVALAELEEADAL 128
Query: 120 QPPFPCFQEILCDVPGSEYIIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSA 179
PPFP +E+L DV GS +++ P+ L+QVTRLKC GFI AL NHTM DG+GL F+ +
Sbjct: 129 LPPFPFLEELLFDVEGSSDVLNTPLLLVQVTRLKCCGFIFALRFNHTMTDGAGLSLFLKS 188
Query: 180 WAEIARGANQPSIQPVWNREIL-MARDPPYITCNHREYEQILPPNTYIKEEDTTIVVHQS 238
E+A G + PS+ PVWNR +L ++ +T HREY+ + + +V +S
Sbjct: 189 LCELACGLHAPSVPPVWNRHLLTVSASEARVTHTHREYDDQVGIDVVATGHP---LVSRS 245
Query: 239 FFFTSAHIAAIRRLVPLHLSRCTAYDLITACYWCCRTKALQVEPDEDVRMMCIVNARSRF 298
FFF + I+AIR+L+P L T+++ +++ W CRT AL +P+ ++R+ CI+N+RS+
Sbjct: 246 FFFRAEEISAIRKLLPPDLHN-TSFEALSSFLWRCRTIALNPDPNTEMRLTCIINSRSKL 304
Query: 299 SANNRSSLIGYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAKAQVTEKYMHSLADFMV 358
N GYYGN AA+ TA++L L +A+ LI++ K+ VTE Y+ S+ M
Sbjct: 305 --RNPPLEPGYYGNVFVIPAAIATARDLIEKPLEFALRLIQETKSSVTEDYVRSVTALMA 362
Query: 359 IKERCLFTKGRTCMVSDWTRAKFSEVNFG-WGEAVYGGAAKGGIGSFLGGTFLVTHKNAK 417
+ R +F ++SD +++FG WG+ VYGG AK GI F G +F V KN K
Sbjct: 363 TRGRPMFVASGNYIISDLRHFDLGKIDFGPWGKPVYGGTAKAGIALFPGVSFYVPFKNKK 422
Query: 418 GEEGLILPICLPPEDMKRFVKELDDML 444
GE G ++ I LP M+ FV EL+ +L
Sbjct: 423 GETGTVVAISLPVRAMETFVAELNGVL 449
>At5g41040 N-hydroxycinnamoyl/benzoyltransferase-like protein
Length = 457
Score = 220 bits (561), Expect = 1e-57
Identities = 133/390 (34%), Positives = 214/390 (54%), Gaps = 13/390 (3%)
Query: 11 VRRSQPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSMKEKDPVKVLKHAL 70
V + +P LV P + T + LS++D + + + I+ F+ E E + V+V+K AL
Sbjct: 33 VHQKEPALVKPESETRKGLYFLSNLD--QNIAVIVRTIYCFKSEERGNE-EAVQVIKKAL 89
Query: 71 SRTLVYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQFGDALQPPFPCFQEIL 130
S+ LV+YYP AGR+ KL VDCT EGV+F+EAEA+ +D+ GD +P +++
Sbjct: 90 SQVLVHYYPLAGRLTISPEGKLTVDCTEEGVVFVEAEANCKMDEIGDITKPDPETLGKLV 149
Query: 131 CDVPGSEYIIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSAWAEIARGANQP 190
DV ++ I++ P QVT+ KCGGF+L L +NH M DG G +FV++W ++ARG
Sbjct: 150 YDVVDAKNILEIPPVTAQVTKFKCGGFVLGLCMNHCMFDGIGAMEFVNSWGQVARGL-PL 208
Query: 191 SIQPVWNREILMARDPPYITCNHREYEQILPPNTYIKEEDTTIVVHQSFFFTSAHI---- 246
+ P +R IL AR+PP I H+E+E+I + +++SF F I
Sbjct: 209 TTPPFSDRTILNARNPPKIENLHQEFEEIEDKSNINSLYTKEPTLYRSFCFDPEKIKKLK 268
Query: 247 -AAIRRLVPLHLSRCTAYDLITACYWCCRTKALQVEPDEDVRMMCIVNARSRFSANNRSS 305
A L + CT+++ ++A W RTK+L++ D+ +++ V+ R++F
Sbjct: 269 LQATENSESLLGNSCTSFEALSAFVWRARTKSLKMLSDQKTKLLFAVDGRAKFEPQLPK- 327
Query: 306 LIGYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAKAQVTEKYMHSLADFMVIKERCLF 365
GY+GN I + ++ A EL L +AV L+R+A VT+ YM S D+ + R
Sbjct: 328 --GYFGNGIVLTNSICEAGELIEKPLSFAVGLVREAIKMVTDGYMRSAIDYFEV-TRARP 384
Query: 366 TKGRTCMVSDWTRAKFSEVNFGWGEAVYGG 395
+ T +++ W+R F +FGWGE + G
Sbjct: 385 SLSSTLLITTWSRLGFHTTDFGWGEPILSG 414
>At5g63560 acyltransferase-like protein
Length = 426
Score = 219 bits (557), Expect = 3e-57
Identities = 137/393 (34%), Positives = 213/393 (53%), Gaps = 25/393 (6%)
Query: 11 VRRSQPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVI---FVFRHEPSMKEKDPVKVLK 67
V R +P LV PA+ TP + LS++D N+ +I F + S ++ +V+K
Sbjct: 9 VTRKEPVLVSPASETPKGLHYLSNLDQ------NIAIIVKTFYYFKSNSRSNEESYEVIK 62
Query: 68 HALSRTLVYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQFGDAL----QPPF 123
+LS LV+YYP AGR+ K+ VDCTGEGV+ +EAEA+ +++ A+ QP
Sbjct: 63 KSLSEVLVHYYPAAGRLTISPEGKIAVDCTGEGVVVVEAEANCGIEKIKKAISEIDQPE- 121
Query: 124 PCFQEILCDVPGSEYIIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSAWAEI 183
++++ DVPG+ I++ P ++QVT KCGGF+L L +NH M DG +F+++WAE
Sbjct: 122 -TLEKLVYDVPGARNILEIPPVVVQVTNFKCGGFVLGLGMNHNMFDGIAAMEFLNSWAET 180
Query: 184 ARGANQPSIQPVWNREILMARDPPYITCNHREYEQILPPNTYIKEEDTTIVVHQSFFFTS 243
ARG S+ P +R +L R PP I H E+E + + K +V++SF F
Sbjct: 181 ARGL-PLSVPPFLDRTLLRPRTPPKIEFPHNEFEDLEDISGTGKLYSDEKLVYKSFLFGP 239
Query: 244 AHIAAIRRLVPLHLSRCTAYDLITACYWCCRTKALQVEPDEDVRMMCIVNARSRFSANNR 303
+ RL + +R T + +T W R +AL ++PD+ ++++ + RSRF
Sbjct: 240 ---EKLERLKIMAETRSTTFQTLTGFLWRARCQALGLKPDQRIKLLFAADGRSRFVPELP 296
Query: 304 SSLIGYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAKAQVTEKYMHSLAD-FMVIKER 362
GY GN I F+ VTTA E+ N L ++V L+++A V + +M S D F V + R
Sbjct: 297 K---GYSGNGIVFTYCVTTAGEVTLNPLSHSVCLVKRAVEMVNDGFMRSAIDYFEVTRAR 353
Query: 363 CLFTKGRTCMVSDWTRAKFSEVNFGWGEAVYGG 395
T T +++ W + F +FGWGE V G
Sbjct: 354 PSLT--ATLLITSWAKLSFHTKDFGWGEPVVSG 384
>At3g48720 unknown protein
Length = 430
Score = 219 bits (557), Expect = 3e-57
Identities = 123/390 (31%), Positives = 220/390 (55%), Gaps = 11/390 (2%)
Query: 9 FTVRRSQPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSMKEKDPVKVLKH 68
F V R PEL+ P + TP+ LS++D + + + ++ ++ E S ++ V+K
Sbjct: 7 FIVTRKNPELIPPVSETPNGHYYLSNLD--QNIAIIVKTLYYYKSE-SRTNQESYNVIKK 63
Query: 69 ALSRTLVYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQFGDAL-QPPFPCFQ 127
+LS LV+YYP AGR+ K+ V+CTGEGV+ +EAEA+ +D +A+ + +
Sbjct: 64 SLSEVLVHYYPVAGRLTISPEGKIAVNCTGEGVVVVEAEANCGIDTIKEAISENRMETLE 123
Query: 128 EILCDVPGSEYIIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSAWAEIARGA 187
+++ DVPG+ I++ P ++QVT KCGGF+L L ++H M DG +F+++W E+A+G
Sbjct: 124 KLVYDVPGARNILEIPPVVVQVTNFKCGGFVLGLGMSHNMFDGVAAAEFLNSWCEMAKGL 183
Query: 188 NQPSIQPVWNREILMARDPPYITCNHREYEQILPPNTYIKEEDTTIVVHQSFFFTSAHIA 247
S+ P +R IL +R+PP I H E+++I + K D ++++SF F +
Sbjct: 184 -PLSVPPFLDRTILRSRNPPKIEFPHNEFDEIEDISDTGKIYDEEKLIYKSFLFEPEKLE 242
Query: 248 AIR--RLVPLHLSRCTAYDLITACYWCCRTKALQVEPDEDVRMMCIVNARSRFSANNRSS 305
++ + + ++ + + +T W R +AL+ +PD+ V+++ + RSRF
Sbjct: 243 KLKIMAIEENNNNKVSTFQALTGFLWKSRCEALRFKPDQRVKLLFAADGRSRFIPRLPQ- 301
Query: 306 LIGYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAKAQVTEKYMHSLADFMVIKERCLF 365
GY GN I + VT++ EL GN L ++V L+++ VT+ +M S D+ + R
Sbjct: 302 --GYCGNGIVLTGLVTSSGELVGNPLSHSVGLVKRLVELVTDGFMRSAMDYFEV-NRTRP 358
Query: 366 TKGRTCMVSDWTRAKFSEVNFGWGEAVYGG 395
+ T +++ W++ +++FGWGE V+ G
Sbjct: 359 SMNATLLITSWSKLTLHKLDFGWGEPVFSG 388
>At1g27620 putative hypersensitivity-related protein
Length = 442
Score = 188 bits (477), Expect = 6e-48
Identities = 130/439 (29%), Positives = 224/439 (50%), Gaps = 31/439 (7%)
Query: 14 SQPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSMKEKDPVKVLKHALSRT 73
+QP L+ P +PTP+ LS++DD LRF++ +++F+ K P+ LK +LSR
Sbjct: 12 NQPTLITPLSPTPNHSLYLSNLDDHHFLRFSIKYLYLFQ-----KSISPL-TLKDSLSRV 65
Query: 74 LVYYYPGAGRIR-EGAGRKLMVDCTGEGVMFIEAEADITLDQFGDALQPPFPCFQEILCD 132
LV YYP AGRIR G KL VDC GEG +F EA DIT F P ++++L
Sbjct: 66 LVDYYPFAGRIRVSDEGSKLEVDCNGEGAVFAEAFMDITCQDFVQLSPKPNKSWRKLLFK 125
Query: 133 VPGSEYIIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSAWAEIARGANQPSI 192
V ++ D P +IQVT L+CGG IL ++NH + DG G QF+ AWA
Sbjct: 126 VQAQSFL-DIPPLVIQVTYLRCGGMILCTAINHCLCDGIGTSQFLHAWAHATTSQAHLPT 184
Query: 193 QPVWNREILMARDPPYITCNHREYEQ---ILPPNTY--IKEEDTTIVVHQSFFFTSAHIA 247
+P +R +L R+PP +T +H + + + +T+ K + + + F +H+
Sbjct: 185 RPFHSRHVLDPRNPPRVTHSHPGFTRTTTVDKSSTFDISKYLQSQPLAPATLTFNQSHLL 244
Query: 248 AIRRLVPLHLSRCTAYDLITACYWCCRTKALQVEPDEDVRMMCIVNARSRFSANNRSSLI 307
+++ L +CT ++ + A W ++L + V+++ VN R R +
Sbjct: 245 RLKKTCAPSL-KCTTFEALAANTWRSWAQSLDLPMTMLVKLLFSVNMRKRLTPELPQ--- 300
Query: 308 GYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAKAQVTEKYMHSLADFMVIKERCLFTK 367
GYYGN + A + ++L + +AV+ I++AK+++T++Y+ S D +++++ + T
Sbjct: 301 GYYGNGFVLACAESKVQDLVNGNIYHAVKSIQEAKSRITDEYVRSTID--LLEDKTVKTD 358
Query: 368 -GRTCMVSDWTRAKFSEVNFGWGEAVYGGAAKGGIGSFLGGTFLVTHKNAKGEEGLILPI 426
+ ++S W + E++ G G+ +Y +G + + A + + + +
Sbjct: 359 VSCSLVISQWAKLGLEELDLGGGKPMY-------MGPLTSDIYCLFLPVASDNDAIRVQM 411
Query: 427 CLPPEDMKR----FVKELD 441
LP E +KR VK LD
Sbjct: 412 SLPEEVVKRLEYCMVKFLD 430
>At1g03390 hypothetical protein
Length = 461
Score = 179 bits (455), Expect = 2e-45
Identities = 132/453 (29%), Positives = 214/453 (47%), Gaps = 43/453 (9%)
Query: 14 SQPELVLPAAPTP-----HEVKLLSDIDDQEGLRFNLPVIFVFRHEPSMKEKDPVKVLKH 68
+Q LV P++PTP H LS++DD G R P ++ F + +E +K L+
Sbjct: 20 NQQFLVHPSSPTPANQSPHHSLYLSNLDDIIGARVFTPSVY-FYPSTNNRESFVLKRLQD 78
Query: 69 ALSRTLVYYYPGAGRIREGAGRKLMVDCTGE-GVMFIEAEADITLDQFGDALQPPFPCFQ 127
ALS LV YYP +GR+RE KL V E GV+ + A + + L GD L P P +
Sbjct: 79 ALSEVLVPYYPLSGRLREVENGKLEVFFGEEQGVLMVSANSSMDLADLGD-LTVPNPAWL 137
Query: 128 EILCDVPGSEY--IIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSAWAEIAR 185
++ PG E I++ P+ + QVT CGGF L + L H + DG G QF+ +WA A+
Sbjct: 138 PLIFRNPGEEAYKILEMPLLIAQVTFFTCGGFSLGIRLCHCICDGFGAMQFLGSWAATAK 197
Query: 186 -GANQPSIQPVWNREILMARDPPYITCNHREYEQILPPNTYIKEEDTTIVVHQSFFFTSA 244
G +PVW+RE R+PP + H EY I + T + + + +
Sbjct: 198 TGKLIADPEPVWDRETFKPRNPPMVKYPHHEYLPIEERSNLTNSLWDTKPLQKCYRISKE 257
Query: 245 HIAAIRRLVPLHLSR--CTAYDLITACYWCCRTKALQVEP-DEDVRMMCIVNARSRFSAN 301
++ + C+ +D + A W KAL V+P D ++R+ VN R+R
Sbjct: 258 FQCRVKSIAQGEDPTLVCSTFDAMAAHIWRSWVKALDVKPLDYNLRLTFSVNVRTRLETL 317
Query: 302 NRSSLIGYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAKAQVTEKYMHSLADFMVIKE 361
G+YGN + + A+++ + L + L L++ A+ +V+E Y+ S+ D++ +K
Sbjct: 318 KLRK--GFYGNVVCLACAMSSVESLINDSLSKTTRLVQDARLRVSEDYLRSMVDYVDVKR 375
Query: 362 RCLFTKGRTCMVSDWTRAKFSE-VNFGWGEAVYGG-------------AAKGGIGSFLGG 407
G ++ WTR + E +FGWG+ VY G +GG+ S
Sbjct: 376 PKRLEFGGKLTITQWTRFEMYETADFGWGKPVYAGPIDLRPTPQVCVLLPQGGVES---- 431
Query: 408 TFLVTHKNAKGEEGLILPICLPPEDMKRFVKEL 440
++ +++ +CLPP + F + L
Sbjct: 432 ---------GNDQSMVVCLCLPPTAVHTFTRLL 455
>At2g19070 putative anthranilate
N-hydroxycinnamoyl/benzoyltransferase
Length = 451
Score = 157 bits (397), Expect = 1e-38
Identities = 131/449 (29%), Positives = 208/449 (46%), Gaps = 44/449 (9%)
Query: 19 VLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSMKEKDPVKVLKHALSRTLVYYY 78
++PA PT L++ D Q G ++P ++ + + + V++LK +LSR LV++Y
Sbjct: 12 IVPAEPTWSGRFPLAEWD-QVGTITHIPTLYFYDKPSESFQGNVVEILKTSLSRVLVHFY 70
Query: 79 PGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQFGDALQPPFPCFQEILCDVPGSEY 138
P AGR+R + ++C EGV FIEAE++ L F D P P F+ ++ V
Sbjct: 71 PMAGRLRWLPRGRFELNCNAEGVEFIEAESEGKLSDFKDF--SPTPEFENLMPQVNYKNP 128
Query: 139 IIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSAWAEIARGANQPSIQPVWNR 198
I P+ L QVT+ KCGG L+++++H + DG +S W +ARG ++ P +R
Sbjct: 129 IETIPLFLAQVTKFKCGGISLSVNVSHAIVDGQSALHLISEWGRLARGEPLETV-PFLDR 187
Query: 199 EILMARD--PPYIT---CNHREYEQILPPNTYIKEEDT-------TIVVHQSFFFTSAHI 246
+IL A + PP+++ +H+E++Q P I E D TIVV +
Sbjct: 188 KILWAGEPLPPFVSPPKFDHKEFDQ---PPFLIGETDNVEERKKKTIVVMLPLSTSQLQK 244
Query: 247 AAIRRLVPLHLSRC---TAYDLITACYWCCRTKALQVEPDEDVRMMCIVNARSRFSANNR 303
+ H T Y+ +T W C KA P++ + ++ RSR
Sbjct: 245 LRSKANGSKHSDPAKGFTRYETVTGHVWRCACKARGHSPEQPTALGICIDTRSRMEPPLP 304
Query: 304 SSLIGYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAKAQVTEKYMHSLADFM-----V 358
GY+GN A +T+ EL N+LG+A LI KA VT +Y+ +++ +
Sbjct: 305 R---GYFGNATLDVVAASTSGELISNELGFAASLISKAIKNVTNEYVMIGIEYLKNQKDL 361
Query: 359 IKERCLFTKGRT---------CMVSDWTRAKFSEVNFGWGEAVYGGAAKGGIGSFLGGTF 409
K + L G T V W ++FGWG+ Y G G F G +
Sbjct: 362 KKFQDLHALGSTEGPFYGNPNLGVVSWLTLPMYGLDFGWGKEFYTGP---GTHDFDGDSL 418
Query: 410 LVTHKNAKGEEGLILPICLPPEDMKRFVK 438
++ +N G +IL CL M+ F K
Sbjct: 419 ILPDQNEDG--SVILATCLQVAHMEAFKK 445
>At3g62160 unknown protein
Length = 428
Score = 156 bits (395), Expect = 2e-38
Identities = 117/416 (28%), Positives = 196/416 (46%), Gaps = 59/416 (14%)
Query: 7 LLFTVRRSQPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSMKEKDPVKVL 66
+ F++ R L+ TP V LS ID+ LR I VF H P D + +
Sbjct: 1 MAFSLTRVNRVLIQACETTPSVVLDLSLIDNIPVLRCFARTIHVFTHGP-----DAARAI 55
Query: 67 KHALSRTLVYYYPGAGRIREGAGRKLMVDCTGE-GVMFIEAEADITLDQFGDALQPPFPC 125
+ AL++ LV YYP +GR++E KL +DCTG+ G+ F++A D+ L+ G
Sbjct: 56 QEALAKALVSYYPLSGRLKELNQGKLQIDCTGKTGIWFVDAVTDVKLESVG--------- 106
Query: 126 FQEILCDVPGSEYIIDR--------PIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFV 177
+ + L ++P + + D+ P+ +QVT+ CGGF++ L H + DG G QF+
Sbjct: 107 YFDNLMEIPYDDLLPDQIPKDDDAEPLVQMQVTQFSCGGFVMGLRFCHAICDGLGAAQFL 166
Query: 178 SAWAEIARGANQPSIQPVWNREILMARDPPYITCNHREYEQILP--PNTYIKEEDTTIVV 235
+A EIA G + PVW+R+ +I HR + I P P + +
Sbjct: 167 TAVGEIACGQTNLGVSPVWHRD--------FIPQQHRANDVISPAIPPPFPPAFPEYKLE 218
Query: 236 HQSFFFTSAHIAAIRRLVPLHLSR-CTAYDLITACYWCCRTKALQVEPDEDVRMMCIVNA 294
H SF S I +R C+A+++I AC+W RT+A+ + D +V+++ N
Sbjct: 219 HLSFDIPSDLIERFKREFRASTGEICSAFEVIAACFWKLRTEAINLREDTEVKLVFFANC 278
Query: 295 RSRFSANNRSSLIGYYGNCIAFSAAVTTAKELCGNQLGY-AVELIRKAKAQVTEKYMHSL 353
R N IG+YGNC + E+ + + V+LI++AK ++ E++
Sbjct: 279 RHLV---NPPLPIGFYGNCFFPVKISAESHEISKEKSVFDVVKLIKQAKTKLPEEF---- 331
Query: 354 ADFMVIKERCLFTKG----------RTCMVSDWTRAKFSEVNFGWGEAVYGGAAKG 399
E+ + G T +S+W + F++V++GWG ++ +G
Sbjct: 332 -------EKFVSGDGDDPFAPAVGYNTLFLSEWGKLGFNQVDYGWGPPLHVAPIQG 380
>At5g48930 anthranilate N-benzoyltransferase
Length = 433
Score = 151 bits (381), Expect = 8e-37
Identities = 122/438 (27%), Positives = 209/438 (46%), Gaps = 32/438 (7%)
Query: 18 LVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSMKEKDPVKVLKHALSRTLVYY 77
+V PA TP S++D RF+ P ++ +R + DP +V+K ALS+ LV +
Sbjct: 10 MVRPATETPITNLWNSNVDLVIP-RFHTPSVYFYRPTGASNFFDP-QVMKEALSKALVPF 67
Query: 78 YPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQFGDALQPPFPCFQEILCDVPGSE 137
YP AGR++ ++ +DC G GV+F+ A+ +D FGD P ++++ +V S
Sbjct: 68 YPMAGRLKRDDDGRIEIDCNGAGVLFVVADTPSVIDDFGDF--APTLNLRQLIPEVDHSA 125
Query: 138 YIIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSAWAEIARGANQPSIQPVWN 197
I P+ ++QVT KCGG L + + H DG F++ W+++ARG + +I P +
Sbjct: 126 GIHSFPLLVLQVTFFKCGGASLGVGMQHHAADGFSGLHFINTWSDMARGLDL-TIPPFID 184
Query: 198 REILMARDPPYITCNHREYE-----QILPPNTYIKEEDTTIVVHQSFFFTSAHIAAIRRL 252
R +L ARDPP +H EY+ +I + E+TT+ + F T + A++
Sbjct: 185 RTLLRARDPPQPAFHHVEYQPAPSMKIPLDPSKSGPENTTVSI---FKLTRDQLVALKAK 241
Query: 253 VPL--HLSRCTAYDLITACYWCCRTKALQVEPDEDVRMMCIVNARSRFSANNRSSL-IGY 309
+ ++Y+++ W KA + D++ ++ + RSR R L GY
Sbjct: 242 SKEDGNTVSYSSYEMLAGHVWRSVGKARGLPNDQETKLYIATDGRSRL----RPQLPPGY 297
Query: 310 YGNCIAFSAAVTTAKELCGNQLGYAVELIRKAKAQVTEKYMHSLADFMVIK-ERCLFTKG 368
+GN I + + A +L YA I ++ + Y+ S D++ ++ + +G
Sbjct: 298 FGNVIFTATPLAVAGDLLSKPTWYAAGQIHDFLVRMDDNYLRSALDYLEMQPDLSALVRG 357
Query: 369 R------TCMVSDWTRAKFSEVNFGWGEAVYGGAAKGGIGSFLGGTFLVTHKNAKGEEGL 422
++ W R + +FGWG ++ G GGI + G +F++ G L
Sbjct: 358 AHTYKCPNLGITSWVRLPIYDADFGWGRPIFMG--PGGI-PYEGLSFVLPSPTNDG--SL 412
Query: 423 ILPICLPPEDMKRFVKEL 440
+ I L E MK F K L
Sbjct: 413 SVAIALQSEHMKLFEKFL 430
>At2g40230 putative anthranilate
N-hydroxycinnamoyl/benzoyltransferase
Length = 433
Score = 150 bits (378), Expect = 2e-36
Identities = 114/400 (28%), Positives = 208/400 (51%), Gaps = 27/400 (6%)
Query: 11 VRRSQPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSMKEKDPVKVLKHAL 70
V + ++ P+ TP V LS +D Q LRF + + V+ S E LK AL
Sbjct: 5 VHVKEATVITPSDQTPSSVLSLSALDSQLFLRFTIEYLLVYP-PVSDPEYSLSGRLKSAL 63
Query: 71 SRTLVYYYPGAGRIRE--GAGRKLMVDCTGEGVMFIEAEADITLDQFGDALQPP--FPCF 126
SR LV Y+P +GR+RE G L V+C G+G +F+EA +DI D +PP +
Sbjct: 64 SRALVPYFPFSGRVREKPDGGGGLEVNCRGQGALFLEAVSDILTCL--DFQKPPRHVTSW 121
Query: 127 QEILCDVPGSEYIID----RPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSAWAE 182
+++L S ++ID P ++Q+T L+ GG LA+ +NH + DG G +F++ +AE
Sbjct: 122 RKLL-----SLHVIDVLAGAPPLVVQLTWLRDGGAALAVGVNHCVSDGIGSAEFLTLFAE 176
Query: 183 IARGA---NQPSIQPVWNREILMARDPPYITCNHREYEQILPPNTYIKEEDTTIVVHQSF 239
+++ + + + +W+R++LM P + +H E+ ++ ++ + +V S
Sbjct: 177 LSKDSLSQTELKRKHLWDRQLLMP-SPIRDSLSHPEFNRVPDLCGFVNRFNAERLVPTSV 235
Query: 240 FFTSAHIAAIRRLVPL---HLSRCTAYDLITACYWCCRTKALQVEPDEDVRMMCIVNARS 296
F + +++L S+ T++++++A W ++L + ++ ++++ VN R
Sbjct: 236 VFERQKLNELKKLASRLGEFNSKHTSFEVLSAHVWRSWARSLNLPSNQVLKLLFSVNIRD 295
Query: 297 RFSANNRSSLIGYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAKAQVTEKYMHSLADF 356
R + S G+YGN A TT K+L L YA L+++AK +V ++Y+ S+ +
Sbjct: 296 RVKPSLPS---GFYGNAFVVGCAQTTVKDLTEKGLSYATMLVKQAKERVGDEYVRSVVE- 351
Query: 357 MVIKERCLFTKGRTCMVSDWTRAKFSEVNFGWGEAVYGGA 396
V KER ++S W+R +++FG G+ V+ G+
Sbjct: 352 AVSKERASPDSVGVLILSQWSRLGLEKLDFGLGKPVHVGS 391
>At5g07080 unknown protein
Length = 450
Score = 146 bits (368), Expect = 3e-35
Identities = 124/416 (29%), Positives = 187/416 (44%), Gaps = 42/416 (10%)
Query: 1 MTSSSPLLFTVRRSQPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRH-EPSMKE 59
M+S SPL+ V++SQ +V P+ TP LS +D+ L I+V+ P+
Sbjct: 6 MSSPSPLV--VKKSQVVIVKPSKATPDVSLSLSTLDNDPYLETLAKTIYVYAPPSPNDHV 63
Query: 60 KDPVKVLKHALSRTLVYYYPGAGRIREGA-GRKLMVDCT-GEGVMFIEAEADITLDQFGD 117
DP + + ALS LVYYYP AG++ G+ +L + C+ EGV F+ A AD TL
Sbjct: 64 HDPASLFQQALSDALVYYYPLAGKLHRGSHDHRLELRCSPAEGVPFVRATADCTLSSLNY 123
Query: 118 ALQPPFPCFQEILCDV--PGSEYIIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQ 175
+Q + CDV P +Y P+ L Q+T CGG LA +L+H++ DG G Q
Sbjct: 124 LKDMDTDLYQLVPCDVAAPSGDY---NPLAL-QITLFACGGLTLATALSHSLCDGFGASQ 179
Query: 176 FVSAWAEIARGANQPSIQPVWNREILMARDPPYITCNHREYEQILP-------------P 222
F E+A G QPSI PVW+R L + + + N + E P
Sbjct: 180 FFKTLTELAAGKTQPSIIPVWDRHRLTSNN---FSLNDQVEEGQAPKLVDFGEACSSAAT 236
Query: 223 NTYIKEEDTTIVVHQSFFFTSAHIAAIRRLVPLHLSRCTAYDLITACYWCCRTKALQVEP 282
+ Y D +V + TS I ++ V T +++ A W R +AL++ P
Sbjct: 237 SPYTPSND---MVCEILNVTSEDITQLKEKV---AGVVTTLEILAAHVWRARCRALKLSP 290
Query: 283 DEDVRMMCIVNARSRFSANNRSSLIGYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAK 342
D V R GYYGN + A EL + L + V+LI++AK
Sbjct: 291 DGTSLFGMAVGIRRIVEPPLPE---GYYGNAFVKANVAMKAGELSNSPLSHVVQLIKEAK 347
Query: 343 AQVTEK-----YMHSLADFMVIKERCLFTKGRTCMVSDWTR-AKFSEVNFGWGEAV 392
EK + + + C KG +++DW + E++FG+G +V
Sbjct: 348 KAAQEKRYVLEQLRETEKTLKMNVACEGGKGAFMLLTDWRQLGLLDEIDFGYGGSV 403
>At5g57840 N-hydroxycinnamoyl/benzoyltransferase
Length = 443
Score = 142 bits (357), Expect = 5e-34
Identities = 120/452 (26%), Positives = 209/452 (45%), Gaps = 46/452 (10%)
Query: 11 VRRSQPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSMKEKDPVKVLKHAL 70
+R Q +V PA TP LS++D + +R ++ ++ ++ S ++ + L AL
Sbjct: 3 IRVKQATIVKPAEETPTHNLWLSNLDLIQ-VRLHMGTLYFYK-PCSSSDRPNTQSLIEAL 60
Query: 71 SRTLVYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQFGDALQPPFPCFQEIL 130
S+ LV++YP AGR+++ +L V C GEGV+F+EAE+D T+ G Q ++
Sbjct: 61 SKVLVFFYPAAGRLQKNTNGRLEVQCNGEGVLFVEAESDSTVQDIGLLTQS-----LDLS 115
Query: 131 CDVPGSEY---IIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSAWAEIARGA 187
VP +Y I P+ L QVT KCG + S++HT G+ + L + AW+ ARG
Sbjct: 116 QLVPTVDYAGDISSYPLLLFQVTYFKCGTICVGSSIHHTFGEATSLGYIMEAWSLTARGL 175
Query: 188 NQPSIQPVWNREILMARDPPYITCNHREYEQILPPNTYIK--------EEDTTIVVHQSF 239
+ P +R +L AR+PP H EY+ N +K E D+ I +
Sbjct: 176 -LVKLTPFLDRTVLHARNPPSSVFPHTEYQSPPFHNHPMKSLAYRSNPESDSAIA---TL 231
Query: 240 FFTSAHIAAIRRLVPLHLSRCTAYDLITACYWCCRTKALQ-VEPDEDVRMMCIVNARSRF 298
T + A++ + S+ + Y+++ A W C + A + + + R+ I++ R R
Sbjct: 232 KLTRLQLKALKARAEIADSKFSTYEVLVAHTWRCASFANEDLSEEHSTRLHIIIDGRPRL 291
Query: 299 SANNRSSLIGYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAKAQVTEKYMHSLADFMV 358
GY GN + + V+ VE + + ++ +Y+ S D++
Sbjct: 292 QPKLPQ---GYIGNTLFHARPVSPLGAFHRESFSETVERVHREIRKMDNEYLRSAIDYLE 348
Query: 359 IK---ERCLFTKGRT--------CMVSDWTRAKFSEVNFGWGEAVYGGAAKGGIG-SFLG 406
++ + +G T C+VS + + E++FGWG+A Y A+ G F+
Sbjct: 349 RHPDLDQLVPGEGNTIFSCAANFCIVSLIKQTAY-EMDFGWGKAYYKRASHLNEGKGFVT 407
Query: 407 GTFLVTHKNAKGEEGLILPICLPPEDMKRFVK 438
G N E ++L +CL + +F K
Sbjct: 408 G-------NPDEEGSMMLTMCLKKTQLSKFRK 432
>At1g28680 anthranilate N-hydroxycinnamoyl/benzoyltransferase like
protein
Length = 451
Score = 137 bits (346), Expect = 1e-32
Identities = 108/372 (29%), Positives = 180/372 (48%), Gaps = 22/372 (5%)
Query: 32 LSDIDDQEGLRFNLPVIFVFRHEPS-MKEKDPVKVLKHALSRTLVYYYPGAGRIREGAG- 89
LS +D+ L + + V+ S + + P V+ +L+ LV+YYP AG +R A
Sbjct: 27 LSHLDNDNNLHVSFRYLRVYSSSSSTVAGESPSAVVSASLATALVHYYPLAGSLRRSASD 86
Query: 90 -RKLMVDCTGEGVMFIEAEADITLDQFGDALQPPFPCFQEILCDVPGSEYIIDRPIRLIQ 148
R ++ G+ V + A + TL+ G L P P F E L P E + P ++Q
Sbjct: 87 NRFELLCSAGQSVPLVNATVNCTLESVG-YLDGPDPGFVERLVPDPTREEGMVNPC-ILQ 144
Query: 149 VTRLKCGGFILALSLNHTMGDGSGLRQFVSAWAEIARGANQPSIQPVWNREILMA-RDPP 207
VT +CGG++L S++H + DG G F +A AE+ARGA + SI+PVW+RE L+ R+ P
Sbjct: 145 VTMFQCGGWVLGASIHHAICDGLGASLFFNAMAELARGATKISIEPVWDRERLLGPREKP 204
Query: 208 YITCNHREY----EQILPPNTYIKEEDTTIVVHQSFFFTSAHIAAIR-RLVPLHLSRCTA 262
++ R++ + P I + V FF T + ++ +L+ T
Sbjct: 205 WVGAPVRDFLSLDKDFDPYGQAIGD-----VKRDCFFVTDDSLDQLKAQLLEKSGLNFTT 259
Query: 263 YDLITACYWCCRTKALQVEPDEDVRMMCIVNARSRFSANNRSSLIGYYGN-CIAFSAAVT 321
++ + A W + +A + E E+V+ + +N R N GY+GN C+ A +
Sbjct: 260 FEALGAYIWRAKVRAAKTEEKENVKFVYSINIRRLM---NPPLPKGYWGNGCVPMYAQI- 315
Query: 322 TAKELCGNQLGYAVELIRKAKAQVTEKYMHSLADFMVIKERCLFTKGR-TCMVSDWTRAK 380
A EL + ELI+++K+ +++Y+ S DF + + G +DW
Sbjct: 316 KAGELIEQPIWKTAELIKQSKSNTSDEYVRSFIDFQELHHKDGINAGTGVTGFTDWRYLG 375
Query: 381 FSEVNFGWGEAV 392
S ++FGWG V
Sbjct: 376 HSTIDFGWGGPV 387
>At3g47170 hypersensitivity-related protein-like protein
Length = 447
Score = 134 bits (336), Expect = 1e-31
Identities = 125/473 (26%), Positives = 208/473 (43%), Gaps = 74/473 (15%)
Query: 9 FTVRRSQPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSMKEKD---PVKV 65
F V +S+ LV P+ PTP LS ID+ L ++ VF P +++ P
Sbjct: 10 FVVEKSEVVLVKPSKPTPDVTLSLSTIDNDPNNEVILDILCVFSPNPYVRDHTNYHPASF 69
Query: 66 LKHALSRTLVYYYPGAGRI-REGAGRKLMVDCT-GEGVMFIEAEADITLDQF-----GDA 118
++ ALS LVYYYP AG++ R + + L +DCT G+GV I+A A TL G+
Sbjct: 70 IQLALSNALVYYYPLAGKLHRRTSDQTLQLDCTQGDGVPLIKATASCTLSSLNYLESGNH 129
Query: 119 LQPPFPCFQEILCDVPGSEYIID-----RPIRLIQVTRLKCGGFILALSLNHTMGDGSGL 173
L+ + VP + + RP+ L Q+T+ CGG + +HT+ DG G+
Sbjct: 130 LEATYQL-------VPSHDTLKGCNLGYRPLAL-QITKFTCGGMTIGSVHSHTVCDGIGM 181
Query: 174 RQFVSAWAEIARGANQPSIQPVWNREILMARDPPYI--TCNHREYEQILPPNTYIKEEDT 231
QF A E+A G QP++ PVW+R ++ + + N + +++ DT
Sbjct: 182 AQFSQAILELAAGRAQPTVIPVWDRHMITSNQISTLCKLGNDKNNPKLVDVEKDCSSPDT 241
Query: 232 TI--VVHQSFFFTSAHIAAIRRLV---------PLHLSRCTAYDLITACYWCCRTKALQV 280
+V + TS I ++ ++ + T +++ A W R +A+++
Sbjct: 242 PTEDMVREILNITSEDITKLKNIIIEDENLTNEKEKNMKITTVEVLAAYVWRARCRAMKL 301
Query: 281 EPDEDVRMMCIVNARSRFSANNRSSLIGYYGNCIAFSAAVTTAKELCGNQLGYAVELIRK 340
PD ++ V+ RS GYYGN ++ TAKEL + V LI+
Sbjct: 302 NPDTITDLVISVSIRSSIEPPLPE---GYYGNAFTHASVALTAKELSKTPISRLVRLIKD 358
Query: 341 AKAQVTE-----KYMHSLADFMVIKERCLFTK----GRTCMVSDWTRAKFSEVNFGWGEA 391
AK + + + + + M +K L +K G M++DW + +G +A
Sbjct: 359 AKRAALDNGYVCEQLREMENTMKLK---LASKEIHGGVFMMLTDWRHLGLDQDVWGASKA 415
Query: 392 VYGGAAKGGIGSFLGGTFLVTHKNAKGEEGLILPICLPPEDMKRFVKELDDML 444
V G + GG ++T LP + M +F +E+D +L
Sbjct: 416 VPGKS---------GGVRVLT--------------TLPRDAMAKFKEEIDAVL 445
>At2g25150 unknown protein
Length = 461
Score = 121 bits (304), Expect = 7e-28
Identities = 129/474 (27%), Positives = 202/474 (42%), Gaps = 64/474 (13%)
Query: 6 PLLFTVRRSQP-ELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSMKEKDPVK 64
P+L + +P ELV P+ T E LS +D+ I+VF+ + DPV
Sbjct: 7 PILPLLLEKKPVELVKPSKHTHCETLSLSTLDNDPFNEVMYATIYVFKANGKNLD-DPVS 65
Query: 65 VLKHALSRTLVYYYPGAGRI-REGAGRKLMVDCTGEGVMFIEAEADITL----------D 113
+L+ ALS LV+YYP +G++ R + KL + GEGV F A + + L D
Sbjct: 66 LLRKALSELLVHYYPLSGKLMRSESNGKLQLVYLGEGVPFEVATSTLDLSSLNYIENLDD 125
Query: 114 QFGDALQPPFPCFQEILCDVPGSEYIIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGL 173
Q L P EI D + + P+ L QVT+ CGGF + +L H + DG G+
Sbjct: 126 QVALRLVP------EIEIDYESN--VCYHPLAL-QVTKFACGGFTIGTALTHAVCDGYGV 176
Query: 174 RQFVSAWAEIARGANQPSIQPVWNREILMAR---DPPYITCNHREYEQILPPNTYIKEED 230
Q + A E+A G +PS++ VW RE L+ + P + +H + L + Y+
Sbjct: 177 AQIIHALTELAAGKTEPSVKSVWQRERLVGKIDNKPGKVPGSH--IDGFLATSAYL---P 231
Query: 231 TTIVVHQSFFFTSAHIAAIRRLVPLHLSRC-------TAYDLITACYWCCRTKALQVEPD 283
TT VV ++ + I+RL + C T Y+++++ W R++AL++ PD
Sbjct: 232 TTDVVTETI---NIRAGDIKRLKDSMMKECEYLKESFTTYEVLSSYIWKLRSRALKLNPD 288
Query: 284 EDVRMMCIVNARSRFSANNRSSLIGYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAKA 343
+ V R GYYGN T +EL + + ++KAK
Sbjct: 289 GITVLGVAVGIRHVLDPPLPK---GYYGNAYIDVYVELTVRELEESSISNIANRVKKAKK 345
Query: 344 QV------------TEKYMHSLADFMVIKERCLFTKGRTCMVSDWTR-AKFSEVNFGWGE 390
TE+ M + F + + F ++DW F ++FGW E
Sbjct: 346 TAYEKGYIEEELKNTERLMRDDSMFEGVSDGLFF-------LTDWRNIGWFGSMDFGWNE 398
Query: 391 AVYGGAAKGGIGSFLGGTFLVTHKNAKGEEGLILPIC-LPPEDMKRFVKELDDM 443
V + G L K+ EG + I LP + M F +E+ M
Sbjct: 399 PVNLRPLTQRESTVHVGMILKPSKSDPSMEGGVKVIMKLPRDAMVEFKREMATM 452
>At2g23510 putative anthranilate
N-hydroxycinnamoyl/benzoyltransferase
Length = 451
Score = 121 bits (304), Expect = 7e-28
Identities = 117/413 (28%), Positives = 183/413 (43%), Gaps = 44/413 (10%)
Query: 3 SSSPLLFTVRRSQPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSMKEKD- 61
SS PL+ V + E+V P+ P + LS +D+ +VF+ + + +
Sbjct: 7 SSIPLM--VEKMLTEMVKPSKHIPQQTLNLSTLDNDPYNEVIYKACYVFKAKNVADDDNR 64
Query: 62 PVKVLKHALSRTLVYYYPGAGRI-REGAGRKLMVDCTGEG--VMFIEAEADITLDQFGDA 118
P +L+ ALS L YYYP +G + R+ + RKL + C G+G V F A A++ L +
Sbjct: 65 PEALLREALSDLLGYYYPLSGSLKRQESDRKLQLSCGGDGGGVPFTVATANVELSSLKNL 124
Query: 119 LQPPFPCFQEILCDVPGSEYIID--RPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQF 176
L P ID RP L QVT+ +CGGFIL ++++H M DG G
Sbjct: 125 ENIDSDTALNFL---PVLHVDIDGYRPFAL-QVTKFECGGFILGMAMSHAMCDGYGEGHI 180
Query: 177 VSAWAEIARGANQPSIQPVWNREILMAR----DPPYITCNHREYEQILPPNTYIKEEDTT 232
+ A ++A G +P + P+W RE L+ + PP++ + LP + ++ E+ T
Sbjct: 181 MCALTDLAGGKKKPMVTPIWERERLVGKPEDDQPPFVPGDDTAASPYLPTDDWVTEKIT- 239
Query: 233 IVVHQSFFFTSAHIAAIRRLVPLHLSR-------CTAYDLITACYWCCRTKALQVEPDED 285
+IRRL L T +++I A W R KAL ++ D
Sbjct: 240 -----------IRADSIRRLKEATLKEYDFSNETITTFEVIGAYLWKSRVKALNLDRDGV 288
Query: 286 VRMMCIVNARSRFSANNRSSLIGYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAKAQV 345
+ V R+ GYYGN TA+E+ + V+LI++AK
Sbjct: 289 TVLGLSVGIRNVVDPPLPD---GYYGNAYIDMYVPLTAREVEEFTISDIVKLIKEAKRNA 345
Query: 346 TEK--YMHSLADFMVIKERCLFTKGR---TCMVSDWTR-AKFSEVNFGWGEAV 392
+K LA+ I + L KG+ ++DW F ++FGW E V
Sbjct: 346 HDKDYLQEELANTEKIIKMNLTIKGKKDGLFCLTDWRNIGIFGSMDFGWDEPV 398
>At1g31490 putative protein
Length = 444
Score = 105 bits (261), Expect = 7e-23
Identities = 99/409 (24%), Positives = 181/409 (44%), Gaps = 47/409 (11%)
Query: 8 LFTVRRSQPELVLPAAPTPHEVKLL--SDIDDQEGLRFNLPVIFVFRHEPSMKEKDPVKV 65
+F V + +V + P +V L S+ D G RF + + + +P + + VK
Sbjct: 3 IFDVSFTGEFIVKASGDQPEKVNSLNLSNFDLLSG-RFPVTYFYFYPKQPQLSFETIVKS 61
Query: 66 LKHALSRTLVYYYPGAGRI--REGAGRKLMVDCTGEGVMFIEAEADITLDQFGDALQPPF 123
L+ +LS+TL Y+YP AG+I E + + M+ C G +F+EA A + L F
Sbjct: 62 LQSSLSQTLSYFYPFAGQIVPNETSQEEPMIICNNNGALFVEARAHVDLKSLDFYNLDAF 121
Query: 124 PCFQEILCDVPGSEYIIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSAWAEI 183
Q L V ++ + IQ T +CGG L + +H +GD S +F++ W+EI
Sbjct: 122 --LQSKLVQV-NPDFSLQ-----IQATEFECGGLALTFTFDHALGDASSFGKFLTLWSEI 173
Query: 184 ARGANQP-SIQPVWNREILMARDPP---------YITCNHREYEQILPPNTYIKEEDTTI 233
+R N+P S P R +L AR PP +I C+ + + I T IK
Sbjct: 174 SR--NKPVSCVPDHRRNLLRARSPPRYDPHLDKTFIKCSEEDIKNIPMSKTLIK------ 225
Query: 234 VVHQSFFFTSAHIAAIRRLVPLHLSRCTAYDLITACYWCCRTKALQVEPDEDVRMMCIVN 293
+ + ++ + A++ L ++ T + +A W +++ + +M +V+
Sbjct: 226 ---RLYHIGASSLDALQALATVNGESRTKIEAFSAYVWKKMVDSIE-SGHKTCKMGWLVD 281
Query: 294 ARSRFSANNRSSLIGYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAKAQVT-EKYMHS 352
R R S Y GN ++ + + + L N + ++ K+ +VT + +
Sbjct: 282 GRGRLETVTSS----YIGNVLSIAVGEASIENLKKNHVSDIANIVHKSITEVTNDTHFTD 337
Query: 353 LADF-------MVIKERCLFTKGRTCMVSDWTRAKFSEVNFGWGEAVYG 394
L D+ +++ L +G ++S R +E++FG+G G
Sbjct: 338 LIDWIESHRPGLMLARVVLGQEGPALVLSSGRRFPVAELDFGFGAPFLG 386
>At4g31910 unknown protein
Length = 458
Score = 103 bits (258), Expect = 2e-22
Identities = 105/416 (25%), Positives = 177/416 (42%), Gaps = 37/416 (8%)
Query: 48 IFVFRHEPSMKEKDPVKVLKHALSRTLVYYYPGAGRIR-EGAGRKLMVDCTGEGVMFIEA 106
+F +++ + LK L T+ +YP AGR+ +G G KL + C G + +EA
Sbjct: 46 VFFYKNTTTRDFDSVFSNLKLGLEETMSVWYPAAGRLGLDGGGCKLNIRCNDGGAVMVEA 105
Query: 107 EAD-ITLDQFGDALQPPFPCFQEILCDVPGSEYIID-RPIRLIQVTRLKCGGFILALSLN 164
A + L + GD Q + F E L P + P+ + QVTR CGG+ + + +
Sbjct: 106 VATGVKLSELGDLTQ--YNEFYENLVYKPSLDGDFSVMPLVVAQVTRFACGGYSIGIGTS 163
Query: 165 HTMGDGSGLRQFVSAWAEIARGANQPS-----------IQPVWNREILMARDPPYITCNH 213
H++ DG +F+ AWA + N+ + I+PV +R L+ N
Sbjct: 164 HSLFDGISAYEFIHAWASNSHIHNKSNSKITNKKEDVVIKPVHDRRNLLVNRDAVRETNA 223
Query: 214 RE----YEQILPPNTYIKEEDTTIVVHQSFFFTSA---HIAAIRRLVPLHLS--RCTAYD 264
Y+ I +E++ + + S F + AI + L C++++
Sbjct: 224 AAICHLYQLIKQAMMTYQEQNRNLELPDSGFVIKTFELNGDAIESMKKKSLEGFMCSSFE 283
Query: 265 LITACYWCCRTKALQVEPDEDVRMMCIVNARSRFSANNRSSLIGYYGNCIAFSAAVTTAK 324
+ A W RT+AL + D V + V+ R R G+ GN ++ +TA+
Sbjct: 284 FLAAHLWKARTRALGLRRDAMVCLQFAVDIRKRTETPLPE---GFSGNAYVLASVASTAR 340
Query: 325 ELCGN-QLGYAVELIRKAKAQVTEKYMHSLADFM-VIKERCLFTKGRTCMVSDWTRAKFS 382
EL L V IR+AK + + Y++S + + + L ++SDWT+ F
Sbjct: 341 ELLEELTLESIVNKIREAKKSIDQGYINSYMEALGGSNDGNLPPLKELTLISDWTKMPFH 400
Query: 383 EVNFGWGEAVYGGAAKGGIGSFLGGTFLVTH--KNAKGEEGLILPICLPPEDMKRF 436
V FG GG + V + KN K +G+++ I L P D+ F
Sbjct: 401 NVGFG-----NGGEPADYMAPLCPPVPQVAYFMKNPKDAKGVLVRIGLDPRDVNGF 451
>At1g24420 unknown protein
Length = 436
Score = 101 bits (252), Expect = 8e-22
Identities = 114/439 (25%), Positives = 190/439 (42%), Gaps = 50/439 (11%)
Query: 17 ELVLPAAPTPHEVKLL--SDIDDQEGLRFNLPVIFVFRHEPSM----KEKDPVKVLKHAL 70
E+V P++PTP + ++L S +D + ++F ++ E+ +K LK +L
Sbjct: 11 EIVKPSSPTPDDKRILNLSLLDILSSPMYTGALLFYAADPQNLLGFSTEETSLK-LKKSL 69
Query: 71 SRTLVYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQFGDALQPPFPCFQEIL 130
S+TL +YP AGRI V+C EG +FIEA D L +F L+ P P E+L
Sbjct: 70 SKTLPIFYPLAGRIIGS-----FVECNDEGAVFIEARVDHLLSEF---LKCPVPESLELL 121
Query: 131 CDVPG-SEYIIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSAWAEIARGANQ 189
V S + P+ LIQ CGG ++ + ++H + D + L F+ WAE +RG
Sbjct: 122 IPVEAKSREAVTWPVLLIQANFFSCGGLVITICVSHKITDATSLAMFIRGWAESSRGLGI 181
Query: 190 PSIQPVWNREILMARDPPYITCNHREYEQILPPNTYIKEEDTTIVVHQSFFFTSAHIAAI 249
I E+ P + E P +E + V + F F ++ I +
Sbjct: 182 TLIPSFTASEVF----PKPLD------ELPSKPMDRKEEVEEMSCVTKRFVFDASKIKKL 231
Query: 250 RRLVPLHL-SRCTAYDLITACYWCCRTKALQVE---PDEDVRMMCIVNARSRFSANNRSS 305
R +L T + +TA +W C TK ++ P V + +VN R + + ++
Sbjct: 232 RAKASRNLVKNPTRVEAVTALFWRCVTKVSRLSSLTPRTSV-LQILVNLRGKVDSLCENT 290
Query: 306 LIGYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAK----AQVTEKYMHSLADFMVIKE 361
+ GN + S + +E ++ V+ IR+AK E S F +++E
Sbjct: 291 I----GNML--SLMILKNEEAAIERIQDVVDEIRRAKEIFSLNCKEMSKSSSRIFELLEE 344
Query: 362 -RCLFTKGRTC---MVSDWTRAKFSEVNFGWGEAVYGGAAKGGIGSFLGGTFLVTHKNAK 417
++ +G M + W + + +FGWG+ V+ G F L+ K+ +
Sbjct: 345 IGKVYGRGNEMDLWMSNSWCKLGLYDADFGWGKPVW--VTGRGTSHFKNLMLLIDTKDGE 402
Query: 418 GEEGLILPICLPPEDMKRF 436
G E I L E M F
Sbjct: 403 GIEAW---ITLTEEQMSLF 418
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.139 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,850,289
Number of Sequences: 26719
Number of extensions: 476918
Number of successful extensions: 1263
Number of sequences better than 10.0: 65
Number of HSP's better than 10.0 without gapping: 59
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 1037
Number of HSP's gapped (non-prelim): 78
length of query: 462
length of database: 11,318,596
effective HSP length: 103
effective length of query: 359
effective length of database: 8,566,539
effective search space: 3075387501
effective search space used: 3075387501
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 62 (28.5 bits)
Medicago: description of AC148345.12


