

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148343.5 - phase: 0
(443 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
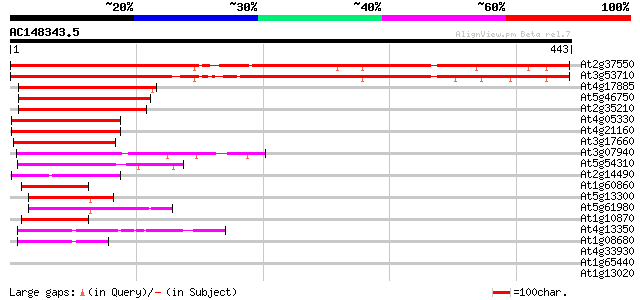
Score E
Sequences producing significant alignments: (bits) Value
At2g37550 Asp1 (asp1) 543 e-155
At3g53710 ARF GAP-like zinc finger-containing protein ZIGA2 526 e-150
At4g17885 unknown protein 122 3e-28
At5g46750 zinc finger protein Glo3-like 121 9e-28
At2g35210 unknown protein 120 2e-27
At4g05330 unknown protein 97 2e-20
At4g21160 unknown protein 93 3e-19
At3g17660 unknown protein 89 4e-18
At3g07940 putative GTPase activating protein 89 4e-18
At5g54310 unknown protein 89 5e-18
At2g14490 unknown protein 73 3e-13
At1g60860 GCN4-complementing like protein 72 5e-13
At5g13300 putative protein 72 6e-13
At5g61980 GCN4-complementing protein - like 71 1e-12
At1g10870 BRCA1-associated RING domain protein isolog 68 9e-12
At4g13350 unknown protein 55 6e-08
At1g08680 unknown protein 52 7e-07
At4g33930 putative protein 37 0.022
At1g65440 35 0.064
At1g13020 eukaryotic initiation factor 4B (EIF4B5) 35 0.064
>At2g37550 Asp1 (asp1)
Length = 456
Score = 543 bits (1400), Expect = e-155
Identities = 287/469 (61%), Positives = 340/469 (72%), Gaps = 41/469 (8%)
Query: 1 MAASRRLRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVT 60
MAA+RRLR LQS+P NK+CVDCSQKNPQWAS+SYG+FMCLECSGKHRGLGVHISFVRSVT
Sbjct: 1 MAAARRLRTLQSQPENKVCVDCSQKNPQWASISYGIFMCLECSGKHRGLGVHISFVRSVT 60
Query: 61 MDSWSDLQIKKMEAGGNRNLNTFLSQYGISKETDIITKYNSNAASIYRDRIQAIAEGRSW 120
MDSWS++QIKKM+AGGN LN FL+QYGISKETDII+KYNSNAAS+YRDRIQA+AEGR W
Sbjct: 61 MDSWSEIQIKKMDAGGNERLNNFLAQYGISKETDIISKYNSNAASVYRDRIQALAEGRQW 120
Query: 121 RDPPVVKENASTRAGKGKPPLAAA----SNGGGWDDNWDNDDGDSYGYGSRGGGDIRRNQ 176
RDPP+VKE+ KPPL+ S GGW DNWDNDD S D+RRNQ
Sbjct: 121 RDPPIVKESVGGGLMNKKPPLSQGGGRDSGNGGW-DNWDNDD-------SFRSTDMRRNQ 172
Query: 177 STGDVIGFVGNGVTPSSRSKSTEDIYTRTQLEASAANKEGFFAKKMAENESRPEGLPPSQ 236
S GD G G ++SKS+EDIY+R+QLEASAANKE FFAK+MAENES+PEGLPPSQ
Sbjct: 173 SAGDFRSSGGRGA--PAKSKSSEDIYSRSQLEASAANKESFFAKRMAENESKPEGLPPSQ 230
Query: 237 GGKYVGFGSSPGPAQRISPQN---DYLSVVSEGIGKLSMVAQSA-------TKEITAKVK 286
GGKYVGFGSSPGPA R + Q+ D SV+SEG G+LS+VA SA T E T+KVK
Sbjct: 231 GGKYVGFGSSPGPAPRSNQQSGGGDVFSVMSEGFGRLSLVAASAANVVQTGTMEFTSKVK 290
Query: 287 DGGYDHKVNETVNIVTQKTSEIGQRTWGIMKGVMALASQKVEEFTKDYPDGNSDNWPRNE 346
+GG D V+ETVN+V KT+EIGQRTWGIMKGVMA+ASQKVEEFTK+ + W +
Sbjct: 291 EGGLDQTVSETVNVVASKTTEIGQRTWGIMKGVMAIASQKVEEFTKE----EASTWNQQN 346
Query: 347 NDRHDFNQENKGWNSSTSTRE---GQPSSGGQTNTY-HSNSWDDW-DNQDTRKEVPAKGS 401
+ +N G + T+ Q SS G N+Y +SNSWDDW + +++KE K S
Sbjct: 347 KTEGNGYYQNSGIGNKTANSSFGGSQSSSSGHNNSYRNSNSWDDWGEENNSKKEAAPKVS 406
Query: 402 APHNNDD--WAGWDDAKDDDDEF------DDKSFGNNGTSGSGWTGGGF 442
+++DD WAGWDD DD+F D KS G+NG S + WTGGGF
Sbjct: 407 TSNDDDDGGWAGWDDNDAKDDDFYYQPASDKKSVGHNGKSDTAWTGGGF 455
>At3g53710 ARF GAP-like zinc finger-containing protein ZIGA2
Length = 459
Score = 526 bits (1356), Expect = e-150
Identities = 288/478 (60%), Positives = 336/478 (70%), Gaps = 56/478 (11%)
Query: 1 MAASRRLRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVT 60
MAA+R+LR LQS+P NK+CVDC+QKNPQWASVSYG+FMCLECSGKHRGLGVHISFVRSVT
Sbjct: 1 MAATRQLRTLQSQPENKVCVDCAQKNPQWASVSYGIFMCLECSGKHRGLGVHISFVRSVT 60
Query: 61 MDSWSDLQIKKMEAGGNRNLNTFLSQYGISKETDIITKYNSNAASIYRDRIQAIAEGRSW 120
MDSWS +QIKKMEAGGN LN F +QYGI+KETDII+KYNSNAAS+YRDRIQA+AEGR W
Sbjct: 61 MDSWSAIQIKKMEAGGNERLNKFFAQYGIAKETDIISKYNSNAASVYRDRIQALAEGRPW 120
Query: 121 RDPPVVKENASTRAGKGKPPLAAA-------SNGGGWDDNWDNDDGDSYGYGSRGGGDIR 173
DPPVVKE KPPLA +N GGW D+WDND DSY D+R
Sbjct: 121 NDPPVVKE------ANKKPPLAQGGYGNNNNNNNGGW-DSWDND--DSY------KSDMR 165
Query: 174 RNQSTGDVIGFVGNGVTPSSRSKSTEDIYTRTQLEASAANKEGFFAKKMAENESRPEGLP 233
RNQS D GN +SKS+EDIYTR+QLEASAA KE FFA++MAENES+PEGLP
Sbjct: 166 RNQSAND-FRASGNREGAHVKSKSSEDIYTRSQLEASAAGKESFFARRMAENESKPEGLP 224
Query: 234 PSQGGKYVGFGSSPGPAQRISPQNDYLSVVSEGIGKLSMVAQSA-----------TKEIT 282
PSQGGKYVGFGSS P R + Q+D SVVS+G G+LS+VA SA TKE T
Sbjct: 225 PSQGGKYVGFGSSSAPPPRNNQQDDVFSVVSQGFGRLSLVAASAAQSAASVVQTGTKEFT 284
Query: 283 AKVKDGGYDHKVNETVNIVTQKTSEIGQRTWGIMKGVMALASQKVEEFTKDYPDGNSDNW 342
+KVK+GGYDHKV+ETVN+V KT+EIG RTWGIMKGVMA+A+QKVEEFTK+ S +W
Sbjct: 285 SKVKEGGYDHKVSETVNVVANKTTEIGHRTWGIMKGVMAMATQKVEEFTKE----GSTSW 340
Query: 343 -PRNENDRH----DFNQENKGWNSSTSTREGQPS--SGGQTNTYHSNSWDDWDNQDTRK- 394
++EN+ + +F NK NSS Q S SG N+ +SNSWD W + +K
Sbjct: 341 NQQSENEGNGYYQNFGNGNKAANSSVGGGRPQSSSTSGHYNNSQNSNSWDSWGENENKKT 400
Query: 395 -EVPAKGSAPHNNDD-WAGWDDAKDDDDEF--------DDKSFGNNGTSGSGWTGGGF 442
V KGS+ N+DD W GWDD DD F D KS G+NG S + WTGGGF
Sbjct: 401 EAVAPKGSSASNDDDGWTGWDDHDAKDDGFDGHYQSAGDKKSAGHNGKSDTAWTGGGF 458
>At4g17885 unknown protein
Length = 413
Score = 122 bits (307), Expect = 3e-28
Identities = 58/114 (50%), Positives = 75/114 (64%), Gaps = 5/114 (4%)
Query: 8 RELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMDSWSDL 67
R+L+S+ NK+C DCS KNP WASV+YG+F+C++CS HR LGVHISFVRS +DSWS
Sbjct: 17 RKLKSKSENKVCFDCSAKNPTWASVTYGIFLCIDCSATHRNLGVHISFVRSTNLDSWSPE 76
Query: 68 QIKKMEAGGNRNLNTFLSQYGISKETDIITKYNSNAASIYRDRI-----QAIAE 116
Q++ M GGN F Q+G + I KY S AA +YR + +AIAE
Sbjct: 77 QLRTMMFGGNNRAQVFFKQHGWTDGGKIEAKYTSRAADLYRQILAKEVAKAIAE 130
>At5g46750 zinc finger protein Glo3-like
Length = 402
Score = 121 bits (303), Expect = 9e-28
Identities = 54/104 (51%), Positives = 69/104 (65%)
Query: 8 RELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMDSWSDL 67
R+L+S+ NK+C DCS KNP WASV YG+F+C++CS HR LGVHISFVRS +DSWS
Sbjct: 14 RKLKSKSENKVCFDCSAKNPTWASVPYGIFLCIDCSAVHRSLGVHISFVRSTNLDSWSPE 73
Query: 68 QIKKMEAGGNRNLNTFLSQYGISKETDIITKYNSNAASIYRDRI 111
Q++ M GGN F Q+G + I KY S AA +YR +
Sbjct: 74 QLRTMMFGGNNRAQVFFKQHGWNDGGKIEAKYTSRAADMYRQTL 117
>At2g35210 unknown protein
Length = 395
Score = 120 bits (300), Expect = 2e-27
Identities = 53/101 (52%), Positives = 68/101 (66%)
Query: 8 RELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMDSWSDL 67
++L+++ NKIC DC+ KNP WASV+YG+F+C++CS HR LGVHISFVRS +DSWS
Sbjct: 14 KKLKAKSDNKICFDCNAKNPTWASVTYGIFLCIDCSAVHRSLGVHISFVRSTNLDSWSSE 73
Query: 68 QIKKMEAGGNRNLNTFLSQYGISKETDIITKYNSNAASIYR 108
Q+K M GGN F QYG S KY S AA +Y+
Sbjct: 74 QLKMMIYGGNNRAQVFFKQYGWSDGGKTEAKYTSRAADLYK 114
>At4g05330 unknown protein
Length = 336
Score = 97.1 bits (240), Expect = 2e-20
Identities = 41/87 (47%), Positives = 59/87 (67%), Gaps = 1/87 (1%)
Query: 2 AASRRLRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTM 61
+ RR+R+L ++P N++C DC +P+WAS + GVF+CL+C G HR LG HIS V SVT+
Sbjct: 13 SGKRRIRDLLNQPDNRVCADCGASDPKWASANIGVFICLKCCGVHRSLGTHISKVLSVTL 72
Query: 62 DSWSDLQIKKM-EAGGNRNLNTFLSQY 87
D WSD ++ M E GGN + N+ +
Sbjct: 73 DEWSDEEVDSMIEIGGNASANSIYEAF 99
>At4g21160 unknown protein
Length = 337
Score = 92.8 bits (229), Expect = 3e-19
Identities = 40/87 (45%), Positives = 58/87 (65%), Gaps = 1/87 (1%)
Query: 2 AASRRLRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTM 61
+ RR+R+L ++ N++C DC +P+WAS + GVF+CL+C G HR LG HIS V SVT+
Sbjct: 13 SGKRRIRDLLTQSDNRVCADCGAPDPKWASANIGVFICLKCCGVHRSLGSHISKVLSVTL 72
Query: 62 DSWSDLQIKKM-EAGGNRNLNTFLSQY 87
D WSD ++ M E GGN + N+ +
Sbjct: 73 DEWSDEEVDSMIEIGGNASANSIYEAF 99
>At3g17660 unknown protein
Length = 117
Score = 89.4 bits (220), Expect = 4e-18
Identities = 39/80 (48%), Positives = 54/80 (66%)
Query: 4 SRRLRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMDS 63
S+ L L P N+ C DC K P+WASV+ G+F+C++CSG HR LGVHIS VRS+T+D+
Sbjct: 16 SKILEALLKHPDNRECADCRSKAPRWASVNLGIFICMQCSGIHRSLGVHISQVRSITLDT 75
Query: 64 WSDLQIKKMEAGGNRNLNTF 83
W Q+ M++ GN N +
Sbjct: 76 WLPDQVAFMKSTGNAKGNEY 95
>At3g07940 putative GTPase activating protein
Length = 385
Score = 89.4 bits (220), Expect = 4e-18
Identities = 66/216 (30%), Positives = 97/216 (44%), Gaps = 31/216 (14%)
Query: 6 RLRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMDSWS 65
RL +L +P NK C DC P+W S+S GVF+C++CSG HR LGVHIS V SV +D W+
Sbjct: 49 RLEKLLKQPGNKYCADCGSPEPKWVSLSLGVFICIKCSGVHRSLGVHISKVLSVKLDEWT 108
Query: 66 DLQIKKMEA-GGNRNLNTFLSQYGISKETDIITKYNSNAASIYR-DRIQAIAEGRSWRDP 123
D Q+ + GGN +N I D K ++ + R D I+ E + DP
Sbjct: 109 DDQVDMLVGYGGNTAVNERFEACNI----DQSKKPKPDSTNEERNDFIRKKYEQHQFMDP 164
Query: 124 ---PVVKENASTRAGKGKPPLAAASN-------GGGWDDNWDNDDGDSYGYGSRGGGDIR 173
+ +R P L +AS+ G + ++W + D G +
Sbjct: 165 KDGALCTYQQPSRTNTSPPSLCSASHRSTKNRIGHAFRNSWGRRESDHKG--------PK 216
Query: 174 RNQSTGDVIGFVG-------NGVTPSSRSKSTEDIY 202
++ S ++ FVG G + R T D Y
Sbjct: 217 KSNSMAGMVEFVGLIKVNVVKGTNLAVRDVMTSDPY 252
>At5g54310 unknown protein
Length = 483
Score = 89.0 bits (219), Expect = 5e-18
Identities = 48/138 (34%), Positives = 74/138 (52%), Gaps = 14/138 (10%)
Query: 7 LRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMDSWSD 66
L L P N+ C DC K P+WASV+ G+F+C++CSG HR LGVHIS VRS T+D+W
Sbjct: 19 LEGLLKHPENRECADCKTKGPRWASVNLGIFICMQCSGIHRSLGVHISKVRSATLDTWLP 78
Query: 67 LQIKKMEAGGNRNLNTFLSQYGISKETDIITKYN----SNAASIYRDRIQAIAEGRSWRD 122
Q+ +++ GN N++ E ++ Y+ N + + ++ G R
Sbjct: 79 EQVAFIQSMGNDKANSYW-------EAELPPNYDRVGIENFIRAKYEEKRWVSRGEKARS 131
Query: 123 PPVVKE---NASTRAGKG 137
PP V++ + R+G G
Sbjct: 132 PPRVEQERRKSVERSGPG 149
>At2g14490 unknown protein
Length = 119
Score = 73.2 bits (178), Expect = 3e-13
Identities = 35/87 (40%), Positives = 50/87 (57%), Gaps = 3/87 (3%)
Query: 2 AASRRLRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTM 61
+ RR+R+L ++P N++C DC P+WA Y L+ G HR LG HI V SVT+
Sbjct: 13 SGKRRIRDLLNQPDNRVCADCGASVPKWAK--YQSIHLLKSCGVHRSLGTHILKVLSVTL 70
Query: 62 DSWSDLQIKKM-EAGGNRNLNTFLSQY 87
D WSD ++ M E GGN + N+ +
Sbjct: 71 DEWSDEEVDSMIETGGNASANSIYEAF 97
>At1g60860 GCN4-complementing like protein
Length = 776
Score = 72.4 bits (176), Expect = 5e-13
Identities = 30/53 (56%), Positives = 39/53 (72%)
Query: 10 LQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMD 62
L+ P N C +C+ +P WAS++ GV MC+ECSG HR LGVHIS VRS+T+D
Sbjct: 473 LREIPGNNTCAECNAPDPDWASLNLGVLMCIECSGVHRNLGVHISKVRSLTLD 525
>At5g13300 putative protein
Length = 750
Score = 72.0 bits (175), Expect = 6e-13
Identities = 35/69 (50%), Positives = 42/69 (60%), Gaps = 2/69 (2%)
Query: 16 NKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMD--SWSDLQIKKME 73
N C DC P WAS++ GV +C+ECSG HR LGVHIS VRS+T+D W I +
Sbjct: 436 NDKCADCGAPEPDWASLNLGVLVCIECSGVHRNLGVHISKVRSLTLDVKVWEPSVISLFQ 495
Query: 74 AGGNRNLNT 82
A GN NT
Sbjct: 496 ALGNTFANT 504
>At5g61980 GCN4-complementing protein - like
Length = 850
Score = 70.9 bits (172), Expect = 1e-12
Identities = 41/115 (35%), Positives = 59/115 (50%), Gaps = 3/115 (2%)
Query: 16 NKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMD--SWSDLQIKKME 73
N+ C DC P WAS++ GV +C+ECSG HR LGVHIS VRS+T+D W + +
Sbjct: 532 NERCADCGAPEPDWASLNLGVLICIECSGIHRNLGVHISKVRSLTLDVKVWEPSVLTLFQ 591
Query: 74 AGGNRNLNTFLSQYGISKETDIITKYNSNAASIYRDRIQAIAEGRSWRDPPVVKE 128
+ GN +N+ + S+ +S R R + + + DP VKE
Sbjct: 592 SLGNVYVNSVWEELLNSESRTSSASRSSGTPKSDRPR-KLLVRKPGFNDPISVKE 645
>At1g10870 BRCA1-associated RING domain protein isolog
Length = 531
Score = 68.2 bits (165), Expect = 9e-12
Identities = 27/53 (50%), Positives = 38/53 (70%)
Query: 10 LQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMD 62
L+ P N C +C+ P WAS++ GV +C++CSG HR LGVHIS VRS+++D
Sbjct: 229 LRGLPGNNACAECNAPEPDWASLNLGVLLCIQCSGVHRNLGVHISKVRSLSLD 281
>At4g13350 unknown protein
Length = 602
Score = 55.5 bits (132), Expect = 6e-08
Identities = 41/165 (24%), Positives = 74/165 (44%), Gaps = 14/165 (8%)
Query: 7 LRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMDSWSD 66
+R L P NK C++C+ PQ+ ++ F+C CSG HR V+S++M ++
Sbjct: 15 IRSLLKLPENKRCINCNSLGPQYVCTTFWTFVCTNCSGIHREF---THRVKSISMAKFTS 71
Query: 67 LQIKKMEAGGNRNLNTFLSQYGISKETDIITKYNSNAASIYRDRIQAI-AEGRSWRDPPV 125
++ ++ GGN++ + G+ ++ + SN + RD I+ + R +
Sbjct: 72 QEVTALKEGGNQHAKDIYFK-GLDQQRQSVPD-GSNVERL-RDFIRHVYVNKRYTNEKND 128
Query: 126 VKENASTRAGKGKPPLAAASNGGGWDDNWDNDDGDSYGYGSRGGG 170
K + TR+ G S ++D +D GD G R G
Sbjct: 129 DKSPSETRSSSG-------SRSPPYEDGYDRRYGDRSSPGGRSPG 166
>At1g08680 unknown protein
Length = 159
Score = 52.0 bits (123), Expect = 7e-07
Identities = 22/72 (30%), Positives = 42/72 (57%), Gaps = 3/72 (4%)
Query: 7 LRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMDSWSD 66
+R L P N+ C++C+ PQ+ ++ F+C+ CSG HR V+SV+M ++
Sbjct: 15 IRGLMKLPPNRRCINCNSLGPQYVCTTFWTFVCMACSGIHREF---THRVKSVSMSKFTS 71
Query: 67 LQIKKMEAGGNR 78
+++ ++ GGN+
Sbjct: 72 KEVEVLQNGGNQ 83
>At4g33930 putative protein
Length = 343
Score = 37.0 bits (84), Expect = 0.022
Identities = 30/116 (25%), Positives = 44/116 (37%), Gaps = 8/116 (6%)
Query: 117 GRSWRDPPVVKENASTRAGKGKPPLAAASNGGGWDDNWDNDDGD-SYGYGSRGGGDIRRN 175
G WR N+ +G G + S G W W +D D ++G+GS G + +
Sbjct: 42 GWGWRSG-----NSGGSSGSGSGGSDSNSGGSSWGWGWSSDGTDTNWGWGSSSGSN--HS 94
Query: 176 QSTGDVIGFVGNGVTPSSRSKSTEDIYTRTQLEASAANKEGFFAKKMAENESRPEG 231
TG +G SS + ST + +T T S+ N N S G
Sbjct: 95 SGTGSTHNGHSSGSNHSSATGSTHNGHTSTGSNHSSGNGSRHNGYSSGSNHSSSTG 150
>At1g65440
Length = 1703
Score = 35.4 bits (80), Expect = 0.064
Identities = 26/111 (23%), Positives = 40/111 (35%), Gaps = 8/111 (7%)
Query: 328 EEFTKDYPDGNSDNWPRNENDRHDFNQENKGWNSSTSTREGQPSSGGQTNTYHSNSWDDW 387
++ D DGN D W N+ D N G S G+ + GG T + S S +
Sbjct: 1554 DDMNSDRQDGNGD-WGNNDTGTADGGWGNSGGGGWGSESAGKKTGGGSTGGWGSESGGN- 1611
Query: 388 DNQDTRKEVPAKGSAPHNNDDWAGWDDAKDDDDEFDDKSFGN-NGTSGSGW 437
+ + + GW + +D FG+ +G GS W
Sbjct: 1612 -----KSDGAGSWGSGSGGGGSGGWGNDSGGKKSSEDGGFGSGSGGGGSDW 1657
>At1g13020 eukaryotic initiation factor 4B (EIF4B5)
Length = 549
Score = 35.4 bits (80), Expect = 0.064
Identities = 18/52 (34%), Positives = 26/52 (49%), Gaps = 7/52 (13%)
Query: 324 SQKVEEFTKDYPDGNSDNWPRNENDRHDFNQ-------ENKGWNSSTSTREG 368
S+ VE + G DNWPR +DR +F N+ ++ S+S REG
Sbjct: 497 SRSVESMERPRSHGTGDNWPRPVDDRRNFQGSKERGFFNNRNFDRSSSAREG 548
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.309 0.129 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,631,309
Number of Sequences: 26719
Number of extensions: 578620
Number of successful extensions: 2510
Number of sequences better than 10.0: 92
Number of HSP's better than 10.0 without gapping: 35
Number of HSP's successfully gapped in prelim test: 61
Number of HSP's that attempted gapping in prelim test: 2248
Number of HSP's gapped (non-prelim): 200
length of query: 443
length of database: 11,318,596
effective HSP length: 102
effective length of query: 341
effective length of database: 8,593,258
effective search space: 2930300978
effective search space used: 2930300978
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC148343.5


