

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148342.7 - phase: 0
(780 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
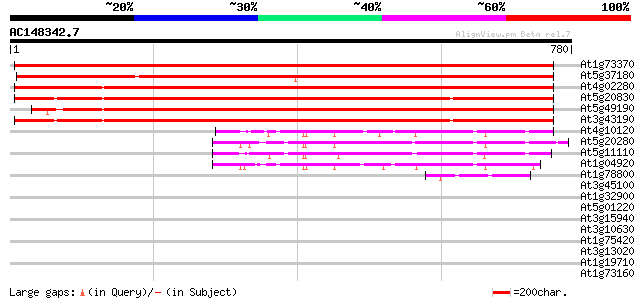
Score E
Sequences producing significant alignments: (bits) Value
At1g73370 1142 0.0
At5g37180 sucrose synthase-like protein 1103 0.0
At4g02280 putative sucrose synthetase 889 0.0
At5g20830 sucrose-UDP glucosyltransferase 857 0.0
At5g49190 sucrose synthase 843 0.0
At3g43190 sucrose synthase like protein 841 0.0
At4g10120 sucrose-phosphate synthase - like protein 139 8e-33
At5g20280 sucrose-phosphate synthase-like protein 138 1e-32
At5g11110 sucrose-phosphate synthase -like protein 135 1e-31
At1g04920 sucrose-phosphate synthase, putative 129 5e-30
At1g78800 unknown protein 54 4e-07
At3g45100 n-acetylglucosaminyl-phosphatidylinositol-like protein 42 0.001
At1g32900 granule-bound starch synthase like protein 42 0.001
At5g01220 unknown protein 41 0.002
At3g15940 unknown protein 36 0.095
At3g10630 unknown protein 36 0.095
At1g75420 unknown protein 35 0.16
At3g13020 hypothetical protein 35 0.21
At1g19710 unknown protein 34 0.36
At1g73160 putative glycosyl transferase 32 1.4
>At1g73370
Length = 942
Score = 1142 bits (2953), Expect = 0.0
Identities = 550/750 (73%), Positives = 638/750 (84%)
Query: 7 LKRTNSIADNMPDALRKSRYHMKKCFAKYLEKGRRIMKLHELMEEVERTIDDINERNYIL 66
L++++SIA+ MPDAL++SRYHMK+CFA ++ G+++MK LM E+E+ I+D ER+ IL
Sbjct: 9 LQKSDSIAEKMPDALKQSRYHMKRCFASFVGGGKKLMKREHLMNEIEKCIEDSRERSKIL 68
Query: 67 EGNLGFILSSTQEAVVDPPYVAFAIRPNPGVWEYVRVNSEDLSVEPITPTDYLKFKERVY 126
EG G+IL+ TQEA V PP+VA A RPNPG WEYV+VNS DL+V+ IT TDYLK KE V+
Sbjct: 69 EGLFGYILTCTQEAAVVPPFVALAARPNPGFWEYVKVNSGDLTVDEITATDYLKLKESVF 128
Query: 127 DQKWANDENAFEADFGAFDIGIPKLTLSSSIGNGLHFVSKFLTSRTTGKLAKAQTIVDYL 186
D+ W+ DENA E DFGA D P+L+LSSSIG G ++SKF++S+ GK K + +++YL
Sbjct: 129 DESWSKDENALEIDFGAIDFTSPRLSLSSSIGKGADYISKFISSKLGGKSDKLEPLLNYL 188
Query: 187 LKLNHHGESLMINDTLSSAAKLQMALIVADVFLSAIPKDTSYQKFELRLKEWGFEKGWGD 246
L+LNHHGE+LMIND L++ AKLQ +L++A + +S K T Y+ F RLKE GFEKGWGD
Sbjct: 189 LRLNHHGENLMINDDLNTVAKLQKSLMLAVIVVSTYSKHTPYETFAQRLKEMGFEKGWGD 248
Query: 247 NAGRVKETMRTLSEVLQAPDPVNLEIFFSRIPTIFKVVIFSVHGYFGQADVLGLPDTGGQ 306
A RVKETM LSEVL+APD L++ FSR+PT+F VVIFSVHGYFGQ DVLGLPDTGGQ
Sbjct: 249 TAERVKETMIILSEVLEAPDNGKLDLLFSRLPTVFNVVIFSVHGYFGQQDVLGLPDTGGQ 308
Query: 307 VVYILDQVKALEEELILRIKQQGLNYKPQILVVTRLIPDARGTKCHQEFEPINDTKHSHI 366
VVYILDQV+ALEEEL++RI QQGL +KPQILVVTRLIP+ARGTKC QE E I TKHSHI
Sbjct: 309 VVYILDQVRALEEELLIRINQQGLGFKPQILVVTRLIPEARGTKCDQELEAIEGTKHSHI 368
Query: 367 LRVPFHTEKGILPQWVSRFDIYPYLERFTQDATTKILDLMEGKPDLVIGNYTDGNLVASL 426
LRVPF T KG+L QWVSRFDIYPYLERFTQDAT+KIL ++ KPDL+IGNYTDGNLVASL
Sbjct: 369 LRVPFVTNKGVLRQWVSRFDIYPYLERFTQDATSKILQRLDCKPDLIIGNYTDGNLVASL 428
Query: 427 MARKLGITQATIAHALEKTKYEDSDVKWKELDPKYHFSCQFMADTVAMNSSDFIITSTYQ 486
MA KLG+TQ TIAHALEKTKYEDSD KWKELDPKYHFSCQF AD +AMN +DFIITSTYQ
Sbjct: 429 MATKLGVTQGTIAHALEKTKYEDSDAKWKELDPKYHFSCQFTADLIAMNVTDFIITSTYQ 488
Query: 487 EIAGSKDRPGQYESHAAFTLPGLCRVVSGINVFDPKFNIAAPGADQSIYFPYTEKDQRHS 546
EIAGSKDRPGQYESH AFT+PGLCRVVSGI+VFDPKFNIAAPGADQS+YFPYTEKD+R +
Sbjct: 489 EIAGSKDRPGQYESHTAFTMPGLCRVVSGIDVFDPKFNIAAPGADQSVYFPYTEKDKRFT 548
Query: 547 QFHPAIEDLLFNKVDNNEHIGYLADKRKPIIFSMARLDVVKNLSGLVEWYGKNKRLRNLV 606
+FHP+I++LL+N+ DN EH+GYLAD+ KPIIFSMARLD VKN++GLVEWYGK+KRLR +
Sbjct: 549 KFHPSIQELLYNEKDNAEHMGYLADREKPIIFSMARLDTVKNITGLVEWYGKDKRLREMA 608
Query: 607 NLVIVGGFFDPSKSKDREEMAEIKKMHDLIEKYQLKGQFRWIAAQTDRYRNGELYRCIAD 666
NLV+V GFFD SKS DREE AEIKKMHDLIEKY+LKG+FRWIAAQTDRYRN ELYRCIAD
Sbjct: 609 NLVVVAGFFDMSKSNDREEKAEIKKMHDLIEKYKLKGKFRWIAAQTDRYRNSELYRCIAD 668
Query: 667 TKGAFVQPALYEAFGLTVIEAMNCGLPTFATNQGGPAEIIVDGVSGFHIDPLNGDESSNK 726
TKG FVQPALYEAFGLTVIEAMNCGLPTFATNQGGPAEIIVDGVSGFHIDP NGDES K
Sbjct: 669 TKGVFVQPALYEAFGLTVIEAMNCGLPTFATNQGGPAEIIVDGVSGFHIDPNNGDESVTK 728
Query: 727 ISDFFEKCKVDPSYWNVISMAGLQRINEWY 756
I DFF KC+ D YW+ IS GL+RI E Y
Sbjct: 729 IGDFFSKCRSDGLYWDNISKGGLKRIYECY 758
>At5g37180 sucrose synthase-like protein
Length = 843
Score = 1103 bits (2854), Expect = 0.0
Identities = 543/754 (72%), Positives = 641/754 (84%), Gaps = 11/754 (1%)
Query: 10 TNSIADNMPDALRKSRYHMKKCFAKYLEKGRRIMKLHELMEEVERTIDDINERNYILEGN 69
+ S+ + +P+A+ ++R ++K+C KY+E GRR+MKL+ELM+E+E I+D+ +R ++EG+
Sbjct: 5 SGSLGNGIPEAMGQNRGNIKRCLEKYIENGRRVMKLNELMDEMEIVINDVTQRRRVMEGD 64
Query: 70 LGFILSSTQEAVVDPPYVAFAIRPNPGVWEYVRVNSEDLSVEPITPTDYLKFKERVYDQK 129
LG IL TQ AVV PP VAFA+R PG W+YV+VNS +LSVE ++ T YLK KE ++D+
Sbjct: 65 LGKILCFTQ-AVVIPPNVAFAVRGTPGNWQYVKVNSSNLSVEALSSTQYLKLKEFLFDEN 123
Query: 130 WANDENAFEADFGAFDIGIPKLTLSSSIGNGLHFVSKFLTSRTTGKLAKAQTIVDYLLKL 189
WANDENA E DFGA D +P L+LSSSIGNGL FVS L R Q++VDYLL L
Sbjct: 124 WANDENALEVDFGALDFTLPWLSLSSSIGNGLSFVSSKLGGRLNDN---PQSLVDYLLSL 180
Query: 190 NHHGESLMINDTLSSAAKLQMALIVADVFLSAIPKDTSYQKFELRLKEWGFEKGWGDNAG 249
H GE LM+N+TL++A KL+M+LI+ADVFLS +PKDT +Q FELR KE GFEKGWG++AG
Sbjct: 181 EHQGEKLMMNETLNTARKLEMSLILADVFLSELPKDTPFQAFELRFKECGFEKGWGESAG 240
Query: 250 RVKETMRTLSEVLQAPDPVNLEIFFSRIPTIFKVVIFSVHGYFGQADVLGLPDTGGQVVY 309
RVKETMR LSE+LQAPDP N++ FF+R+P IF VVIFSVHGYFGQ DVLGLPDTGGQVVY
Sbjct: 241 RVKETMRILSEILQAPDPQNIDRFFARVPRIFNVVIFSVHGYFGQTDVLGLPDTGGQVVY 300
Query: 310 ILDQVKALEEELILRIKQQGLNYKPQILVVTRLIPDARGTKCHQEFEPINDTKHSHILRV 369
ILDQVKALE+EL+ RI QGLN+KPQILVVTRLIPDA+ TKC+QE EPI TK+S+ILR+
Sbjct: 301 ILDQVKALEDELLQRINSQGLNFKPQILVVTRLIPDAKKTKCNQELEPIFGTKYSNILRI 360
Query: 370 PFHTEKGILPQWVSRFDIYPYLERFTQ-------DATTKILDLMEGKPDLVIGNYTDGNL 422
PF TE GIL +WVSRFDIYPYLERFT+ DATTKILD++EGKPDL+IGNYTDGNL
Sbjct: 361 PFVTENGILRRWVSRFDIYPYLERFTKVKSYIRMDATTKILDILEGKPDLIIGNYTDGNL 420
Query: 423 VASLMARKLGITQATIAHALEKTKYEDSDVKWKELDPKYHFSCQFMADTVAMNSSDFIIT 482
VASLMA KLGITQATIAHALEKTKYEDSD+KWKE DPKYHFS QF AD ++MNS+DFII
Sbjct: 421 VASLMANKLGITQATIAHALEKTKYEDSDIKWKEFDPKYHFSSQFTADLISMNSADFIIA 480
Query: 483 STYQEIAGSKDRPGQYESHAAFTLPGLCRVVSGINVFDPKFNIAAPGADQSIYFPYTEKD 542
STYQEIAGSK+R GQYESH +FT+PGL RVVSGINVFDP+FNIAAPGAD SIYFP+T +D
Sbjct: 481 STYQEIAGSKERAGQYESHMSFTVPGLYRVVSGINVFDPRFNIAAPGADDSIYFPFTAQD 540
Query: 543 QRHSQFHPAIEDLLFNKVDNNEHIGYLADKRKPIIFSMARLDVVKNLSGLVEWYGKNKRL 602
+R ++F+ +I++LL+++ +N+EHIGYL DK+KPIIFSMARLDVVKNL+GL EWY KNKRL
Sbjct: 541 RRFTKFYTSIDELLYSQSENDEHIGYLVDKKKPIIFSMARLDVVKNLTGLTEWYAKNKRL 600
Query: 603 RNLVNLVIVGGFFDPSKSKDREEMAEIKKMHDLIEKYQLKGQFRWIAAQTDRYRNGELYR 662
R+LVNLVIVGGFFD SKSKDREE++EIKKMH LIEKYQLKGQFRWI AQTDR RNGELYR
Sbjct: 601 RDLVNLVIVGGFFDASKSKDREEISEIKKMHSLIEKYQLKGQFRWITAQTDRTRNGELYR 660
Query: 663 CIADTKGAFVQPALYEAFGLTVIEAMNCGLPTFATNQGGPAEIIVDGVSGFHIDPLNGDE 722
IADT+GAFVQPA YEAFGLTVIEAM+CGL TFATNQGGPAEIIVDGVSGFHIDP NG+E
Sbjct: 661 SIADTRGAFVQPAHYEAFGLTVIEAMSCGLVTFATNQGGPAEIIVDGVSGFHIDPSNGEE 720
Query: 723 SSNKISDFFEKCKVDPSYWNVISMAGLQRINEWY 756
SS+KI+DFFEK +DP YWN+ S GLQRINE Y
Sbjct: 721 SSDKIADFFEKSGMDPDYWNMFSNEGLQRINECY 754
>At4g02280 putative sucrose synthetase
Length = 809
Score = 889 bits (2296), Expect = 0.0
Identities = 430/750 (57%), Positives = 566/750 (75%), Gaps = 1/750 (0%)
Query: 7 LKRTNSIADNMPDALRKSRYHMKKCFAKYLEKGRRIMKLHELMEEVERTIDDINERNYIL 66
L R S D + D L R + ++Y+++G+ I++ H L++E+E I D + +
Sbjct: 6 LTRVLSTRDRVQDTLSAHRNELVALLSRYVDQGKGILQPHNLIDELESVIGDDETKKSLS 65
Query: 67 EGNLGFILSSTQEAVVDPPYVAFAIRPNPGVWEYVRVNSEDLSVEPITPTDYLKFKERVY 126
+G G IL S EA+V PP+VA A+RP PGVWEYVRVN +LSVE +T ++YL+FKE +
Sbjct: 66 DGPFGEILKSAMEAIVVPPFVALAVRPRPGVWEYVRVNVFELSVEQLTVSEYLRFKEELV 125
Query: 127 DQKWANDENAFEADFGAFDIGIPKLTLSSSIGNGLHFVSKFLTSRTTGKLAKAQTIVDYL 186
D +D E DF F+ +P+ + SSSIGNG+ F+++ L+S + ++D+L
Sbjct: 126 DGP-NSDPFCLELDFEPFNANVPRPSRSSSIGNGVQFLNRHLSSVMFRNKDCLEPLLDFL 184
Query: 187 LKLNHHGESLMINDTLSSAAKLQMALIVADVFLSAIPKDTSYQKFELRLKEWGFEKGWGD 246
+ G LM+ND + S ++LQ+ L A+ +S + ++T + +FE L+ GFEKGWGD
Sbjct: 185 RVHKYKGHPLMLNDRIQSISRLQIQLSKAEDHISKLSQETPFSEFEYALQGMGFEKGWGD 244
Query: 247 NAGRVKETMRTLSEVLQAPDPVNLEIFFSRIPTIFKVVIFSVHGYFGQADVLGLPDTGGQ 306
AGRV E M LS++LQAPDP +LE F +P +F VVI S HGYFGQA+VLGLPDTGGQ
Sbjct: 245 TAGRVLEMMHLLSDILQAPDPSSLEKFLGMVPMVFNVVILSPHGYFGQANVLGLPDTGGQ 304
Query: 307 VVYILDQVKALEEELILRIKQQGLNYKPQILVVTRLIPDARGTKCHQEFEPINDTKHSHI 366
VVYILDQV+ALE E++LRIK+QGL+ P IL+VTRLIPDA+GT C+Q E ++ T+H+HI
Sbjct: 305 VVYILDQVRALETEMLLRIKRQGLDISPSILIVTRLIPDAKGTTCNQRLERVSGTEHTHI 364
Query: 367 LRVPFHTEKGILPQWVSRFDIYPYLERFTQDATTKILDLMEGKPDLVIGNYTDGNLVASL 426
LRVPF +EKGIL +W+SRFD++PYLE + QDA ++I+ ++G PD +IGNY+DGNLVASL
Sbjct: 365 LRVPFRSEKGILRKWISRFDVWPYLENYAQDAASEIVGELQGVPDFIIGNYSDGNLVASL 424
Query: 427 MARKLGITQATIAHALEKTKYEDSDVKWKELDPKYHFSCQFMADTVAMNSSDFIITSTYQ 486
MA ++G+TQ TIAHALEKTKY DSD+ WK+ D KYHFSCQF AD +AMN++DFIITSTYQ
Sbjct: 425 MAHRMGVTQCTIAHALEKTKYPDSDIYWKDFDNKYHFSCQFTADLIAMNNADFIITSTYQ 484
Query: 487 EIAGSKDRPGQYESHAAFTLPGLCRVVSGINVFDPKFNIAAPGADQSIYFPYTEKDQRHS 546
EIAG+K+ GQYESH AFTLPGL RVV GI+VFDPKFNI +PGAD +IYFPY+E+ +R +
Sbjct: 485 EIAGTKNTVGQYESHGAFTLPGLYRVVHGIDVFDPKFNIVSPGADMTIYFPYSEETRRLT 544
Query: 547 QFHPAIEDLLFNKVDNNEHIGYLADKRKPIIFSMARLDVVKNLSGLVEWYGKNKRLRNLV 606
H +IE++L++ +EH+G L+D+ KPI+FSMARLD VKN+SGLVE Y KN +LR LV
Sbjct: 545 ALHGSIEEMLYSPDQTDEHVGTLSDRSKPILFSMARLDKVKNISGLVEMYSKNTKLRELV 604
Query: 607 NLVIVGGFFDPSKSKDREEMAEIKKMHDLIEKYQLKGQFRWIAAQTDRYRNGELYRCIAD 666
NLV++ G D +KSKDREE+ EI+KMH+L++ Y+L GQFRWI AQT+R RNGELYR IAD
Sbjct: 605 NLVVIAGNIDVNKSKDREEIVEIEKMHNLMKNYKLDGQFRWITAQTNRARNGELYRYIAD 664
Query: 667 TKGAFVQPALYEAFGLTVIEAMNCGLPTFATNQGGPAEIIVDGVSGFHIDPLNGDESSNK 726
T+GAF QPA YEAFGLTV+EAM CGLPTFAT GGPAEII G+SGFHIDP + +++ N
Sbjct: 665 TRGAFAQPAFYEAFGLTVVEAMTCGLPTFATCHGGPAEIIEHGLSGFHIDPYHPEQAGNI 724
Query: 727 ISDFFEKCKVDPSYWNVISMAGLQRINEWY 756
++DFFE+CK DP++W +S AGLQRI E Y
Sbjct: 725 MADFFERCKEDPNHWKKVSDAGLQRIYERY 754
>At5g20830 sucrose-UDP glucosyltransferase
Length = 808
Score = 857 bits (2214), Expect = 0.0
Identities = 428/752 (56%), Positives = 557/752 (73%), Gaps = 8/752 (1%)
Query: 7 LKRTNSIADNMPDALRKSRYHMKKCFAKYLEKGRRIMKLHELMEEVERTIDDINERNYIL 66
+ R +S + + + L R + ++ KG+ I++ ++++ E E + ++ L
Sbjct: 8 ITRVHSQRERLNETLVSERNEVLALLSRVEAKGKGILQQNQIIAEFEALPEQTRKK---L 64
Query: 67 EGNLGF-ILSSTQEAVVDPPYVAFAIRPNPGVWEYVRVNSEDLSVEPITPTDYLKFKERV 125
EG F +L STQEA+V PP+VA A+RP PGVWEY+RVN L VE + P ++L FKE +
Sbjct: 65 EGGPFFDLLKSTQEAIVLPPWVALAVRPRPGVWEYLRVNLHALVVEELQPAEFLHFKEEL 124
Query: 126 YDQKWANDENAFEADFGAFDIGIPKLTLSSSIGNGLHFVSKFLTSRTTGKLAKAQTIVDY 185
D N E DF F+ IP+ TL IGNG+ F+++ L+++ ++ +
Sbjct: 125 VDGV-KNGNFTLELDFEPFNASIPRPTLHKYIGNGVDFLNRHLSAKLFHDKESLLPLLKF 183
Query: 186 LLKLNHHGESLMINDTLSSAAKLQMALIVADVFLSAIPKDTSYQKFELRLKEWGFEKGWG 245
L +H G++LM+++ + + LQ L A+ +L+ + +T Y++FE + +E G E+GWG
Sbjct: 184 LRLHSHQGKNLMLSEKIQNLNTLQHTLRKAEEYLAELKSETLYEEFEAKFEEIGLERGWG 243
Query: 246 DNAGRVKETMRTLSEVLQAPDPVNLEIFFSRIPTIFKVVIFSVHGYFGQADVLGLPDTGG 305
DNA RV + +R L ++L+APDP LE F R+P +F VVI S HGYF Q +VLG PDTGG
Sbjct: 244 DNAERVLDMIRLLLDLLEAPDPCTLETFLGRVPMVFNVVILSPHGYFAQDNVLGYPDTGG 303
Query: 306 QVVYILDQVKALEEELILRIKQQGLNYKPQILVVTRLIPDARGTKCHQEFEPINDTKHSH 365
QVVYILDQV+ALE E++ RIKQQGLN KP+IL++TRL+PDA GT C + E + D+++
Sbjct: 304 QVVYILDQVRALEIEMLQRIKQQGLNIKPRILILTRLLPDAVGTTCGERLERVYDSEYCD 363
Query: 366 ILRVPFHTEKGILPQWVSRFDIYPYLERFTQDATTKILDLMEGKPDLVIGNYTDGNLVAS 425
ILRVPF TEKGI+ +W+SRF+++PYLE +T+DA ++ + GKPDL+IGNY+DGNLVAS
Sbjct: 364 ILRVPFRTEKGIVRKWISRFEVWPYLETYTEDAAVELSKELNGKPDLIIGNYSDGNLVAS 423
Query: 426 LMARKLGITQATIAHALEKTKYEDSDVKWKELDPKYHFSCQFMADTVAMNSSDFIITSTY 485
L+A KLG+TQ TIAHALEKTKY DSD+ WK+LD KYHFSCQF AD AMN +DFIITST+
Sbjct: 424 LLAHKLGVTQCTIAHALEKTKYPDSDIYWKKLDDKYHFSCQFTADIFAMNHTDFIITSTF 483
Query: 486 QEIAGSKDRPGQYESHAAFTLPGLCRVVSGINVFDPKFNIAAPGADQSIYFPYTEKDQRH 545
QEIAGSK+ GQYESH AFTLPGL RVV GI+VFDPKFNI +PGAD SIYFPYTE+ +R
Sbjct: 484 QEIAGSKETVGQYESHTAFTLPGLYRVVHGIDVFDPKFNIVSPGADMSIYFPYTEEKRRL 543
Query: 546 SQFHPAIEDLLFNKVDNNEHIGYLADKRKPIIFSMARLDVVKNLSGLVEWYGKNKRLRNL 605
++FH IE+LL++ V+N EH+ L DK+KPI+F+MARLD VKNLSGLVEWYGKN RLR L
Sbjct: 544 TKFHSEIEELLYSDVENKEHLCVLKDKKKPILFTMARLDRVKNLSGLVEWYGKNTRLREL 603
Query: 606 VNLVIVGGFFDPSK-SKDREEMAEIKKMHDLIEKYQLKGQFRWIAAQTDRYRNGELYRCI 664
NLV+VGG D K SKD EE AE+KKM+DLIE+Y+L GQFRWI++Q DR RNGELYR I
Sbjct: 604 ANLVVVGG--DRRKESKDNEEKAEMKKMYDLIEEYKLNGQFRWISSQMDRVRNGELYRYI 661
Query: 665 ADTKGAFVQPALYEAFGLTVIEAMNCGLPTFATNQGGPAEIIVDGVSGFHIDPLNGDESS 724
DTKGAFVQPALYEAFGLTV+EAM CGLPTFAT +GGPAEIIV G SGFHIDP +GD+++
Sbjct: 662 CDTKGAFVQPALYEAFGLTVVEAMTCGLPTFATCKGGPAEIIVHGKSGFHIDPYHGDQAA 721
Query: 725 NKISDFFEKCKVDPSYWNVISMAGLQRINEWY 756
+ ++DFF KCK DPS+W+ IS GLQRI E Y
Sbjct: 722 DTLADFFTKCKEDPSHWDEISKGGLQRIEEKY 753
>At5g49190 sucrose synthase
Length = 805
Score = 843 bits (2178), Expect = 0.0
Identities = 416/732 (56%), Positives = 546/732 (73%), Gaps = 16/732 (2%)
Query: 31 CFAKYLEKGRRIMKLHELMEE------VERTIDDINERNYILEGNLGFILSSTQEAVVDP 84
C +Y+ +G+ I++ H+L++E V+ T++D+N+ + + QEA+V P
Sbjct: 28 CLVRYVAQGKGILQSHQLIDEFLKTVKVDGTLEDLNKSPF---------MKVLQEAIVLP 78
Query: 85 PYVAFAIRPNPGVWEYVRVNSEDLSVEPITPTDYLKFKERVYDQKWANDENAFEADFGAF 144
P+VA AIRP PGV EYVRVN +LSV+ +T ++YL+FKE + + AN + E DF F
Sbjct: 79 PFVALAIRPRPGVREYVRVNVYELSVDHLTVSEYLRFKEELVNGH-ANGDYLLELDFEPF 137
Query: 145 DIGIPKLTLSSSIGNGLHFVSKFLTSRTTGKLAKAQTIVDYLLKLNHHGESLMINDTLSS 204
+ +P+ T SSSIGNG+ F+++ L+S + ++++L H G +M+ND + +
Sbjct: 138 NATLPRPTRSSSIGNGVQFLNRHLSSIMFRNKESMEPLLEFLRTHKHDGRPMMLNDRIQN 197
Query: 205 AAKLQMALIVADVFLSAIPKDTSYQKFELRLKEWGFEKGWGDNAGRVKETMRTLSEVLQA 264
LQ AL A+ FLS +P T Y +FE L+ GFE+GWGD A +V E + L ++LQA
Sbjct: 198 IPILQGALARAEEFLSKLPLATPYSEFEFELQGMGFERGWGDTAQKVSEMVHLLLDILQA 257
Query: 265 PDPVNLEIFFSRIPTIFKVVIFSVHGYFGQADVLGLPDTGGQVVYILDQVKALEEELILR 324
PDP LE F RIP +F VVI S HGYFGQA+VLGLPDTGGQVVYILDQV+ALE E++LR
Sbjct: 258 PDPSVLETFLGRIPMVFNVVILSPHGYFGQANVLGLPDTGGQVVYILDQVRALENEMLLR 317
Query: 325 IKQQGLNYKPQILVVTRLIPDARGTKCHQEFEPINDTKHSHILRVPFHTEKGILPQWVSR 384
I++QGL P+IL+VTRL+P+A+GT C+Q E ++ T+H+HILR+PF TEKGIL +W+SR
Sbjct: 318 IQKQGLEVIPKILIVTRLLPEAKGTTCNQRLERVSGTEHAHILRIPFRTEKGILRKWISR 377
Query: 385 FDIYPYLERFTQDATTKILDLMEGKPDLVIGNYTDGNLVASLMARKLGITQATIAHALEK 444
FD++PYLE F +DA+ +I ++G P+L+IGNY+DGNLVASL+A KLG+ Q IAHALEK
Sbjct: 378 FDVWPYLETFAEDASNEISAELQGVPNLIIGNYSDGNLVASLLASKLGVIQCNIAHALEK 437
Query: 445 TKYEDSDVKWKELDPKYHFSCQFMADTVAMNSSDFIITSTYQEIAGSKDRPGQYESHAAF 504
TKY +SD+ W+ + KYHFS QF AD +AMN++DFIITSTYQEIAGSK+ GQYESH AF
Sbjct: 438 TKYPESDIYWRNHEDKYHFSSQFTADLIAMNNADFIITSTYQEIAGSKNNVGQYESHTAF 497
Query: 505 TLPGLCRVVSGINVFDPKFNIAAPGADQSIYFPYTEKDQRHSQFHPAIEDLLFNKVDNNE 564
T+PGL RVV GI+VFDPKFNI +PGAD +IYFPY++K++R + H +IE+LLF+ N+E
Sbjct: 498 TMPGLYRVVHGIDVFDPKFNIVSPGADMTIYFPYSDKERRLTALHESIEELLFSAEQNDE 557
Query: 565 HIGYLADKRKPIIFSMARLDVVKNLSGLVEWYGKNKRLRNLVNLVIVGGFFDPSKSKDRE 624
H+G L+D+ KPIIFSMARLD VKNL+GLVE Y KN +LR L NLVIVGG+ D ++S+DRE
Sbjct: 558 HVGLLSDQSKPIIFSMARLDRVKNLTGLVECYAKNSKLRELANLVIVGGYIDENQSRDRE 617
Query: 625 EMAEIKKMHDLIEKYQLKGQFRWIAAQTDRYRNGELYRCIADTKGAFVQPALYEAFGLTV 684
EMAEI+KMH LIE+Y L G+FRWIAAQ +R RNGELYR IADTKG FVQPA YEAFGLTV
Sbjct: 618 EMAEIQKMHSLIEQYDLHGEFRWIAAQMNRARNGELYRYIADTKGVFVQPAFYEAFGLTV 677
Query: 685 IEAMNCGLPTFATNQGGPAEIIVDGVSGFHIDPLNGDESSNKISDFFEKCKVDPSYWNVI 744
+E+M C LPTFAT GGPAEII +GVSGFHIDP + D+ + + FFE C +P++W I
Sbjct: 678 VESMTCALPTFATCHGGPAEIIENGVSGFHIDPYHPDQVAATLVSFFETCNTNPNHWVKI 737
Query: 745 SMAGLQRINEWY 756
S GL+RI E Y
Sbjct: 738 SEGGLKRIYERY 749
>At3g43190 sucrose synthase like protein
Length = 808
Score = 841 bits (2172), Expect = 0.0
Identities = 424/752 (56%), Positives = 549/752 (72%), Gaps = 8/752 (1%)
Query: 7 LKRTNSIADNMPDALRKSRYHMKKCFAKYLEKGRRIMKLHELMEEVERTIDDINERNYIL 66
+ R +S + + L + + ++ KG+ I++ H+++ E E + ++ L
Sbjct: 8 ITRVHSQRERLDATLVAQKNEVFALLSRVEAKGKGILQHHQIIAEFEAMPLETQKK---L 64
Query: 67 EGNLGF-ILSSTQEAVVDPPYVAFAIRPNPGVWEYVRVNSEDLSVEPITPTDYLKFKERV 125
+G F L S QEA+V PP+VA A+RP PGVWEYVRVN DL VE + ++YL+FKE +
Sbjct: 65 KGGAFFEFLRSAQEAIVLPPFVALAVRPRPGVWEYVRVNLHDLVVEELQASEYLQFKEEL 124
Query: 126 YDQKWANDENAFEADFGAFDIGIPKLTLSSSIGNGLHFVSKFLTSRTTGKLAKAQTIVDY 185
D N E DF F+ P+ TL+ IG+G+ F+++ L+++ ++ +
Sbjct: 125 VDGI-KNGNFTLELDFEPFNAAFPRPTLNKYIGDGVEFLNRHLSAKLFHDKESLHPLLKF 183
Query: 186 LLKLNHHGESLMINDTLSSAAKLQMALIVADVFLSAIPKDTSYQKFELRLKEWGFEKGWG 245
L +H G++LM+N+ + + LQ L A+ +L + +T Y +FE + +E G E+GWG
Sbjct: 184 LRLHSHEGKTLMLNNRIQNLNTLQHNLRKAEEYLMELKPETLYSEFEHKFQEIGLERGWG 243
Query: 246 DNAGRVKETMRTLSEVLQAPDPVNLEIFFSRIPTIFKVVIFSVHGYFGQADVLGLPDTGG 305
D A RV +R L ++L+APDP LE F RIP +F VVI S HGYF Q +VLG PDTGG
Sbjct: 244 DTAERVLNMIRLLLDLLEAPDPCTLENFLGRIPMVFNVVILSPHGYFAQDNVLGYPDTGG 303
Query: 306 QVVYILDQVKALEEELILRIKQQGLNYKPQILVVTRLIPDARGTKCHQEFEPINDTKHSH 365
QVVYILDQV+ALE E++ RIKQQGLN P+IL++TRL+PDA GT C Q E + +++
Sbjct: 304 QVVYILDQVRALETEMLQRIKQQGLNITPRILIITRLLPDAAGTTCGQRLEKVYGSQYCD 363
Query: 366 ILRVPFHTEKGILPQWVSRFDIYPYLERFTQDATTKILDLMEGKPDLVIGNYTDGNLVAS 425
ILRVPF TEKGI+ +W+SRF+++PYLE FT+D +I ++GKPDL+IGNY+DGNLVAS
Sbjct: 364 ILRVPFRTEKGIVRKWISRFEVWPYLETFTEDVAAEISKELQGKPDLIIGNYSDGNLVAS 423
Query: 426 LMARKLGITQATIAHALEKTKYEDSDVKWKELDPKYHFSCQFMADTVAMNSSDFIITSTY 485
L+A KLG+TQ TIAHALEKTKY DSD+ WK+LD KYHFSCQF AD +AMN +DFIITST+
Sbjct: 424 LLAHKLGVTQCTIAHALEKTKYPDSDIYWKKLDEKYHFSCQFTADLIAMNHTDFIITSTF 483
Query: 486 QEIAGSKDRPGQYESHAAFTLPGLCRVVSGINVFDPKFNIAAPGADQSIYFPYTEKDQRH 545
QEIAGSKD GQYESH +FTLPGL RVV GI+VFDPKFNI +PGAD SIYF YTE+ +R
Sbjct: 484 QEIAGSKDTVGQYESHRSFTLPGLYRVVHGIDVFDPKFNIVSPGADMSIYFAYTEEKRRL 543
Query: 546 SQFHPAIEDLLFNKVDNNEHIGYLADKRKPIIFSMARLDVVKNLSGLVEWYGKNKRLRNL 605
+ FH IE+LL++ V+N EH+ L DK+KPIIF+MARLD VKNLSGLVEWYGKN RLR L
Sbjct: 544 TAFHLEIEELLYSDVENEEHLCVLKDKKKPIIFTMARLDRVKNLSGLVEWYGKNTRLREL 603
Query: 606 VNLVIVGGFFDPSK-SKDREEMAEIKKMHDLIEKYQLKGQFRWIAAQTDRYRNGELYRCI 664
VNLV+VGG D K S+D EE AE+KKM++LIE+Y+L GQFRWI++Q +R RNGELYR I
Sbjct: 604 VNLVVVGG--DRRKESQDNEEKAEMKKMYELIEEYKLNGQFRWISSQMNRVRNGELYRYI 661
Query: 665 ADTKGAFVQPALYEAFGLTVIEAMNCGLPTFATNQGGPAEIIVDGVSGFHIDPLNGDESS 724
DTKGAFVQPALYEAFGLTV+EAM CGLPTFAT GGPAEIIV G SGFHIDP +GD+++
Sbjct: 662 CDTKGAFVQPALYEAFGLTVVEAMTCGLPTFATCNGGPAEIIVHGKSGFHIDPYHGDKAA 721
Query: 725 NKISDFFEKCKVDPSYWNVISMAGLQRINEWY 756
++DFF KCK DPS+W+ IS+ GL+RI E Y
Sbjct: 722 ESLADFFTKCKHDPSHWDQISLGGLERIQEKY 753
>At4g10120 sucrose-phosphate synthase - like protein
Length = 1083
Score = 139 bits (349), Expect = 8e-33
Identities = 134/509 (26%), Positives = 232/509 (45%), Gaps = 70/509 (13%)
Query: 287 SVHGYF-GQADVLGLP-DTGGQVVYILDQVKALEEELILRIKQQGLNYKPQILVVTRLIP 344
S+HG G+ LG DTGGQV Y+++ +AL +G++ ++ ++TR I
Sbjct: 234 SMHGLVRGENMELGRDSDTGGQVKYVVELARALANT-------EGVH---RVDLLTRQIS 283
Query: 345 DARGTKCHQEFEPI---------NDTKHSHILRVPFHTEKGILPQWVSRFDIYPYLERFT 395
+ E P+ +D+ S+I+R+P G +++ + ++P++ F
Sbjct: 284 SPEVDYSYGE--PVEMLSCPPEGSDSCGSYIIRIPC----GSRDKYIPKESLWPHIPEFV 337
Query: 396 QDATTKILDLME--------GKPD---LVIGNYTDGNLVASLMARKLGITQATIAHALEK 444
A I+ + GKP ++ G+Y D VA+ +A L + H+L +
Sbjct: 338 DGALNHIVSIARSLGEQVNGGKPIWPYVIHGHYADAGEVAAHLAGALNVPMVLTGHSLGR 397
Query: 445 TKYED----SDVKWKELDPKYHFSCQFMADTVAMNSSDFIITSTYQEIAGSKDRPGQYES 500
K+E + +++D Y + A+ ++++++ ++TST QEI G Y+
Sbjct: 398 NKFEQLLQQGRITREDIDRTYKIMRRIEAEEQSLDAAEMVVTSTRQEIDAQW---GLYDG 454
Query: 501 HAAFTLPGLCRV-----VSGINVFDPKFNIAAPGADQSIYFPYTEKDQRHSQFHPAIEDL 555
L RV VS + + P+ + PG D S + T+ Q ++
Sbjct: 455 FD-IKLERKLRVRRRRGVSCLGRYMPRMVVIPPGMDFS--YVLTQDSQEPDGDLKSLIGP 511
Query: 556 LFNKVDN------NEHIGYLADKRKPIIFSMARLDVVKNLSGLVEWYGKNKRLRNLVNLV 609
N++ +E + + ++ KP I +++R D KN++ LV+ +G+ + LR L NLV
Sbjct: 512 DRNQIKKPVPPIWSEIMRFFSNPHKPTILALSRPDHKKNVTTLVKAFGECQPLRELANLV 571
Query: 610 IVGGFFDPSKSKDREEMAEIKKMHDLIEKYQLKGQFRWIAAQTDRYRNGE---LYRCIAD 666
++ G D + + + LI++Y L GQ A ++ E +YR A
Sbjct: 572 LILGNRDDIEEMPNSSSVVLMNVLKLIDQYDLYGQ----VAYPKHHKQSEVPDIYRLAAK 627
Query: 667 TKGAFVQPALYEAFGLTVIEAMNCGLPTFATNQGGPAEIIVDGVSGFHIDPLNGDESSNK 726
TKG F+ PAL E FGLT+IEA GLP AT GGP +I+ +G +DP
Sbjct: 628 TKGVFINPALVEPFGLTLIEAAAYGLPIVATRNGGPVDIVKALNNGLLVDP----HDQQA 683
Query: 727 ISDFFEKCKVDPSYWNVISMAGLQRINEW 755
ISD K + W GL+ I+ +
Sbjct: 684 ISDALLKLVANKHLWAECRKNGLKNIHRF 712
>At5g20280 sucrose-phosphate synthase-like protein
Length = 1043
Score = 138 bits (348), Expect = 1e-32
Identities = 133/528 (25%), Positives = 240/528 (45%), Gaps = 59/528 (11%)
Query: 283 VVIFSVHGYF-GQADVLGLP-DTGGQVVYILDQVKALEE-------ELILR-IKQQGLNY 332
+V+ S+HG G+ LG DTGGQV Y+++ +AL +L+ R + ++Y
Sbjct: 170 LVLISLHGLIRGENMELGRDSDTGGQVKYVVELARALGSMPGVYRVDLLTRQVSSPDVDY 229
Query: 333 ---KPQILVVTRLIPDARGTKCHQEFEPINDTKHSHILRVPFHTEKGILPQWVSRFDIYP 389
+P ++ R D + + ++ ++I+R+PF + +P+ + ++P
Sbjct: 230 SYGEPTEMLTPRDSEDFS--------DEMGESSGAYIVRIPFGPKDKYIPKEL----LWP 277
Query: 390 YLERFTQDATTKILDLME--------GKP---DLVIGNYTDGNLVASLMARKLGITQATI 438
++ F A + I+ + GKP + G+Y D +L++ L +
Sbjct: 278 HIPEFVDGAMSHIMQMSNVLGEQVGVGKPIWPSAIHGHYADAGDATALLSGALNVPMLLT 337
Query: 439 AHALEKTKYED----SDVKWKELDPKYHFSCQFMADTVAMNSSDFIITSTYQEIAGSKDR 494
H+L + K E + +E++ Y + + ++++ S+ +ITST QEI
Sbjct: 338 GHSLGRDKLEQLLRQGRLSKEEINSTYKIMRRIEGEELSLDVSEMVITSTRQEIDEQWRL 397
Query: 495 PGQYESHAAFTLPG-LCRVVSGINVFDPKFNIAAPGADQSIYFPY--TEKDQRHSQFHPA 551
++ L + R VS F P+ PG + + P+ +D ++ HP
Sbjct: 398 YDGFDPILERKLRARIKRNVSCYGRFMPRMVKIPPGMEFNHIVPHGGDMEDTDGNEEHPT 457
Query: 552 IEDLLFNKVDNNEHIGYLADKRKPIIFSMARLDVVKNLSGLVEWYGKNKRLRNLVNLVIV 611
D E + + ++ RKP+I ++AR D KN++ LV+ +G+ + LR L NL ++
Sbjct: 458 SPDPPIWA----EIMRFFSNSRKPMILALARPDPKKNITTLVKAFGECRPLRELANLALI 513
Query: 612 GGFFDPSKSKDREEMAEIKKMHDLIEKYQLKGQFRWIAAQTDRYRNGE---LYRCIADTK 668
G D + + + LI+KY L GQ A ++ + +YR A +K
Sbjct: 514 MGNRDGIDEMSSTSSSVLLSVLKLIDKYDLYGQ----VAYPKHHKQSDVPDIYRLAAKSK 569
Query: 669 GAFVQPALYEAFGLTVIEAMNCGLPTFATNQGGPAEIIVDGVSGFHIDPLNGDESSNKIS 728
G F+ PA+ E FGLT+IEA GLP AT GGP +I +G +DP IS
Sbjct: 570 GVFINPAIIEPFGLTLIEAAAHGLPMVATKNGGPVDIHRVLDNGLLVDP----HDQQSIS 625
Query: 729 DFFEKCKVDPSYWNVISMAGLQRINEWYYLRPLLRCFVAKKINFKHVH 776
+ K D W GL+ I+++ + + ++++ +FK H
Sbjct: 626 EALLKLVADKHLWAKCRQNGLKNIHQFSWPEH-CKTYLSRITSFKPRH 672
>At5g11110 sucrose-phosphate synthase -like protein
Length = 1047
Score = 135 bits (339), Expect = 1e-31
Identities = 129/501 (25%), Positives = 228/501 (44%), Gaps = 55/501 (10%)
Query: 283 VVIFSVHGYF-GQADVLGLP-DTGGQVVYILDQVKALEEELILRIKQQGLNYKPQILV-- 338
+V+ S+HG G+ LG DTGGQV Y+++ +AL G+ Y+ +L
Sbjct: 177 IVLISLHGLIRGENMELGRDSDTGGQVKYVVELARALGS-------MPGV-YRVDLLTRQ 228
Query: 339 VTRLIPDARGTKCHQEFEPIN--------DTKHSHILRVPFHTEKGILPQWVSRFDIYPY 390
VT D+ ++ + PI+ ++ ++I+R+PF + +P+ + ++P+
Sbjct: 229 VTAPDVDSSYSEPSEMLNPIDTDIEQENGESSGAYIIRIPFGPKDKYVPKEL----LWPH 284
Query: 391 LERFTQDATTKILDLME--------GK---PDLVIGNYTDGNLVASLMARKLGITQATIA 439
+ F A + I+ + + G+ P + G+Y D +L++ L +
Sbjct: 285 IPEFVDRALSHIMQISKVLGEQIGGGQQVWPVSIHGHYADAGDSTALLSGALNVPMVFTG 344
Query: 440 HALEKTKYEDSDVKWK---ELDPKYHFSCQFMADTVAMNSSDFIITSTYQEIAGSKDRPG 496
H+L + K E + + E++ Y + A+ + +++S+ +ITST QE+
Sbjct: 345 HSLGRDKLEQLLKQGRPKEEINSNYKIWRRIEAEELCLDASEIVITSTRQEVDEQWRLYD 404
Query: 497 QYESHAAFTLPG-LCRVVSGINVFDPKFNIAAPGADQSIYFPYTEKDQRHSQFHPAIEDL 555
++ L + R VS + F P+ + PG + P+ + +
Sbjct: 405 GFDPVLERKLRARMKRGVSCLGRFMPRMVVIPPGMEFHHIVPHDVDADGDDENPQTADPP 464
Query: 556 LFNKVDNNEHIGYLADKRKPIIFSMARLDVVKNLSGLVEWYGKNKRLRNLVNLVIVGGFF 615
+++++ + + ++ RKP+I ++AR D KNL LV+ +G+ + LR L NL ++ G
Sbjct: 465 IWSEI-----MRFFSNPRKPMILALARPDPKKNLVTLVKAFGECRPLRELANLTLIMGNR 519
Query: 616 DPSKSKDREEMAEIKKMHDLIEKYQLKGQFRWIAAQTDRYRNG---ELYRCIADTKGAFV 672
+ + + + LI+KY L GQ A ++ E+YR A TKG F+
Sbjct: 520 NDIDELSSTNSSVLLSILKLIDKYDLYGQ----VAMPKHHQQSDVPEIYRLAAKTKGVFI 575
Query: 673 QPALYEAFGLTVIEAMNCGLPTFATNQGGPAEIIVDGVSGFHIDPLNGDESSNKISDFFE 732
PA E FGLT+IEA GLPT AT GGP +I +G +DP I+D
Sbjct: 576 NPAFIEPFGLTLIEAGAHGLPTVATINGGPVDIHRVLDNGLLVDP----HDQQAIADALL 631
Query: 733 KCKVDPSYWNVISMAGLQRIN 753
K D W GL I+
Sbjct: 632 KLVSDRQLWGRCRQNGLNNIH 652
>At1g04920 sucrose-phosphate synthase, putative
Length = 1062
Score = 129 bits (325), Expect = 5e-30
Identities = 125/504 (24%), Positives = 232/504 (45%), Gaps = 72/504 (14%)
Query: 283 VVIFSVHGYF-GQADVLGLP-DTGGQVVYILDQVKALEE-------ELILR---IKQQGL 330
VV+ S+HG G+ LG DTGGQV Y+++ +AL +L R +
Sbjct: 172 VVLISLHGLVRGENMELGSDSDTGGQVKYVVELARALARMPGVYRVDLFTRQICSSEVDW 231
Query: 331 NYKPQILVVTRLIPDARGTKCHQEFEPINDTKHSHILRVPFHTEKGILPQWVSRFDIYPY 390
+Y ++T D G + ++ ++I+R+PF G +++++ ++P+
Sbjct: 232 SYAEPTEMLTTA-EDCDG-------DETGESSGAYIIRIPF----GPRDKYLNKEILWPF 279
Query: 391 LERFTQDATTKILDLME--------GKPD---LVIGNYTDGNLVASLMARKLGITQATIA 439
++ F A IL++ + GKP ++ G+Y D A+L++ L +
Sbjct: 280 VQEFVDGALAHILNMSKVLGEQIGKGKPVWPYVIHGHYADAGDSAALLSGALNVPMVLTG 339
Query: 440 HALEKTKYED----SDVKWKELDPKYHFSCQFMADTVAMNSSDFIITSTYQEIAGSKDRP 495
H+L + K E ++++ Y + A+ +++++++ +ITST QEI ++
Sbjct: 340 HSLGRNKLEQLLKQGRQSKEDINSTYKIKRRIEAEELSLDAAELVITSTRQEI---DEQW 396
Query: 496 GQYESHAAFTLPGL-CRVVSGINV---FDPKFNIAAPGADQSIYFPYTEKDQRHSQFHPA 551
G Y+ L R G+N F P+ + PG D F E + +
Sbjct: 397 GLYDGFDVKLEKVLRARARRGVNCHGRFMPRMAVIPPGMD----FTNVEVQEDTPEGDGD 452
Query: 552 IEDLLFNKVDNN---------EHIGYLADKRKPIIFSMARLDVVKNLSGLVEWYGKNKRL 602
+ L+ ++ E + + + KP+I +++R D KN++ L++ +G+ + L
Sbjct: 453 LASLVGGTEGSSPKAVPTIWSEVMRFFTNPHKPMILALSRPDPKKNITTLLKAFGECRPL 512
Query: 603 RNLVNLVIVGGFFDPSKSKDREEMAEIKKMHDLIEKYQLKGQFRWIAAQTDRYRNGE--- 659
R L NL ++ G D + + + LI+KY L G A ++ +
Sbjct: 513 RELANLTLIMGNRDDIDELSSGNASVLTTVLKLIDKYDLYGS----VAYPKHHKQSDVPD 568
Query: 660 LYRCIADTKGAFVQPALYEAFGLTVIEAMNCGLPTFATNQGGPAEIIVDGVSGFHIDPLN 719
+YR A+TKG F+ PAL E FGLT+IEA GLP AT GGP +I +G +DP +
Sbjct: 569 IYRLAANTKGVFINPALVEPFGLTLIEAAAHGLPMVATKNGGPVDIHRALHNGLLVDPHD 628
Query: 720 GDESSNKI------SDFFEKCKVD 737
+ +N + + + +C+++
Sbjct: 629 QEAIANALLKLVSEKNLWHECRIN 652
>At1g78800 unknown protein
Length = 403
Score = 53.5 bits (127), Expect = 4e-07
Identities = 41/149 (27%), Positives = 70/149 (46%), Gaps = 12/149 (8%)
Query: 579 SMARLDVVKNLSGLVEWYG---KNKRLRNLVNLVIVGGFFDPSKSKDREEMAEIKKMHDL 635
S+ R + KN+ V + K+K+ + V L + GG+ + K E + ++++ L
Sbjct: 214 SINRFERKKNIDLAVSAFAILCKHKQNLSDVTLTVAGGYDERLK----ENVEYLEELRSL 269
Query: 636 IEKYQLKGQFRWIAAQTDRYRNGELYRCIADTKGAFVQPALYEAFGLTVIEAMNCGLPTF 695
EK + + +I + + RN L C+ + E FG+ +EAM P
Sbjct: 270 AEKEGVSDRVNFITSCSTAERNELLSSCLC-----VLYTPTDEHFGIVPLEAMAAYKPVI 324
Query: 696 ATNQGGPAEIIVDGVSGFHIDPLNGDESS 724
A N GGP E + +GV+G+ +P D SS
Sbjct: 325 ACNSGGPVETVKNGVTGYLCEPTPEDFSS 353
>At3g45100 n-acetylglucosaminyl-phosphatidylinositol-like protein
Length = 447
Score = 42.0 bits (97), Expect = 0.001
Identities = 53/182 (29%), Positives = 77/182 (42%), Gaps = 21/182 (11%)
Query: 531 DQSIYFPYTEKDQR--HSQFHPAIEDLLFNKVDNNEHIGYLADKRKPII--FSMARLDVV 586
DQ+I +T K+ S PA ++ N VD II ++RL
Sbjct: 153 DQAICVSHTSKENTVLRSGLSPAKVFMIPNAVDTAMFKPASVRPSTDIITIVVISRLVYR 212
Query: 587 KNLSGLVEWYGKNKRLRNLVNLVIVGGFFDPSKSKDREEMAEIKKMHDLIEKYQLKGQFR 646
K LVE + RL V V+ G D K EEM E H L ++ ++ G
Sbjct: 213 KGADLLVEVIPEVCRLYPNVRFVVGG---DGPKHVRLEEMRE---KHSLQDRVEMLG--- 263
Query: 647 WIAAQTDRYRNGELYRCIADTKGAFVQPALYEAFGLTVIEAMNCGLPTFATNQGGPAEII 706
A R R+ + T F+ +L EAF + ++EA +CGL T +T GG E++
Sbjct: 264 --AVPHSRVRS------VLVTGHIFLNSSLTEAFCIAILEAASCGLLTVSTRVGGVPEVL 315
Query: 707 VD 708
D
Sbjct: 316 PD 317
>At1g32900 granule-bound starch synthase like protein
Length = 610
Score = 42.0 bits (97), Expect = 0.001
Identities = 58/234 (24%), Positives = 95/234 (39%), Gaps = 34/234 (14%)
Query: 486 QEIAGSKDRPGQYESHAAFTLPGLCRVVSGINVFDPKFNIAAPGADQSIYFPYTEKDQRH 545
QE+ DR E H + + +++G++V ++N P D+ I Y
Sbjct: 328 QELISGVDRG--VELHKYLRMKTVSGIINGMDV--QEWN---PSTDKYIDIKYDITTVTD 380
Query: 546 SQFHPAIEDLLFNKVDNNEHIGYLADKRKPIIFSMARLDVVKNLSGLVEWYGKNKRLRNL 605
++ P I++ L +G D+ P+I + RL+ K LVE K L
Sbjct: 381 AK--PLIKEAL------QAAVGLPVDRDVPVIGFIGRLEEQKGSDILVEAISKFMGLN-- 430
Query: 606 VNLVIVGGFFDPSKSKDREEMAEIKKMHDLIEKYQLKGQFRWIAAQTDRYRNGELYRCIA 665
V +VI+G K K ++ E++ EK+ K A ++ N L I
Sbjct: 431 VQMVILG----TGKKKMEAQILELE------EKFPGK------AVGVAKF-NVPLAHMIT 473
Query: 666 DTKGAFVQPALYEAFGLTVIEAMNCGLPTFATNQGGPAEIIVDGVSGFHIDPLN 719
+ P+ +E GL + AM G + GG + + DG +GFHI N
Sbjct: 474 AGADFIIVPSRFEPCGLIQLHAMRYGTVPIVASTGGLVDTVKDGYTGFHIGRFN 527
>At5g01220 unknown protein
Length = 510
Score = 41.2 bits (95), Expect = 0.002
Identities = 23/60 (38%), Positives = 34/60 (56%), Gaps = 3/60 (5%)
Query: 671 FVQPALYEAFGLTVIEAMNCGLPTFATNQGGPAEII---VDGVSGFHIDPLNGDESSNKI 727
FV P+ E GL V+EAM+ GLP A GG +II +G +GF +P + ++ K+
Sbjct: 380 FVMPSESETLGLVVLEAMSSGLPVVAARAGGIPDIIPEDQEGKTGFLFNPGDVEDCVTKL 439
>At3g15940 unknown protein
Length = 697
Score = 35.8 bits (81), Expect = 0.095
Identities = 17/35 (48%), Positives = 20/35 (56%)
Query: 678 EAFGLTVIEAMNCGLPTFATNQGGPAEIIVDGVSG 712
E FG IEAM GLP T+ GG EI+ V+G
Sbjct: 602 ETFGRVTIEAMAYGLPVLGTDAGGTKEIVEHNVTG 636
>At3g10630 unknown protein
Length = 487
Score = 35.8 bits (81), Expect = 0.095
Identities = 16/39 (41%), Positives = 21/39 (53%)
Query: 670 AFVQPALYEAFGLTVIEAMNCGLPTFATNQGGPAEIIVD 708
AFV P E +G ++EAM LP TN GP E + +
Sbjct: 365 AFVLPTRGEGWGRPIVEAMAMSLPVITTNWSGPTEYLTE 403
>At1g75420 unknown protein
Length = 265
Score = 35.0 bits (79), Expect = 0.16
Identities = 17/35 (48%), Positives = 20/35 (56%)
Query: 678 EAFGLTVIEAMNCGLPTFATNQGGPAEIIVDGVSG 712
E FG IEAM LP T GG EI+V+G +G
Sbjct: 163 ECFGRITIEAMAFKLPVLGTAAGGTMEIVVNGTTG 197
>At3g13020 hypothetical protein
Length = 605
Score = 34.7 bits (78), Expect = 0.21
Identities = 48/167 (28%), Positives = 73/167 (42%), Gaps = 29/167 (17%)
Query: 517 NVFDPKFNIAAPGADQSIYFPYTEKDQRH----SQFHPAIEDLL------------FNKV 560
+VF K N+AA A S++ K + S F A+E++L F+
Sbjct: 354 SVFKAKKNLAAMFAS-SVWKKEEGKSVSNLVNDSSFWEAVEEILKCTSPLTDGLRLFSNA 412
Query: 561 DNNEHIGYLADKRKPIIFSMARLDVVKNLSGLVEW----YGKNKRLRNLVNLVIVGGFFD 616
DNN+H+GY+ D I S+ + + L W NK L N L G + +
Sbjct: 413 DNNQHVGYIYDTLDGIKLSIKKEFNDEKKHYLTLWDVIDDVWNKHLHN--PLHAAGYYLN 470
Query: 617 PSK--SKDREEMAEIKK--MHDLIEKYQLKGQFRWIAAQTDRYRNGE 659
P+ S D E+ H L+ + +GQ + IA+Q DRYR G+
Sbjct: 471 PTSFYSTDFHLDPEVSSGLTHSLVHVAK-EGQIK-IASQLDRYRLGK 515
>At1g19710 unknown protein
Length = 479
Score = 33.9 bits (76), Expect = 0.36
Identities = 22/79 (27%), Positives = 34/79 (42%), Gaps = 2/79 (2%)
Query: 678 EAFGLTVIEAMNCGLPTFATNQGGPAEIIVDGVSGFHIDPLNGDESSNKISDFFEKCKVD 737
E FG IEAM LP T GG EI+V+ +G + G + ++ K +
Sbjct: 372 ECFGRITIEAMAFKLPVLGTAAGGTMEIVVNRTTGLLHN--TGKDGVLPLAKNIVKLATN 429
Query: 738 PSYWNVISMAGLQRINEWY 756
N + G +R+ E +
Sbjct: 430 VKMRNTMGKKGYERVKEMF 448
>At1g73160 putative glycosyl transferase
Length = 486
Score = 32.0 bits (71), Expect = 1.4
Identities = 19/49 (38%), Positives = 24/49 (48%), Gaps = 2/49 (4%)
Query: 671 FVQPALY-EAFGLTVIEAMNCGLPTFATNQGG-PAEIIVDGVSGFHIDP 717
FV P L + LT+IEAM CG P A N ++VD G+ P
Sbjct: 373 FVNPTLRPQGLDLTIIEAMQCGKPVVAPNYPSIVGTVVVDERFGYTFSP 421
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.138 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,888,219
Number of Sequences: 26719
Number of extensions: 793374
Number of successful extensions: 1896
Number of sequences better than 10.0: 29
Number of HSP's better than 10.0 without gapping: 21
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 1843
Number of HSP's gapped (non-prelim): 31
length of query: 780
length of database: 11,318,596
effective HSP length: 107
effective length of query: 673
effective length of database: 8,459,663
effective search space: 5693353199
effective search space used: 5693353199
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 64 (29.3 bits)
Medicago: description of AC148342.7


