

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148342.16 - phase: 0 /pseudo
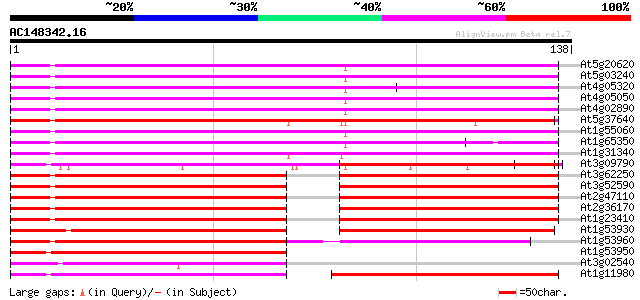
(138 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g20620 polyubiquitin 4 UBQ4 114 2e-26
At5g03240 polyubiquitin (ubq3) 114 2e-26
At4g05320 polyubiquitin (ubq10) 114 2e-26
At4g05050 114 2e-26
At4g02890 polyubiquitin 114 2e-26
At5g37640 polyubiquitin 4 110 2e-25
At1g55060 ubiquitin-like (UBQ12) 108 1e-24
At1g65350 107 3e-24
At1g31340 putative ubiquitin (AtRUB1) 102 6e-23
At3g09790 polyubiquitin 98 1e-21
At3g62250 ubiquitin extension protein (UBQ5) 74 2e-14
At3g52590 ubiquitin / ribosomal protein CEP52 74 2e-14
At2g47110 putative ubiquitin extension protein (UBQ6) 74 2e-14
At2g36170 putative ubiquitin/ribosomal protein CEP52 74 2e-14
At1g23410 ubiquitin extension protein, putative 74 2e-14
At1g53930 hypothetical protein 66 7e-12
At1g53960 unknown protein 62 9e-11
At1g53950 hypothetical protein 49 6e-07
At3g02540 RAD23 like protein 47 2e-06
At1g11980 unknown protein 47 4e-06
>At5g20620 polyubiquitin 4 UBQ4
Length = 382
Score = 114 bits (285), Expect = 2e-26
Identities = 71/151 (47%), Positives = 90/151 (59%), Gaps = 17/151 (11%)
Query: 1 MQIFIKIMYGNKMFPLIVKSSDTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLANY 60
MQIF+K + G K L V+SSDTI +VK KI DKEGIP QQRL F GK ++D +TLA+Y
Sbjct: 153 MQIFVKTLTG-KTITLEVESSDTIDNVKAKIQDKEGIPPDQQRLIFAGKQLEDGRTLADY 211
Query: 61 NIPENSTIDTRSIAKRSRPLF----------------NTILDVKKKIHDKEGIPVHQQRL 104
NI + ST+ + +F +TI +VK KI DKEGIP QQRL
Sbjct: 212 NIQKESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPDQQRL 271
Query: 105 FFDGKPIKDRQTLTNYNIPRTQPLTLVRSLR 135
F GK ++D +TL +YNI + L LV LR
Sbjct: 272 IFAGKQLEDGRTLADYNIQKESTLHLVLRLR 302
Score = 114 bits (285), Expect = 2e-26
Identities = 71/151 (47%), Positives = 90/151 (59%), Gaps = 17/151 (11%)
Query: 1 MQIFIKIMYGNKMFPLIVKSSDTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLANY 60
MQIF+K + G K L V+SSDTI +VK KI DKEGIP QQRL F GK ++D +TLA+Y
Sbjct: 229 MQIFVKTLTG-KTITLEVESSDTIDNVKAKIQDKEGIPPDQQRLIFAGKQLEDGRTLADY 287
Query: 61 NIPENSTIDTRSIAKRSRPLF----------------NTILDVKKKIHDKEGIPVHQQRL 104
NI + ST+ + +F +TI +VK KI DKEGIP QQRL
Sbjct: 288 NIQKESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPDQQRL 347
Query: 105 FFDGKPIKDRQTLTNYNIPRTQPLTLVRSLR 135
F GK ++D +TL +YNI + L LV LR
Sbjct: 348 IFAGKQLEDGRTLADYNIQKESTLHLVLRLR 378
Score = 114 bits (285), Expect = 2e-26
Identities = 71/151 (47%), Positives = 90/151 (59%), Gaps = 17/151 (11%)
Query: 1 MQIFIKIMYGNKMFPLIVKSSDTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLANY 60
MQIF+K + G K L V+SSDTI +VK KI DKEGIP QQRL F GK ++D +TLA+Y
Sbjct: 1 MQIFVKTLTG-KTITLEVESSDTIDNVKAKIQDKEGIPPDQQRLIFAGKQLEDGRTLADY 59
Query: 61 NIPENSTIDTRSIAKRSRPLF----------------NTILDVKKKIHDKEGIPVHQQRL 104
NI + ST+ + +F +TI +VK KI DKEGIP QQRL
Sbjct: 60 NIQKESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPDQQRL 119
Query: 105 FFDGKPIKDRQTLTNYNIPRTQPLTLVRSLR 135
F GK ++D +TL +YNI + L LV LR
Sbjct: 120 IFAGKQLEDGRTLADYNIQKESTLHLVLRLR 150
>At5g03240 polyubiquitin (ubq3)
Length = 306
Score = 114 bits (285), Expect = 2e-26
Identities = 71/151 (47%), Positives = 90/151 (59%), Gaps = 17/151 (11%)
Query: 1 MQIFIKIMYGNKMFPLIVKSSDTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLANY 60
MQIF+K + G K L V+SSDTI +VK KI DKEGIP QQRL F GK ++D +TLA+Y
Sbjct: 1 MQIFVKTLTG-KTITLEVESSDTIDNVKAKIQDKEGIPPDQQRLIFAGKQLEDGRTLADY 59
Query: 61 NIPENSTIDTRSIAKRSRPLF----------------NTILDVKKKIHDKEGIPVHQQRL 104
NI + ST+ + +F +TI +VK KI DKEGIP QQRL
Sbjct: 60 NIQKESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPDQQRL 119
Query: 105 FFDGKPIKDRQTLTNYNIPRTQPLTLVRSLR 135
F GK ++D +TL +YNI + L LV LR
Sbjct: 120 IFAGKQLEDGRTLADYNIQKESTLHLVLRLR 150
Score = 114 bits (285), Expect = 2e-26
Identities = 71/151 (47%), Positives = 90/151 (59%), Gaps = 17/151 (11%)
Query: 1 MQIFIKIMYGNKMFPLIVKSSDTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLANY 60
MQIF+K + G K L V+SSDTI +VK KI DKEGIP QQRL F GK ++D +TLA+Y
Sbjct: 153 MQIFVKTLTG-KTITLEVESSDTIDNVKAKIQDKEGIPPDQQRLIFAGKQLEDGRTLADY 211
Query: 61 NIPENSTIDTRSIAKRSRPLF----------------NTILDVKKKIHDKEGIPVHQQRL 104
NI + ST+ + +F +TI +VK KI DKEGIP QQRL
Sbjct: 212 NIQKESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPDQQRL 271
Query: 105 FFDGKPIKDRQTLTNYNIPRTQPLTLVRSLR 135
F GK ++D +TL +YNI + L LV LR
Sbjct: 272 IFAGKQLEDGRTLADYNIQKESTLHLVLRLR 302
>At4g05320 polyubiquitin (ubq10)
Length = 464
Score = 114 bits (285), Expect = 2e-26
Identities = 71/151 (47%), Positives = 90/151 (59%), Gaps = 17/151 (11%)
Query: 1 MQIFIKIMYGNKMFPLIVKSSDTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLANY 60
MQIF+K + G K L V+SSDTI +VK KI DKEGIP QQRL F GK ++D +TLA+Y
Sbjct: 229 MQIFVKTLTG-KTITLEVESSDTIDNVKAKIQDKEGIPPDQQRLIFAGKQLEDGRTLADY 287
Query: 61 NIPENSTIDTRSIAKRSRPLF----------------NTILDVKKKIHDKEGIPVHQQRL 104
NI + ST+ + +F +TI +VK KI DKEGIP QQRL
Sbjct: 288 NIQKESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPDQQRL 347
Query: 105 FFDGKPIKDRQTLTNYNIPRTQPLTLVRSLR 135
F GK ++D +TL +YNI + L LV LR
Sbjct: 348 IFAGKQLEDGRTLADYNIQKESTLHLVLRLR 378
Score = 114 bits (285), Expect = 2e-26
Identities = 71/151 (47%), Positives = 90/151 (59%), Gaps = 17/151 (11%)
Query: 1 MQIFIKIMYGNKMFPLIVKSSDTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLANY 60
MQIF+K + G K L V+SSDTI +VK KI DKEGIP QQRL F GK ++D +TLA+Y
Sbjct: 1 MQIFVKTLTG-KTITLEVESSDTIDNVKAKIQDKEGIPPDQQRLIFAGKQLEDGRTLADY 59
Query: 61 NIPENSTIDTRSIAKRSRPLF----------------NTILDVKKKIHDKEGIPVHQQRL 104
NI + ST+ + +F +TI +VK KI DKEGIP QQRL
Sbjct: 60 NIQKESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPDQQRL 119
Query: 105 FFDGKPIKDRQTLTNYNIPRTQPLTLVRSLR 135
F GK ++D +TL +YNI + L LV LR
Sbjct: 120 IFAGKQLEDGRTLADYNIQKESTLHLVLRLR 150
Score = 114 bits (285), Expect = 2e-26
Identities = 71/151 (47%), Positives = 90/151 (59%), Gaps = 17/151 (11%)
Query: 1 MQIFIKIMYGNKMFPLIVKSSDTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLANY 60
MQIF+K + G K L V+SSDTI +VK KI DKEGIP QQRL F GK ++D +TLA+Y
Sbjct: 153 MQIFVKTLTG-KTITLEVESSDTIDNVKAKIQDKEGIPPDQQRLIFAGKQLEDGRTLADY 211
Query: 61 NIPENSTIDTRSIAKRSRPLF----------------NTILDVKKKIHDKEGIPVHQQRL 104
NI + ST+ + +F +TI +VK KI DKEGIP QQRL
Sbjct: 212 NIQKESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPDQQRL 271
Query: 105 FFDGKPIKDRQTLTNYNIPRTQPLTLVRSLR 135
F GK ++D +TL +YNI + L LV LR
Sbjct: 272 IFAGKQLEDGRTLADYNIQKESTLHLVLRLR 302
Score = 77.0 bits (188), Expect = 3e-15
Identities = 50/111 (45%), Positives = 64/111 (57%), Gaps = 17/111 (15%)
Query: 1 MQIFIKIMYGNKMFPLIVKSSDTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLANY 60
MQIF+K + G K L V+SSDTI +VK KI DKEGIP QQRL F GK ++D +TLA+Y
Sbjct: 305 MQIFVKTLTG-KTITLEVESSDTIDNVKAKIQDKEGIPPDQQRLIFAGKQLEDGRTLADY 363
Query: 61 NIPENSTIDTRSIAKRSRPLF----------------NTILDVKKKIHDKE 95
NI + ST+ + +F +TI +VK KI DKE
Sbjct: 364 NIQKESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKE 414
>At4g05050
Length = 229
Score = 114 bits (285), Expect = 2e-26
Identities = 71/151 (47%), Positives = 90/151 (59%), Gaps = 17/151 (11%)
Query: 1 MQIFIKIMYGNKMFPLIVKSSDTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLANY 60
MQIF+K + G K L V+SSDTI +VK KI DKEGIP QQRL F GK ++D +TLA+Y
Sbjct: 1 MQIFVKTLTG-KTITLEVESSDTIDNVKAKIQDKEGIPPDQQRLIFAGKQLEDGRTLADY 59
Query: 61 NIPENSTIDTRSIAKRSRPLF----------------NTILDVKKKIHDKEGIPVHQQRL 104
NI + ST+ + +F +TI +VK KI DKEGIP QQRL
Sbjct: 60 NIQKESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPDQQRL 119
Query: 105 FFDGKPIKDRQTLTNYNIPRTQPLTLVRSLR 135
F GK ++D +TL +YNI + L LV LR
Sbjct: 120 IFAGKQLEDGRTLADYNIQKESTLHLVLRLR 150
Score = 114 bits (285), Expect = 2e-26
Identities = 71/151 (47%), Positives = 90/151 (59%), Gaps = 17/151 (11%)
Query: 1 MQIFIKIMYGNKMFPLIVKSSDTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLANY 60
MQIF+K + G K L V+SSDTI +VK KI DKEGIP QQRL F GK ++D +TLA+Y
Sbjct: 77 MQIFVKTLTG-KTITLEVESSDTIDNVKAKIQDKEGIPPDQQRLIFAGKQLEDGRTLADY 135
Query: 61 NIPENSTIDTRSIAKRSRPLF----------------NTILDVKKKIHDKEGIPVHQQRL 104
NI + ST+ + +F +TI +VK KI DKEGIP QQRL
Sbjct: 136 NIQKESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPDQQRL 195
Query: 105 FFDGKPIKDRQTLTNYNIPRTQPLTLVRSLR 135
F GK ++D +TL +YNI + L LV LR
Sbjct: 196 IFAGKQLEDGRTLADYNIQKESTLHLVLRLR 226
>At4g02890 polyubiquitin
Length = 229
Score = 114 bits (285), Expect = 2e-26
Identities = 71/151 (47%), Positives = 90/151 (59%), Gaps = 17/151 (11%)
Query: 1 MQIFIKIMYGNKMFPLIVKSSDTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLANY 60
MQIF+K + G K L V+SSDTI +VK KI DKEGIP QQRL F GK ++D +TLA+Y
Sbjct: 1 MQIFVKTLTG-KTITLEVESSDTIDNVKAKIQDKEGIPPDQQRLIFAGKQLEDGRTLADY 59
Query: 61 NIPENSTIDTRSIAKRSRPLF----------------NTILDVKKKIHDKEGIPVHQQRL 104
NI + ST+ + +F +TI +VK KI DKEGIP QQRL
Sbjct: 60 NIQKESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPDQQRL 119
Query: 105 FFDGKPIKDRQTLTNYNIPRTQPLTLVRSLR 135
F GK ++D +TL +YNI + L LV LR
Sbjct: 120 IFAGKQLEDGRTLADYNIQKESTLHLVLRLR 150
Score = 114 bits (285), Expect = 2e-26
Identities = 71/151 (47%), Positives = 90/151 (59%), Gaps = 17/151 (11%)
Query: 1 MQIFIKIMYGNKMFPLIVKSSDTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLANY 60
MQIF+K + G K L V+SSDTI +VK KI DKEGIP QQRL F GK ++D +TLA+Y
Sbjct: 77 MQIFVKTLTG-KTITLEVESSDTIDNVKAKIQDKEGIPPDQQRLIFAGKQLEDGRTLADY 135
Query: 61 NIPENSTIDTRSIAKRSRPLF----------------NTILDVKKKIHDKEGIPVHQQRL 104
NI + ST+ + +F +TI +VK KI DKEGIP QQRL
Sbjct: 136 NIQKESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPDQQRL 195
Query: 105 FFDGKPIKDRQTLTNYNIPRTQPLTLVRSLR 135
F GK ++D +TL +YNI + L LV LR
Sbjct: 196 IFAGKQLEDGRTLADYNIQKESTLHLVLRLR 226
>At5g37640 polyubiquitin 4
Length = 322
Score = 110 bits (276), Expect = 2e-25
Identities = 70/150 (46%), Positives = 91/150 (60%), Gaps = 17/150 (11%)
Query: 1 MQIFIKIMYGNKMFPLIVKSSDTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLANY 60
MQIF+K + G K L V+SSDTI +VK KI DKEG+P QQRL F GK + D +TLA+Y
Sbjct: 79 MQIFVKTLTG-KTITLEVESSDTIDNVKAKIQDKEGVPPDQQRLIFAGKQLDDGRTLADY 137
Query: 61 NIPENST------------IDTRSIAKRSRPL----FNTILDVKKKIHDKEGIPVHQQRL 104
NI + ST I R++ +++ L +T +VK KI DKEGIP QQRL
Sbjct: 138 NIQKESTLHLVLRLRGGMQIFVRTLTRKTIALEVESSDTTDNVKAKIQDKEGIPPDQQRL 197
Query: 105 FFDGKPIKDRQTLTNYNIPRTQPLTLVRSL 134
F GK ++D +TL +YNI + L LV L
Sbjct: 198 IFAGKQLEDGRTLADYNIQKESTLHLVLRL 227
Score = 99.4 bits (246), Expect = 5e-22
Identities = 64/151 (42%), Positives = 84/151 (55%), Gaps = 17/151 (11%)
Query: 1 MQIFIKIMYGNKMFPLIVKSSDTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLANY 60
MQI K + K + V SSDTI +VK KI D EGIP+ QQRL F GK + D +TLA+Y
Sbjct: 3 MQIHAKTLT-EKTITIDVVSSDTINNVKAKIQDIEGIPLDQQRLIFSGKLLDDGRTLADY 61
Query: 61 NIPENSTIDTRSIAKRSRPLF----------------NTILDVKKKIHDKEGIPVHQQRL 104
+I ++S + + +F +TI +VK KI DKEG+P QQRL
Sbjct: 62 SIQKDSILHLALRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGVPPDQQRL 121
Query: 105 FFDGKPIKDRQTLTNYNIPRTQPLTLVRSLR 135
F GK + D +TL +YNI + L LV LR
Sbjct: 122 IFAGKQLDDGRTLADYNIQKESTLHLVLRLR 152
Score = 94.0 bits (232), Expect = 2e-20
Identities = 65/152 (42%), Positives = 84/152 (54%), Gaps = 18/152 (11%)
Query: 1 MQIFIKIMYGNKMFPLIVKSSDTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLANY 60
MQIF++ + K L V+SSDT +VK KI DKEGIP QQRL F GK ++D +TLA+Y
Sbjct: 155 MQIFVRTLT-RKTIALEVESSDTTDNVKAKIQDKEGIPPDQQRLIFAGKQLEDGRTLADY 213
Query: 61 NIPENSTIDTRSIAKRSRPLF----------------NTILDVKKKIHDKEGIPVHQQRL 104
NI + ST+ +F +TI +VK KI DKE I QQRL
Sbjct: 214 NIQKESTLHLVLRLCGGMQIFVNTLTGKTITLEVESSDTIDNVKAKIQDKERIQPDQQRL 273
Query: 105 FFDGKPIKD-RQTLTNYNIPRTQPLTLVRSLR 135
F G+ ++D TL +YNI + L LV LR
Sbjct: 274 IFAGEQLEDGYYTLADYNIQKESTLHLVLRLR 305
>At1g55060 ubiquitin-like (UBQ12)
Length = 230
Score = 108 bits (269), Expect = 1e-24
Identities = 69/151 (45%), Positives = 88/151 (57%), Gaps = 17/151 (11%)
Query: 1 MQIFIKIMYGNKMFPLIVKSSDTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLANY 60
MQIF+K + G K L V+SSDTI ++K KI DKEGIP QQRL F GK ++D +TLA+Y
Sbjct: 77 MQIFVKTLTG-KTITLEVESSDTIDNLKAKIQDKEGIPPDQQRLIFAGKQLEDGRTLADY 135
Query: 61 NIPENSTIDTRSIAKRSRPLF----------------NTILDVKKKIHDKEGIPVHQQRL 104
NI + ST+ + +F +TI +VK KI DKEGI QQRL
Sbjct: 136 NIQKESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGISPDQQRL 195
Query: 105 FFDGKPIKDRQTLTNYNIPRTQPLTLVRSLR 135
F GK +D +TL +YNI + L LV LR
Sbjct: 196 IFAGKQHEDGRTLADYNIQKESTLHLVLRLR 226
Score = 107 bits (268), Expect = 2e-24
Identities = 68/151 (45%), Positives = 89/151 (58%), Gaps = 17/151 (11%)
Query: 1 MQIFIKIMYGNKMFPLIVKSSDTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLANY 60
MQIF+K + G K L V+SSDTI +VK KI D EGIP Q RL F GK ++D +TLA+Y
Sbjct: 1 MQIFLKTLTG-KTKVLEVESSDTIDNVKAKIQDIEGIPPDQHRLIFAGKQLEDGRTLADY 59
Query: 61 NIPENSTIDTRSIAKRSRPLF----------------NTILDVKKKIHDKEGIPVHQQRL 104
N+ E+ST+ + +F +TI ++K KI DKEGIP QQRL
Sbjct: 60 NVQEDSTLHLLLRFRGGMQIFVKTLTGKTITLEVESSDTIDNLKAKIQDKEGIPPDQQRL 119
Query: 105 FFDGKPIKDRQTLTNYNIPRTQPLTLVRSLR 135
F GK ++D +TL +YNI + L LV LR
Sbjct: 120 IFAGKQLEDGRTLADYNIQKESTLHLVLRLR 150
>At1g65350
Length = 294
Score = 107 bits (266), Expect = 3e-24
Identities = 70/151 (46%), Positives = 89/151 (58%), Gaps = 18/151 (11%)
Query: 1 MQIFIKIMYGNKMFPLIVKSSDTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLANY 60
MQIF+K + G K L V+SSDTI +VK KI DKEGIP QQRL F GK ++D +TLA+Y
Sbjct: 1 MQIFVKTLTG-KTITLEVESSDTIDNVKAKIQDKEGIPPDQQRLIFAGKQLEDGRTLADY 59
Query: 61 NIPENSTIDTRSIAKRSRPLF----------------NTILDVKKKIHDKEGIPVHQQRL 104
NI + ST+ + +F +TI +VK KI DKEGIP QQRL
Sbjct: 60 NIQKESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPDQQRL 119
Query: 105 FFDGKPIKDRQTLTNYNIPRTQPLTLVRSLR 135
F GK ++D +TL + NI + L LV LR
Sbjct: 120 IFAGKQLEDGRTLAD-NIQKESTLHLVLRLR 149
Score = 103 bits (258), Expect = 2e-23
Identities = 69/151 (45%), Positives = 88/151 (57%), Gaps = 18/151 (11%)
Query: 1 MQIFIKIMYGNKMFPLIVKSSDTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLANY 60
MQIF+K + G K L V+SSDTI +VK KI DKEGIP QQRL F GK ++D +TLA+
Sbjct: 77 MQIFVKTLTG-KTITLEVESSDTIDNVKAKIQDKEGIPPDQQRLIFAGKQLEDGRTLAD- 134
Query: 61 NIPENSTIDTRSIAKRSRPLF----------------NTILDVKKKIHDKEGIPVHQQRL 104
NI + ST+ + +F +TI +VK KI DKE IP QQRL
Sbjct: 135 NIQKESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEWIPPDQQRL 194
Query: 105 FFDGKPIKDRQTLTNYNIPRTQPLTLVRSLR 135
F GK ++D +TL +YNI + L LV LR
Sbjct: 195 IFAGKQLEDGRTLADYNIQKESTLHLVLRLR 225
Score = 92.8 bits (229), Expect = 5e-20
Identities = 59/128 (46%), Positives = 74/128 (57%), Gaps = 17/128 (13%)
Query: 1 MQIFIKIMYGNKMFPLIVKSSDTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLANY 60
MQIF+K + G K L V+SSDTI +VK KI DKE IP QQRL F GK ++D +TLA+Y
Sbjct: 152 MQIFVKTLTG-KTITLEVESSDTIDNVKAKIQDKEWIPPDQQRLIFAGKQLEDGRTLADY 210
Query: 61 NIPENSTIDTRSIAKRSRPLF----------------NTILDVKKKIHDKEGIPVHQQRL 104
NI + ST+ + +F TI +VK KI DKEGIP QQRL
Sbjct: 211 NIQKESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSGTIDNVKAKIQDKEGIPPDQQRL 270
Query: 105 FFDGKPIK 112
F GK ++
Sbjct: 271 IFAGKQLE 278
>At1g31340 putative ubiquitin (AtRUB1)
Length = 156
Score = 102 bits (254), Expect = 6e-23
Identities = 65/151 (43%), Positives = 90/151 (59%), Gaps = 17/151 (11%)
Query: 1 MQIFIKIMYGNKMFPLIVKSSDTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLANY 60
MQIF+K + G K L V+SSDTI +VK KI DKEGIP QQRL F GK ++D +TLA+Y
Sbjct: 1 MQIFVKTLTG-KTITLEVESSDTIDNVKAKIQDKEGIPPDQQRLIFAGKQLEDGRTLADY 59
Query: 61 NIPENST------------IDTRSIAKRSRPL----FNTILDVKKKIHDKEGIPVHQQRL 104
NI + ST I +++ + + +TI +K+++ +KEGIP QQRL
Sbjct: 60 NIQKESTLHLVLRLRGGTMIKVKTLTGKEIEIDIEPTDTIDRIKERVEEKEGIPPVQQRL 119
Query: 105 FFDGKPIKDRQTLTNYNIPRTQPLTLVRSLR 135
+ GK + D +T +YNI L LV +LR
Sbjct: 120 IYAGKQLADDKTAKDYNIEGGSVLHLVLALR 150
>At3g09790 polyubiquitin
Length = 631
Score = 98.2 bits (243), Expect = 1e-21
Identities = 63/150 (42%), Positives = 86/150 (57%), Gaps = 17/150 (11%)
Query: 1 MQIFIKIMYGNKMFPLIVKSSDTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLANY 60
+QI+ K + K L V++SD+I +VK KI +KEGIP+ QQRL F GK ++D TLA+Y
Sbjct: 3 IQIYAKTLT-EKTITLDVETSDSIHNVKAKIQNKEGIPLDQQRLIFAGKQLEDGLTLADY 61
Query: 61 NIPENSTIDTRSIAKRSRPLF----------------NTILDVKKKIHDKEGIPVHQQRL 104
NI + ST+ + +F +TI +VK KI DKEGI QQRL
Sbjct: 62 NIQKESTLHLVLRLRGGMQIFVQTLTGKTITLEVKSSDTIDNVKAKIQDKEGILPRQQRL 121
Query: 105 FFDGKPIKDRQTLTNYNIPRTQPLTLVRSL 134
F GK ++D +TL +YNI + L LV L
Sbjct: 122 IFAGKQLEDGRTLADYNIQKESTLHLVLRL 151
Score = 92.0 bits (227), Expect = 9e-20
Identities = 65/161 (40%), Positives = 86/161 (53%), Gaps = 30/161 (18%)
Query: 1 MQIFIKIMYGNKMFPLIVKSSDTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLANY 60
MQIF++ + G K L VKSSDTI +VK KI DKEGI QQRL F GK ++D +TLA+Y
Sbjct: 79 MQIFVQTLTG-KTITLEVKSSDTIDNVKAKIQDKEGILPRQQRLIFAGKQLEDGRTLADY 137
Query: 61 NIPENSTI------------------------DTRSIAKRSRPLFNTILDVKKKIHDKEG 96
NI + ST+ DT ++ S +TI +VK KI D+EG
Sbjct: 138 NIQKESTLHLVLRLCGGMQIFVSTFSGKNFTSDTLTLKVESS---DTIENVKAKIQDREG 194
Query: 97 IPVHQQRLFFDGKPI--KDRQTLTNYNIPRTQPLTLVRSLR 135
+ QRL F G+ + +D +TL +Y I L L LR
Sbjct: 195 LRPDHQRLIFHGEELFTEDNRTLADYGIRNRSTLCLALRLR 235
Score = 86.7 bits (213), Expect = 4e-18
Identities = 60/158 (37%), Positives = 84/158 (52%), Gaps = 22/158 (13%)
Query: 1 MQIFIKIMYGN------KMFPLIVKSSDTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDR 54
MQIF+K + K L V+SSDTI +VK KI K GIP+ +QRL F G+ +
Sbjct: 469 MQIFVKTFSFSGETPTCKTITLEVESSDTIDNVKVKIQHKVGIPLDRQRLIFGGRVLVGS 528
Query: 55 QTLANYNIPENSTIDTRSIAKRSRPLF----------------NTILDVKKKIHDKEGIP 98
+TL +YNI + STI + + +F +TI +VK+KI KEGI
Sbjct: 529 RTLLDYNIQKGSTIHQLFLQRGGMQIFIKTLTGKTIILEVESSDTIANVKEKIQVKEGIK 588
Query: 99 VHQQRLFFDGKPIKDRQTLTNYNIPRTQPLTLVRSLRE 136
QQ L F G+ ++D TL +Y+I + L LV LR+
Sbjct: 589 PDQQMLIFFGQQLEDGVTLGDYDIHKKSTLYLVLRLRQ 626
Score = 85.1 bits (209), Expect = 1e-17
Identities = 61/157 (38%), Positives = 84/157 (52%), Gaps = 22/157 (14%)
Query: 1 MQIFIKIMYGNKM----FPLIVKSSDTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQT 56
M IF+K + N F L V+SSDTI +VK K+ DKE IP+ RL F GKP++ +T
Sbjct: 238 MYIFVKNLPYNSFTGENFILEVESSDTIDNVKAKLQDKERIPMDLHRLIFAGKPLEGGRT 297
Query: 57 LANYNIPENSTID----------------TRSIAKRSRPLFNTILDVKKKIHDKEGI--P 98
LA+YNI + ST+ TR ++TI +VK + DKEGI
Sbjct: 298 LAHYNIQKGSTLYLVTRFRCGMQIFVKTLTRKRINLEVESWDTIENVKAMVQDKEGIQPQ 357
Query: 99 VHQQRLFFDGKPIKDRQTLTNYNIPRTQPLTLVRSLR 135
+ QRL F GK +KD TL +Y+I + L LV ++
Sbjct: 358 PNLQRLIFLGKELKDGCTLADYSIQKESTLHLVLGMQ 394
Score = 85.1 bits (209), Expect = 1e-17
Identities = 56/147 (38%), Positives = 76/147 (51%), Gaps = 24/147 (16%)
Query: 1 MQIFIKIMYGNKMFPLIVKSSDTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLANY 60
MQIF+K+ +G K+ L V SSDTI VK KI DK G P QQ L F G ++D +TL +Y
Sbjct: 393 MQIFVKL-FGGKIITLEVLSSDTIKSVKAKIQDKVGSPPDQQILLFRGGQLQDGRTLGDY 451
Query: 61 NIPENSTIDTRSIAKRSRPLF-----------------------NTILDVKKKIHDKEGI 97
NI ST+ + +F +TI +VK KI K GI
Sbjct: 452 NIRNESTLHLFFHIRHGMQIFVKTFSFSGETPTCKTITLEVESSDTIDNVKVKIQHKVGI 511
Query: 98 PVHQQRLFFDGKPIKDRQTLTNYNIPR 124
P+ +QRL F G+ + +TL +YNI +
Sbjct: 512 PLDRQRLIFGGRVLVGSRTLLDYNIQK 538
Score = 78.2 bits (191), Expect = 1e-15
Identities = 60/149 (40%), Positives = 77/149 (51%), Gaps = 15/149 (10%)
Query: 1 MQIFIKIMYGNKMFPLIVKSSDTILDVKKKIHDKEGIPVHQ--QRLFFDGKPIKDRQTLA 58
MQIF+K + K L V+S DTI +VK + DKEGI QRL F GK +KD TLA
Sbjct: 319 MQIFVKTLT-RKRINLEVESWDTIENVKAMVQDKEGIQPQPNLQRLIFLGKELKDGCTLA 377
Query: 59 NYNIPENSTIDTRSIAKRSRPLF------------NTILDVKKKIHDKEGIPVHQQRLFF 106
+Y+I + ST+ + LF +TI VK KI DK G P QQ L F
Sbjct: 378 DYSIQKESTLHLVLGMQIFVKLFGGKIITLEVLSSDTIKSVKAKIQDKVGSPPDQQILLF 437
Query: 107 DGKPIKDRQTLTNYNIPRTQPLTLVRSLR 135
G ++D +TL +YNI L L +R
Sbjct: 438 RGGQLQDGRTLGDYNIRNESTLHLFFHIR 466
Score = 54.3 bits (129), Expect = 2e-08
Identities = 28/54 (51%), Positives = 37/54 (67%)
Query: 82 NTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLTNYNIPRTQPLTLVRSLR 135
++I +VK KI +KEGIP+ QQRL F GK ++D TL +YNI + L LV LR
Sbjct: 23 DSIHNVKAKIQNKEGIPLDQQRLIFAGKQLEDGLTLADYNIQKESTLHLVLRLR 76
>At3g62250 ubiquitin extension protein (UBQ5)
Length = 157
Score = 73.9 bits (180), Expect = 2e-14
Identities = 40/68 (58%), Positives = 50/68 (72%), Gaps = 1/68 (1%)
Query: 1 MQIFIKIMYGNKMFPLIVKSSDTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLANY 60
MQIF+K + G K L V+SSDTI +VK KI DKEGIP QQRL F GK ++D +TLA+Y
Sbjct: 1 MQIFVKTLTG-KTITLEVESSDTIDNVKAKIQDKEGIPPDQQRLIFAGKQLEDGRTLADY 59
Query: 61 NIPENSTI 68
NI + ST+
Sbjct: 60 NIQKESTL 67
Score = 57.4 bits (137), Expect = 2e-09
Identities = 30/54 (55%), Positives = 37/54 (67%)
Query: 82 NTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLTNYNIPRTQPLTLVRSLR 135
+TI +VK KI DKEGIP QQRL F GK ++D +TL +YNI + L LV LR
Sbjct: 21 DTIDNVKAKIQDKEGIPPDQQRLIFAGKQLEDGRTLADYNIQKESTLHLVLRLR 74
>At3g52590 ubiquitin / ribosomal protein CEP52
Length = 128
Score = 73.9 bits (180), Expect = 2e-14
Identities = 40/68 (58%), Positives = 50/68 (72%), Gaps = 1/68 (1%)
Query: 1 MQIFIKIMYGNKMFPLIVKSSDTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLANY 60
MQIF+K + G K L V+SSDTI +VK KI DKEGIP QQRL F GK ++D +TLA+Y
Sbjct: 1 MQIFVKTLTG-KTITLEVESSDTIDNVKAKIQDKEGIPPDQQRLIFAGKQLEDGRTLADY 59
Query: 61 NIPENSTI 68
NI + ST+
Sbjct: 60 NIQKESTL 67
Score = 57.4 bits (137), Expect = 2e-09
Identities = 30/54 (55%), Positives = 37/54 (67%)
Query: 82 NTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLTNYNIPRTQPLTLVRSLR 135
+TI +VK KI DKEGIP QQRL F GK ++D +TL +YNI + L LV LR
Sbjct: 21 DTIDNVKAKIQDKEGIPPDQQRLIFAGKQLEDGRTLADYNIQKESTLHLVLRLR 74
>At2g47110 putative ubiquitin extension protein (UBQ6)
Length = 157
Score = 73.9 bits (180), Expect = 2e-14
Identities = 40/68 (58%), Positives = 50/68 (72%), Gaps = 1/68 (1%)
Query: 1 MQIFIKIMYGNKMFPLIVKSSDTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLANY 60
MQIF+K + G K L V+SSDTI +VK KI DKEGIP QQRL F GK ++D +TLA+Y
Sbjct: 1 MQIFVKTLTG-KTITLEVESSDTIDNVKAKIQDKEGIPPDQQRLIFAGKQLEDGRTLADY 59
Query: 61 NIPENSTI 68
NI + ST+
Sbjct: 60 NIQKESTL 67
Score = 57.4 bits (137), Expect = 2e-09
Identities = 30/54 (55%), Positives = 37/54 (67%)
Query: 82 NTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLTNYNIPRTQPLTLVRSLR 135
+TI +VK KI DKEGIP QQRL F GK ++D +TL +YNI + L LV LR
Sbjct: 21 DTIDNVKAKIQDKEGIPPDQQRLIFAGKQLEDGRTLADYNIQKESTLHLVLRLR 74
>At2g36170 putative ubiquitin/ribosomal protein CEP52
Length = 128
Score = 73.9 bits (180), Expect = 2e-14
Identities = 40/68 (58%), Positives = 50/68 (72%), Gaps = 1/68 (1%)
Query: 1 MQIFIKIMYGNKMFPLIVKSSDTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLANY 60
MQIF+K + G K L V+SSDTI +VK KI DKEGIP QQRL F GK ++D +TLA+Y
Sbjct: 1 MQIFVKTLTG-KTITLEVESSDTIDNVKAKIQDKEGIPPDQQRLIFAGKQLEDGRTLADY 59
Query: 61 NIPENSTI 68
NI + ST+
Sbjct: 60 NIQKESTL 67
Score = 57.4 bits (137), Expect = 2e-09
Identities = 30/54 (55%), Positives = 37/54 (67%)
Query: 82 NTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLTNYNIPRTQPLTLVRSLR 135
+TI +VK KI DKEGIP QQRL F GK ++D +TL +YNI + L LV LR
Sbjct: 21 DTIDNVKAKIQDKEGIPPDQQRLIFAGKQLEDGRTLADYNIQKESTLHLVLRLR 74
>At1g23410 ubiquitin extension protein, putative
Length = 156
Score = 73.9 bits (180), Expect = 2e-14
Identities = 40/68 (58%), Positives = 50/68 (72%), Gaps = 1/68 (1%)
Query: 1 MQIFIKIMYGNKMFPLIVKSSDTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLANY 60
MQIF+K + G K L V+SSDTI +VK KI DKEGIP QQRL F GK ++D +TLA+Y
Sbjct: 1 MQIFVKTLTG-KTITLEVESSDTIDNVKAKIQDKEGIPPDQQRLIFAGKQLEDGRTLADY 59
Query: 61 NIPENSTI 68
NI + ST+
Sbjct: 60 NIQKESTL 67
Score = 57.4 bits (137), Expect = 2e-09
Identities = 30/54 (55%), Positives = 37/54 (67%)
Query: 82 NTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLTNYNIPRTQPLTLVRSLR 135
+TI +VK KI DKEGIP QQRL F GK ++D +TL +YNI + L LV LR
Sbjct: 21 DTIDNVKAKIQDKEGIPPDQQRLIFAGKQLEDGRTLADYNIQKESTLHLVLRLR 74
>At1g53930 hypothetical protein
Length = 158
Score = 65.9 bits (159), Expect = 7e-12
Identities = 35/68 (51%), Positives = 48/68 (70%), Gaps = 1/68 (1%)
Query: 1 MQIFIKIMYGNKMFPLIVKSSDTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLANY 60
M IFIK + G ++ L V++ DTI ++K KI DK+GIPV QQRL F GK ++D TLA+Y
Sbjct: 86 MFIFIKTLTGRRIV-LEVENCDTIENIKAKIQDKQGIPVDQQRLIFKGKQLEDGLTLADY 144
Query: 61 NIPENSTI 68
NI +S +
Sbjct: 145 NIQNDSIL 152
Score = 55.5 bits (132), Expect = 9e-09
Identities = 27/53 (50%), Positives = 36/53 (66%)
Query: 82 NTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLTNYNIPRTQPLTLVRSL 134
+TI ++K KI DK+GIPV QQRL F GK ++D TL +YNI L L++ L
Sbjct: 106 DTIENIKAKIQDKQGIPVDQQRLIFKGKQLEDGLTLADYNIQNDSILHLIKRL 158
>At1g53960 unknown protein
Length = 144
Score = 62.0 bits (149), Expect = 9e-11
Identities = 32/68 (47%), Positives = 45/68 (66%), Gaps = 1/68 (1%)
Query: 1 MQIFIKIMYGNKMFPLIVKSSDTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLANY 60
M IFIK + G + VK SDTI ++K K +KEGIPV QQRL F G+ ++D + +++Y
Sbjct: 69 MMIFIKTLTG-RTNTYEVKGSDTIRELKAKHEEKEGIPVEQQRLIFQGRVLEDSKAISDY 127
Query: 61 NIPENSTI 68
NI ST+
Sbjct: 128 NIKHESTL 135
Score = 50.1 bits (118), Expect = 4e-07
Identities = 43/128 (33%), Positives = 61/128 (47%), Gaps = 12/128 (9%)
Query: 1 MQIFIKIMYGNKMFPLIVKSSDTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLANY 60
M I IK + G K L V+ S +D+K IH G P + L D P D +
Sbjct: 20 MHISIKTLEG-KRIKLEVEDSSNTIDLK--IH---GEPTRE--LVVDLSPTPDTGAIMMI 71
Query: 61 NIPENSTIDTRSIAKRSRPLFNTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLTNY 120
I + K S +TI ++K K +KEGIPV QQRL F G+ ++D + +++Y
Sbjct: 72 FIKTLTGRTNTYEVKGS----DTIRELKAKHEEKEGIPVEQQRLIFQGRVLEDSKAISDY 127
Query: 121 NIPRTQPL 128
NI L
Sbjct: 128 NIKHESTL 135
>At1g53950 hypothetical protein
Length = 121
Score = 49.3 bits (116), Expect = 6e-07
Identities = 28/68 (41%), Positives = 41/68 (60%), Gaps = 1/68 (1%)
Query: 1 MQIFIKIMYGNKMFPLIVKSSDTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLANY 60
M+IF+ + K F L VK SDTI +K IHD+ G PV QQ L G +K+ T+++Y
Sbjct: 46 MRIFVSTIK-EKTFILEVKGSDTIKKLKSMIHDQGGPPVDQQDLNHLGTKLKNNATISDY 104
Query: 61 NIPENSTI 68
NI ++ +
Sbjct: 105 NIKPDANL 112
Score = 37.7 bits (86), Expect = 0.002
Identities = 41/133 (30%), Positives = 62/133 (45%), Gaps = 16/133 (12%)
Query: 1 MQIFIKIMYGNKMFPLIVKSSDTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLANY 60
MQI IK + G + + SSDTI D+K G P Q L G + R +
Sbjct: 1 MQISIKTVKGKSINLEVADSSDTI-DLKIN-----GEPTRQLVLRL-GPAVLMRIFV--- 50
Query: 61 NIPENSTIDTRSIAKRSRPLFNTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLTNY 120
STI ++ + +TI +K IHD+ G PV QQ L G +K+ T+++Y
Sbjct: 51 -----STIKEKTFILEVKGS-DTIKKLKSMIHDQGGPPVDQQDLNHLGTKLKNNATISDY 104
Query: 121 NIPRTQPLTLVRS 133
NI L ++++
Sbjct: 105 NIKPDANLVIMQN 117
>At3g02540 RAD23 like protein
Length = 419
Score = 47.4 bits (111), Expect = 2e-06
Identities = 27/71 (38%), Positives = 40/71 (56%), Gaps = 4/71 (5%)
Query: 1 MQIFIKIMYGNKMFPLIVKSSDTILDVKKKIHDKEGIPVH---QQRLFFDGKPIKDRQTL 57
M+IF+K + G F + VK D+++DVKK I +G V+ +Q L GK +KD T+
Sbjct: 1 MKIFVKTLKGTH-FEIEVKPEDSVVDVKKNIESVQGADVYPAAKQMLIHQGKVLKDETTI 59
Query: 58 ANYNIPENSTI 68
+ ENS I
Sbjct: 60 EENKVAENSFI 70
Score = 28.5 bits (62), Expect = 1.2
Identities = 14/39 (35%), Positives = 23/39 (58%), Gaps = 3/39 (7%)
Query: 82 NTILDVKKKIHDKEGIPVH---QQRLFFDGKPIKDRQTL 117
++++DVKK I +G V+ +Q L GK +KD T+
Sbjct: 21 DSVVDVKKNIESVQGADVYPAAKQMLIHQGKVLKDETTI 59
>At1g11980 unknown protein
Length = 78
Score = 46.6 bits (109), Expect = 4e-06
Identities = 24/56 (42%), Positives = 35/56 (61%)
Query: 80 LFNTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLTNYNIPRTQPLTLVRSLR 135
L +TI +K++I +KEGIP QR+ + GK + D T +YN+ R L LV +LR
Sbjct: 19 LTDTIERIKERIEEKEGIPPVHQRIVYTGKQLADDLTAKHYNLERGSVLHLVLALR 74
Score = 46.2 bits (108), Expect = 5e-06
Identities = 23/68 (33%), Positives = 40/68 (58%), Gaps = 1/68 (1%)
Query: 1 MQIFIKIMYGNKMFPLIVKSSDTILDVKKKIHDKEGIPVHQQRLFFDGKPIKDRQTLANY 60
M+I +K + K + ++ +DTI +K++I +KEGIP QR+ + GK + D T +Y
Sbjct: 1 MEIKVKTLT-EKQIDIEIELTDTIERIKERIEEKEGIPPVHQRIVYTGKQLADDLTAKHY 59
Query: 61 NIPENSTI 68
N+ S +
Sbjct: 60 NLERGSVL 67
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.141 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,025,976
Number of Sequences: 26719
Number of extensions: 125837
Number of successful extensions: 491
Number of sequences better than 10.0: 66
Number of HSP's better than 10.0 without gapping: 58
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 308
Number of HSP's gapped (non-prelim): 132
length of query: 138
length of database: 11,318,596
effective HSP length: 89
effective length of query: 49
effective length of database: 8,940,605
effective search space: 438089645
effective search space used: 438089645
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 54 (25.4 bits)
Medicago: description of AC148342.16


