

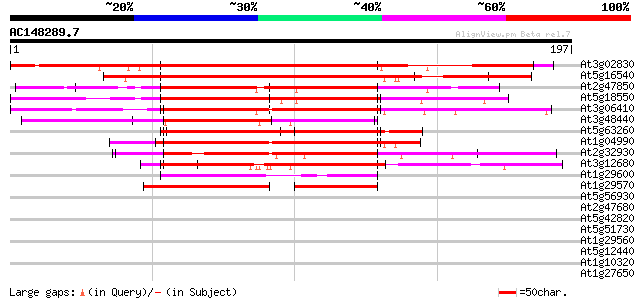
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148289.7 - phase: 0
(197 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g02830 zinc finger protein 1 (zfn1) 210 4e-55
At5g16540 zinc finger protein 3 (ZFN3) 202 1e-52
At2g47850 unknown protein 157 4e-39
At5g18550 zinc finger -like protein 129 8e-31
At3g06410 hypothetical protein 129 1e-30
At3g48440 unknown protein 124 3e-29
At5g63260 unknown protein 114 3e-26
At1g04990 zinc finger protein 2, putative 102 2e-22
At2g32930 putative protein 101 3e-22
At3g12680 floral homeotic protein HUA1 (HUA1) 93 8e-20
At1g29600 zinc finger protein, putative 49 2e-06
At1g29570 zinc finger protein, putative 48 3e-06
At5g56930 unknown protein 37 0.009
At2g47680 putative ATP-dependent RNA helicase A 35 0.020
At5g42820 U2 snRNP auxiliary factor, small subunit 33 0.075
At5g51730 unknown protein 32 0.17
At1g29560 hypothetical protein 32 0.22
At5g12440 unknown protein 32 0.28
At1g10320 unknown protein 32 0.28
At1g27650 U2 snRNP auxiliary factor, putative 30 0.63
>At3g02830 zinc finger protein 1 (zfn1)
Length = 397
Score = 210 bits (534), Expect = 4e-55
Identities = 108/188 (57%), Positives = 129/188 (68%), Gaps = 27/188 (14%)
Query: 1 MGSDSPQQTMRNDQTYGTSHQGDPENAGLQ--GVYSQYRSGS-VPVG-FYALQRENIFPE 56
+GS SP T NDQ Y Q + +G Q G +S Y GS VP+G +YAL REN+FPE
Sbjct: 215 LGSVSPSGT-GNDQNYRNLQQNETIESGSQSQGSFSGYNPGSSVPLGGYYALPRENVFPE 273
Query: 57 RPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGEPLCVFYSRYG 116
RP QPECQFYMKTGDCKFG VC+FHHPR+R P PDC+LS +GLPLRPGEPLCVFY+RYG
Sbjct: 274 RPGQPECQFYMKTGDCKFGTVCKFHHPRDRQAPPPDCLLSSIGLPLRPGEPLCVFYTRYG 333
Query: 117 ICKFGPSCKFDHPMGIFTYNVSASPLAEAAGRRLLGSSSGTAALSLSSEGLVESGSVKPR 176
ICKFGPSCKFDHPM +FTY+ +AS ++ +VE+ + K R
Sbjct: 334 ICKFGPSCKFDHPMRVFTYDNTAS----------------------ETDEVVETSTGKSR 371
Query: 177 RLSLSETR 184
RLS+SETR
Sbjct: 372 RLSVSETR 379
Score = 97.8 bits (242), Expect = 3e-21
Identities = 57/147 (38%), Positives = 77/147 (51%), Gaps = 11/147 (7%)
Query: 54 FPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGEPLCVFYS 113
+PER QPEC++Y+KTG CKFG C+FHHPR + A L+ LG PLR E C ++
Sbjct: 82 YPERIGQPECEYYLKTGTCKFGVTCKFHHPRNKAGIAGRVSLNMLGYPLRSNEVDCAYFL 141
Query: 114 RYGICKFGPSCKFDHP------MGIFTYNVSASPLAEA---AGRRLLGSSSGTAALSLSS 164
R G CKFG +CKF+HP M + T + P + A A R SS A +
Sbjct: 142 RTGHCKFGGTCKFNHPQPQPTNMMVPTSGQQSYPWSRASFIASPRWQDPSS--YASLIMP 199
Query: 165 EGLVESGSVKPRRLSLSETRPNPSGDD 191
+G+V P L P+ +G+D
Sbjct: 200 QGVVPVQGWNPYSGQLGSVSPSGTGND 226
Score = 86.3 bits (212), Expect = 1e-17
Identities = 35/76 (46%), Positives = 51/76 (67%), Gaps = 1/76 (1%)
Query: 54 FPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGEPLCVFYS 113
+PERP +P+C +Y++TG C+FG+ CRF+HPR+R + + P R G+P C +Y
Sbjct: 37 YPERPGEPDCSYYIRTGLCRFGSTCRFNHPRDRELVIATARMRG-EYPERIGQPECEYYL 95
Query: 114 RYGICKFGPSCKFDHP 129
+ G CKFG +CKF HP
Sbjct: 96 KTGTCKFGVTCKFHHP 111
>At5g16540 zinc finger protein 3 (ZFN3)
Length = 375
Score = 202 bits (513), Expect = 1e-52
Identities = 99/152 (65%), Positives = 113/152 (74%), Gaps = 9/152 (5%)
Query: 34 SQYRSG-SVPVGFYALQRENIFPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPD 92
S + SG SVP+GFYAL REN+FPERP QPECQFYMKTGDCKFG VC+FHHPR+R P PD
Sbjct: 219 SGFHSGNSVPLGFYALPRENVFPERPGQPECQFYMKTGDCKFGTVCKFHHPRDRQTPPPD 278
Query: 93 CVLSPLGLPLRPGEPLCVFYSRYGICKFGPSCKFDHPMGIFTY-NVSASPLAEAAGRRLL 151
CVLS +GLPLRPGEPLCVFYSRYGICKFGPSCKFDHPM +FTY N +ASP +
Sbjct: 279 CVLSSVGLPLRPGEPLCVFYSRYGICKFGPSCKFDHPMRVFTYNNNTASPSPSS------ 332
Query: 152 GSSSGTAALSLSSEGLVESGSVKPRRLSLSET 183
S A++ L+ S SV+ + SL ET
Sbjct: 333 -SLHQETAITTELRNLLVSSSVEAKPTSLPET 363
Score = 97.1 bits (240), Expect = 6e-21
Identities = 51/118 (43%), Positives = 66/118 (55%), Gaps = 8/118 (6%)
Query: 54 FPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGEPLCVFYS 113
+PER QPEC+FY+KTG CKFG C+FHHPR + ++ L PLRP E C ++
Sbjct: 84 YPERIGQPECEFYLKTGTCKFGVTCKFHHPRNKAGIDGSVSVNVLSYPLRPNEDDCSYFL 143
Query: 114 RYGICKFGPSCKFDHPMGIFT---YNVSASPLAEAAGRRLLGSSSGTAALSLSSEGLV 168
R G CKFG +CKF+HP T +V SP+ A L S +G + S S V
Sbjct: 144 RIGQCKFGGTCKFNHPQTQSTNLMVSVRGSPVYSA-----LQSLTGQPSYSWSRTSFV 196
Score = 83.2 bits (204), Expect = 8e-17
Identities = 39/95 (41%), Positives = 56/95 (58%), Gaps = 7/95 (7%)
Query: 54 FPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGEPLCVFYS 113
+PER +P+C +Y++TG C+FG+ CRF+HP +R + + P R G+P C FY
Sbjct: 39 YPERHGEPDCAYYIRTGLCRFGSTCRFNHPHDRKLVIATARIKG-EYPERIGQPECEFYL 97
Query: 114 RYGICKFGPSCKFDHPM------GIFTYNVSASPL 142
+ G CKFG +CKF HP G + NV + PL
Sbjct: 98 KTGTCKFGVTCKFHHPRNKAGIDGSVSVNVLSYPL 132
>At2g47850 unknown protein
Length = 553
Score = 157 bits (396), Expect = 4e-39
Identities = 86/175 (49%), Positives = 104/175 (59%), Gaps = 13/175 (7%)
Query: 3 SDSPQQTMRNDQTYGTSHQGDPENAGLQGVYSQYRSGSVPVGFYALQRENIFPERPDQPE 62
S Q + YG + Q L GVY S P G +Q+E FPERP +PE
Sbjct: 326 SPGAQHAVGATSLYGVT-QLTSTTPSLPGVYPSLSS---PTG--VIQKEQAFPERPGEPE 379
Query: 63 CQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGEPLCVFYSRYGICKFGP 122
CQ+Y+KTGDCKFG C+FHHPR+R P +CVLSP+GLPLRPG C FY + G CKFG
Sbjct: 380 CQYYLKTGDCKFGTSCKFHHPRDRVPPRANCVLSPIGLPLRPGVQRCTFYVQNGFCKFGS 439
Query: 123 SCKFDHPMGIFTYNVSASPLAEA-----AGRRLLGSSSGTAALSLSSEGLVESGS 172
+CKFDHPMG YN SAS LA+A LLG+ + AA S SS L+ G+
Sbjct: 440 TCKFDHPMGTIRYNPSASSLADAPVAPYPVSSLLGALA--AAPSSSSTELIAGGA 492
Score = 89.4 bits (220), Expect = 1e-18
Identities = 38/76 (50%), Positives = 49/76 (64%)
Query: 54 FPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGEPLCVFYS 113
+PER +P CQFY+KTG CKFGA C+FHHP+ L+ G P+R G+ C +Y
Sbjct: 161 YPERFGEPPCQFYLKTGTCKFGASCKFHHPKNAGGSMSHVPLNIYGYPVREGDNECSYYL 220
Query: 114 RYGICKFGPSCKFDHP 129
+ G CKFG +CKF HP
Sbjct: 221 KTGQCKFGITCKFHHP 236
Score = 84.7 bits (208), Expect = 3e-17
Identities = 38/77 (49%), Positives = 50/77 (64%), Gaps = 3/77 (3%)
Query: 54 FPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLG-LPLRPGEPLCVFY 112
+PERP P+C +YM+TG C +G CR++HPR+R + + + G P R GEP C FY
Sbjct: 116 YPERPGAPDCAYYMRTGVCGYGNRCRYNHPRDRA--SVEATVRATGQYPERFGEPPCQFY 173
Query: 113 SRYGICKFGPSCKFDHP 129
+ G CKFG SCKF HP
Sbjct: 174 LKTGTCKFGASCKFHHP 190
Score = 53.5 bits (127), Expect = 7e-08
Identities = 29/70 (41%), Positives = 37/70 (52%), Gaps = 15/70 (21%)
Query: 24 PENAGLQGVYSQYRSGSVPVGFYALQRENIFPERPDQPECQFYMKTGDCKFGAVCRFHHP 83
P+NAG G S VP+ Y +P R EC +Y+KTG CKFG C+FHHP
Sbjct: 190 PKNAG--GSMSH-----VPLNIYG------YPVREGDNECSYYLKTGQCKFGITCKFHHP 236
Query: 84 RE--RTIPAP 91
+ T+P P
Sbjct: 237 QPAGTTVPPP 246
>At5g18550 zinc finger -like protein
Length = 456
Score = 129 bits (325), Expect = 8e-31
Identities = 75/179 (41%), Positives = 98/179 (53%), Gaps = 15/179 (8%)
Query: 1 MGSDSPQQTMRNDQTYGTSHQGDPENAGLQGVYSQYRSGSVPVGFYALQRENIFPERPDQ 60
M S Q +M YG + A Y+SG G +E FP+RP+Q
Sbjct: 245 MPSPGTQPSMGTSSVYGITPLSPSAPA--------YQSGPSSTG--VSNKEQTFPQRPEQ 294
Query: 61 PECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGEPLCVFYSRYGICKF 120
PECQ++M+TGDCKFG CRFHHP E P LS +GLPLRPG C ++++GICKF
Sbjct: 295 PECQYFMRTGDCKFGTSCRFHHPMEAASPEAS-TLSHIGLPLRPGAVPCTHFAQHGICKF 353
Query: 121 GPSCKFDHPMGIFTYNVSASPLA-EAAGRRLLGSSSGTAALSLSSE---GLVESGSVKP 175
GP+CKFDH +G + + S SP + SS GT A S SS+ L+ S S++P
Sbjct: 354 GPACKFDHSLGSSSLSYSPSPSSLTDMPVAPYPSSLGTLAPSSSSDQCTELISSSSIEP 412
Score = 90.5 bits (223), Expect = 5e-19
Identities = 41/81 (50%), Positives = 54/81 (66%), Gaps = 6/81 (7%)
Query: 54 FPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCV----LSPLGLPLRPGEPLC 109
FPER QP CQ +M+TG CKFGA C++HHPR+ D V L+ +G PLRPGE C
Sbjct: 85 FPERMGQPVCQHFMRTGTCKFGASCKYHHPRQG--GGGDSVTPVSLNYMGFPLRPGEKEC 142
Query: 110 VFYSRYGICKFGPSCKFDHPM 130
++ R G CKFG +C++ HP+
Sbjct: 143 SYFMRTGQCKFGSTCRYHHPV 163
Score = 87.0 bits (214), Expect = 6e-18
Identities = 38/77 (49%), Positives = 52/77 (67%), Gaps = 2/77 (2%)
Query: 54 FPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLG-LPLRPGEPLCVFY 112
FPERPD+P+C +Y++TG C +G+ CRF+HPR R P + + G P R G+P+C +
Sbjct: 39 FPERPDEPDCIYYLRTGVCGYGSRCRFNHPRNRA-PVLGGLRTEAGEFPERMGQPVCQHF 97
Query: 113 SRYGICKFGPSCKFDHP 129
R G CKFG SCK+ HP
Sbjct: 98 MRTGTCKFGASCKYHHP 114
Score = 55.5 bits (132), Expect = 2e-08
Identities = 20/38 (52%), Positives = 27/38 (70%)
Query: 54 FPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAP 91
FP RP + EC ++M+TG CKFG+ CR+HHP + AP
Sbjct: 133 FPLRPGEKECSYFMRTGQCKFGSTCRYHHPVPPGVQAP 170
>At3g06410 hypothetical protein
Length = 437
Score = 129 bits (324), Expect = 1e-30
Identities = 77/198 (38%), Positives = 106/198 (52%), Gaps = 17/198 (8%)
Query: 1 MGSDSPQQTMRNDQTYGTSHQGDPENAGLQGVYSQYRSGSVPVGFYALQRENIFPERPDQ 60
M S Q ++ + YG + P G Y S + FP+RPDQ
Sbjct: 235 MPSPGTQPSIGSSSIYGLTPLS-PSATAYTGTYQSVPSSN--------STSKEFPQRPDQ 285
Query: 61 PECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGEPLCVFYSRYGICKF 120
PECQ++M+TGDCKFG+ CR+HHP + P VLS +GLPLRPG C ++++GICKF
Sbjct: 286 PECQYFMRTGDCKFGSSCRYHHPVDAVPPKTGIVLSSIGLPLRPGVAQCTHFAQHGICKF 345
Query: 121 GPSCKFDHPM-GIFTYNVSASPLAE-AAGRRLLGSSS---GTAALSLSSEGLVESGSVKP 175
GP+CKFDH M +Y+ SAS L + +GSSS +A +S S+E E+ +
Sbjct: 346 GPACKFDHSMSSSLSYSPSASSLTDMPVAPYPIGSSSLSGSSAPVSSSNEPTKEAVTPAV 405
Query: 176 RRLSLSETRPNP---SGD 190
+ +RP P SGD
Sbjct: 406 SSMVSGLSRPEPAETSGD 423
Score = 89.7 bits (221), Expect = 9e-19
Identities = 43/79 (54%), Positives = 53/79 (66%), Gaps = 4/79 (5%)
Query: 55 PERPDQPECQFYMKTGDCKFGAVCRFHHPRE---RTIPAPDCVLSPLGLPLRPGEPLCVF 111
PER P CQ +M+TG CKFGA C++HHPR+ AP LS LG PLRPGE C +
Sbjct: 73 PERMGHPVCQHFMRTGTCKFGASCKYHHPRQGGGGGSVAP-VSLSYLGYPLRPGEKECSY 131
Query: 112 YSRYGICKFGPSCKFDHPM 130
Y R G CKFG +C+F+HP+
Sbjct: 132 YLRTGQCKFGLTCRFNHPV 150
Score = 86.7 bits (213), Expect = 7e-18
Identities = 36/76 (47%), Positives = 49/76 (64%)
Query: 54 FPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGEPLCVFYS 113
+PERPD+P+C +Y++TG C +G+ CRF+HPR+R LP R G P+C +
Sbjct: 26 YPERPDEPDCIYYLRTGVCGYGSRCRFNHPRDRGAVIGGVRGEAGALPERMGHPVCQHFM 85
Query: 114 RYGICKFGPSCKFDHP 129
R G CKFG SCK+ HP
Sbjct: 86 RTGTCKFGASCKYHHP 101
Score = 51.6 bits (122), Expect = 3e-07
Identities = 18/38 (47%), Positives = 25/38 (65%)
Query: 54 FPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAP 91
+P RP + EC +Y++TG CKFG CRF+HP + P
Sbjct: 120 YPLRPGEKECSYYLRTGQCKFGLTCRFNHPVPLAVQGP 157
>At3g48440 unknown protein
Length = 448
Score = 124 bits (311), Expect = 3e-29
Identities = 61/141 (43%), Positives = 76/141 (53%), Gaps = 17/141 (12%)
Query: 5 SPQQTMRNDQTYGTSHQGDPENAGLQGVYSQYRSGSVPVGFYALQRENI----------- 53
SP + QT+G + Q N VYS R P Y + +
Sbjct: 277 SPFIPVMLSQTHGVTSQNPEWNGYQASVYSSERGVFSPSTTYLMNNSSAETSMLLSQYRH 336
Query: 54 ------FPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGEP 107
FPERPDQPEC +YMKTGDCKF C++HHP+ R P L+ GLPLRP +
Sbjct: 337 QMPAEEFPERPDQPECSYYMKTGDCKFKFNCKYHHPKNRLPKLPPYALNDKGLPLRPDQN 396
Query: 108 LCVFYSRYGICKFGPSCKFDH 128
+C +YSRYGICKFGP+C+FDH
Sbjct: 397 ICTYYSRYGICKFGPACRFDH 417
Score = 84.3 bits (207), Expect = 4e-17
Identities = 46/134 (34%), Positives = 59/134 (43%), Gaps = 48/134 (35%)
Query: 44 GFYALQRENIFPERPDQPECQFYMKTGDCKFGAVCRFHHPRER----------------- 86
G+ + EN++P RP +C FYM+TG CKFG+ C+F+HP R
Sbjct: 97 GWSENESENVYPVRPGAEDCSFYMRTGSCKFGSSCKFNHPLARKFQIARDNKVREKEDDG 156
Query: 87 ---------------------------TIPAPDCVLSP----LGLPLRPGEPLCVFYSRY 115
TIP +P LGLPLRPGE C +Y R
Sbjct: 157 GKLGLIDCKYYFRTGGCKYGETCRFNHTIPKSGLASAPELNFLGLPLRPGEVECPYYMRN 216
Query: 116 GICKFGPSCKFDHP 129
G CK+G CKF+HP
Sbjct: 217 GSCKYGAECKFNHP 230
Score = 49.7 bits (117), Expect = 1e-06
Identities = 19/38 (50%), Positives = 25/38 (65%)
Query: 55 PERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPD 92
P RP + EC +YM+ G CK+GA C+F+HP TI D
Sbjct: 202 PLRPGEVECPYYMRNGSCKYGAECKFNHPDPTTIGGTD 239
Score = 37.7 bits (86), Expect = 0.004
Identities = 14/29 (48%), Positives = 19/29 (65%)
Query: 101 PLRPGEPLCVFYSRYGICKFGPSCKFDHP 129
P RP +P C +Y + G CKF +CK+ HP
Sbjct: 344 PERPDQPECSYYMKTGDCKFKFNCKYHHP 372
>At5g63260 unknown protein
Length = 435
Score = 114 bits (285), Expect = 3e-26
Identities = 50/92 (54%), Positives = 64/92 (69%), Gaps = 1/92 (1%)
Query: 54 FPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGEPLCVFYS 113
FPERPDQPEC +Y+KTGDCKF C++HHP+ R + GLPLRP + +C YS
Sbjct: 331 FPERPDQPECTYYLKTGDCKFKYKCKYHHPKNRLPKQAAFSFNDKGLPLRPDQSMCTHYS 390
Query: 114 RYGICKFGPSCKFDHPMGIFTYNVSASPLAEA 145
RYGICKFGP+C+FDH + T++ S+S EA
Sbjct: 391 RYGICKFGPACRFDHSIPP-TFSPSSSQTVEA 421
Score = 91.7 bits (226), Expect = 2e-19
Identities = 40/74 (54%), Positives = 48/74 (64%)
Query: 56 ERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGEPLCVFYSRY 115
E P EC++Y +TG CK+G CRF H +E PA L+ LGLP+RPGE C FY R
Sbjct: 147 ENPKLMECKYYFRTGGCKYGESCRFSHMKEHNSPASVPELNFLGLPIRPGEKECPFYMRN 206
Query: 116 GICKFGPSCKFDHP 129
G CKFG CKF+HP
Sbjct: 207 GSCKFGSDCKFNHP 220
Score = 48.9 bits (115), Expect = 2e-06
Identities = 20/41 (48%), Positives = 25/41 (60%)
Query: 55 PERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVL 95
P RP + EC FYM+ G CKFG+ C+F+HP I D L
Sbjct: 192 PIRPGEKECPFYMRNGSCKFGSDCKFNHPDPTAIGGVDSPL 232
Score = 44.3 bits (103), Expect = 4e-05
Identities = 17/30 (56%), Positives = 21/30 (69%)
Query: 101 PLRPGEPLCVFYSRYGICKFGPSCKFDHPM 130
P+RP C FY R G CK+G SCKF+HP+
Sbjct: 99 PVRPDSEDCSFYMRTGSCKYGSSCKFNHPV 128
>At1g04990 zinc finger protein 2, putative
Length = 404
Score = 102 bits (253), Expect = 2e-22
Identities = 45/95 (47%), Positives = 57/95 (59%)
Query: 36 YRSGSVPVGFYALQRENIFPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVL 95
Y SGS A+ E DQPEC+F+M TG CK+G C++ HP R P ++
Sbjct: 240 YYSGSSASMAMAVALNRGLSESSDQPECRFFMNTGTCKYGDDCKYSHPGVRISQPPPSLI 299
Query: 96 SPLGLPLRPGEPLCVFYSRYGICKFGPSCKFDHPM 130
+P LP RPG+P C + YG CKFGP+CKFDHPM
Sbjct: 300 NPFVLPARPGQPACGNFRSYGFCKFGPNCKFDHPM 334
Score = 92.4 bits (228), Expect = 1e-19
Identities = 42/94 (44%), Positives = 62/94 (65%), Gaps = 5/94 (5%)
Query: 55 PERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGEPLCVFYSR 114
PER QP+C++++KTG CK+G C++HHP++R P + + +GLP+R GE C +Y R
Sbjct: 88 PERIGQPDCEYFLKTGACKYGPTCKYHHPKDRNGAQP-VMFNVIGLPMRLGEKPCPYYLR 146
Query: 115 YGICKFGPSCKFDHPM---GIFT-YNVSASPLAE 144
G C+FG +CKF HP G T Y +S+ P A+
Sbjct: 147 TGTCRFGVACKFHHPQPDNGHSTAYGMSSFPAAD 180
Score = 78.6 bits (192), Expect = 2e-15
Identities = 33/78 (42%), Positives = 50/78 (63%), Gaps = 3/78 (3%)
Query: 52 NIFPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGEPLCVF 111
N +P+RP + +CQFY++TG C +G+ CR++HP T D LP R G+P C +
Sbjct: 42 NPYPDRPGERDCQFYLRTGLCGYGSSCRYNHP---THLPQDVAYYKEELPERIGQPDCEY 98
Query: 112 YSRYGICKFGPSCKFDHP 129
+ + G CK+GP+CK+ HP
Sbjct: 99 FLKTGACKYGPTCKYHHP 116
>At2g32930 putative protein
Length = 453
Score = 101 bits (251), Expect = 3e-22
Identities = 59/168 (35%), Positives = 81/168 (48%), Gaps = 16/168 (9%)
Query: 38 SGSVPVGFYALQRENIFPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSP 97
S S V + F ER EC+F+M TG CK+G C++ HP+ER + +P +L+P
Sbjct: 245 SSSASVAVTVTSHHHSFSERA---ECRFFMNTGTCKYGDDCKYSHPKERLLQSPPTLLNP 301
Query: 98 LGLPLRPGEPLCVFYSRYGICKFGPSCKFDHPMGIFTYN---------VSASPLAEAAGR 148
+ LP RPG+P C + YG CKFG +CKFDH M + YN + P A
Sbjct: 302 IVLPARPGQPACGNFKAYGFCKFGANCKFDHSMLLNPYNNTGLAMSSLPTPYPYAPPVST 361
Query: 149 RLLGSS----SGTAALSLSSEGLVESGSVKPRRLSLSETRPNPSGDDD 192
L SS S LS E+ S++ + S T P S +D
Sbjct: 362 NLRISSPPSPSDMTTLSNGKPAAAEAQSLETEKQDDSPTEPEKSEVED 409
Score = 82.8 bits (203), Expect = 1e-16
Identities = 35/75 (46%), Positives = 50/75 (66%), Gaps = 5/75 (6%)
Query: 55 PERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGEPLCVFYSR 114
PER QP+C+ TG CK+G C++HHP++R P + + LGLP+R GE C +Y +
Sbjct: 86 PERVGQPDCE----TGACKYGPTCKYHHPKDRNGAGP-VLFNVLGLPMRQGEKPCPYYMQ 140
Query: 115 YGICKFGPSCKFDHP 129
G+C+FG +CKF HP
Sbjct: 141 TGLCRFGVACKFHHP 155
Score = 50.1 bits (118), Expect = 8e-07
Identities = 42/152 (27%), Positives = 63/152 (40%), Gaps = 32/152 (21%)
Query: 37 RSGSVPVGFYALQRENIFPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPD---- 92
R+G+ PV F L P R + C +YM+TG C+FG C+FHHP + P+
Sbjct: 113 RNGAGPVLFNVLG----LPMRQGEKPCPYYMQTGLCRFGVACKFHHPHPHSQPSNGHSAY 168
Query: 93 --CVLSPLGLPL------------------RPGEPLCVFYSRYGICKFGPSCKFDHPMGI 132
+G P RP P Y Y + PS P G
Sbjct: 169 AMSSFPSVGFPYASGMTMVSLPPATYGAIPRPQVPQSQAYMPYMV---APSQGLLPPQGW 225
Query: 133 FTYNVSASPLAEAAGRRLLGSSSGTAALSLSS 164
TY +++P+ +L SSS + A++++S
Sbjct: 226 ATYMTASNPIYNMK-TQLDSSSSASVAVTVTS 256
>At3g12680 floral homeotic protein HUA1 (HUA1)
Length = 341
Score = 93.2 bits (230), Expect = 8e-20
Identities = 59/180 (32%), Positives = 86/180 (47%), Gaps = 42/180 (23%)
Query: 47 ALQRENIFPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPA--------------PD 92
+++ ++ PERP +P C FYMKTG CKFG C+FHHP++ +P+ PD
Sbjct: 75 SVETQDSLPERPSEPMCTFYMKTGKCKFGLSCKFHHPKDIQLPSSSQDIGSSVGLTSEPD 134
Query: 93 CVLSP------------LGLPLRPGEPLCVFYSRYGICKFGPSCKFDHPMGIFTYNVSAS 140
+P GLP+R GE C FY + G CK+G +C+++HP +
Sbjct: 135 ATNNPHVTFTPALYHNSKGLPVRSGEVDCPFYLKTGSCKYGATCRYNHP-------ERTA 187
Query: 141 PLAEAAGRRLLGSSSGTAALSLSSEGLVESGS------VKPRRLSLSETRPNPSGDDDID 194
+ +AAG SS TA L+L GLV + +P +S T P G + D
Sbjct: 188 FIPQAAGVNYSLVSSNTANLNL---GLVTPATSFYQTLTQPTLGVISATYPQRPGQSECD 244
Score = 93.2 bits (230), Expect = 8e-20
Identities = 41/79 (51%), Positives = 53/79 (66%), Gaps = 3/79 (3%)
Query: 54 FPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGEPLCVFYS 113
+PERP +P+C +Y+KT CK+G+ C+F+HPRE A V + LP RP EP+C FY
Sbjct: 39 YPERPGEPDCPYYIKTQRCKYGSKCKFNHPREE---AAVSVETQDSLPERPSEPMCTFYM 95
Query: 114 RYGICKFGPSCKFDHPMGI 132
+ G CKFG SCKF HP I
Sbjct: 96 KTGKCKFGLSCKFHHPKDI 114
Score = 91.7 bits (226), Expect = 2e-19
Identities = 40/83 (48%), Positives = 51/83 (61%), Gaps = 7/83 (8%)
Query: 54 FPERPDQPECQFYMKTGDCKFGAVCRFHHPRER-------TIPAPDCVLSPLGLPLRPGE 106
+P+RP Q EC +YMKTG+CKFG C+FHHP +R P+ LS G P R G
Sbjct: 234 YPQRPGQSECDYYMKTGECKFGERCKFHHPADRLSAMTKQAPQQPNVKLSLAGYPRREGA 293
Query: 107 PLCVFYSRYGICKFGPSCKFDHP 129
C +Y + G CK+G +CKFDHP
Sbjct: 294 LNCPYYMKTGTCKYGATCKFDHP 316
Score = 67.4 bits (163), Expect = 5e-12
Identities = 34/109 (31%), Positives = 46/109 (42%), Gaps = 34/109 (31%)
Query: 55 PERPDQPECQFYMKTGDCKFGAVCRFHHPRE----------------------------- 85
P R + +C FY+KTG CK+GA CR++HP
Sbjct: 155 PVRSGEVDCPFYLKTGSCKYGATCRYNHPERTAFIPQAAGVNYSLVSSNTANLNLGLVTP 214
Query: 86 -----RTIPAPDCVLSPLGLPLRPGEPLCVFYSRYGICKFGPSCKFDHP 129
+T+ P + P RPG+ C +Y + G CKFG CKF HP
Sbjct: 215 ATSFYQTLTQPTLGVISATYPQRPGQSECDYYMKTGECKFGERCKFHHP 263
Score = 57.8 bits (138), Expect = 4e-09
Identities = 33/70 (47%), Positives = 39/70 (55%), Gaps = 9/70 (12%)
Query: 67 MKTGDCKFGAVCRFHHP---RERTIP----APDCVLSPLGLPLRPGEPLCVFYSRYGICK 119
M+T CKFG CRF HP E IP AP V+ P RPGEP C +Y + CK
Sbjct: 1 MQTRTCKFGESCRFDHPIWVPEGGIPDWKEAP--VVPNEEYPERPGEPDCPYYIKTQRCK 58
Query: 120 FGPSCKFDHP 129
+G CKF+HP
Sbjct: 59 YGSKCKFNHP 68
Score = 31.2 bits (69), Expect = 0.37
Identities = 10/15 (66%), Positives = 13/15 (86%)
Query: 118 CKFGPSCKFDHPMGI 132
CKFG SC+FDHP+ +
Sbjct: 6 CKFGESCRFDHPIWV 20
>At1g29600 zinc finger protein, putative
Length = 287
Score = 48.5 bits (114), Expect = 2e-06
Identities = 26/76 (34%), Positives = 38/76 (49%), Gaps = 15/76 (19%)
Query: 54 FPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGEPLCVFYS 113
+P RP + C FYMK C++G+ C ++HP + IP R G+ L
Sbjct: 128 YPVRPGEDNCLFYMKNHLCEWGSECCYNHPPLQEIPC------------RIGKKL---DC 172
Query: 114 RYGICKFGPSCKFDHP 129
+ G CK G +C F+HP
Sbjct: 173 KAGACKRGSNCPFNHP 188
>At1g29570 zinc finger protein, putative
Length = 321
Score = 48.1 bits (113), Expect = 3e-06
Identities = 17/44 (38%), Positives = 30/44 (67%)
Query: 48 LQRENIFPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAP 91
+++ + +P RP + +CQFY+K G C++ + CRF+HP +R P
Sbjct: 46 MRQSSPYPVRPGKKDCQFYLKNGLCRYRSSCRFNHPTQRPQELP 89
Score = 41.6 bits (96), Expect = 3e-04
Identities = 14/29 (48%), Positives = 22/29 (75%)
Query: 101 PLRPGEPLCVFYSRYGICKFGPSCKFDHP 129
P+RPG+ C FY + G+C++ SC+F+HP
Sbjct: 53 PVRPGKKDCQFYLKNGLCRYRSSCRFNHP 81
>At5g56930 unknown protein
Length = 675
Score = 36.6 bits (83), Expect = 0.009
Identities = 26/96 (27%), Positives = 39/96 (40%), Gaps = 18/96 (18%)
Query: 41 VPVGFYALQRENIFPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGL 100
+ +G L+ + + P+ C+ Y+K G C G C+F H TIP C
Sbjct: 334 IALGVKKLKLKPVAPKPKPIKYCRHYLK-GRCHEGDKCKFSHD---TIPETKC------- 382
Query: 101 PLRPGEPLCVFYSRYGICKFGPSCKFDHPMGIFTYN 136
P C F ++ C G C FDH + + N
Sbjct: 383 -----SPCCYFATQS--CMKGDDCPFDHDLSKYPCN 411
>At2g47680 putative ATP-dependent RNA helicase A
Length = 1015
Score = 35.4 bits (80), Expect = 0.020
Identities = 25/87 (28%), Positives = 36/87 (40%), Gaps = 20/87 (22%)
Query: 44 GFYALQRENIFPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLR 103
G AL+ E+ + E + P C +++ G C G C F H + T PA
Sbjct: 715 GHGALEPED-YVEYGEAPVCVYFLN-GYCNRGGQCTFTHTLQSTRPA------------- 759
Query: 104 PGEPLCVFYSRYGICKFGPSCKFDHPM 130
C F++ C+ G SC F H M
Sbjct: 760 -----CKFFASSQGCRNGESCLFSHAM 781
Score = 30.4 bits (67), Expect = 0.63
Identities = 14/39 (35%), Positives = 21/39 (52%), Gaps = 4/39 (10%)
Query: 60 QPECQFYMKTGDCKFGAVCRFHHP-RERT---IPAPDCV 94
+P C+F+ + C+ G C F H R RT +P P C+
Sbjct: 757 RPACKFFASSQGCRNGESCLFSHAMRRRTTSYLPPPQCL 795
>At5g42820 U2 snRNP auxiliary factor, small subunit
Length = 283
Score = 33.5 bits (75), Expect = 0.075
Identities = 21/67 (31%), Positives = 27/67 (39%), Gaps = 12/67 (17%)
Query: 52 NIFPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPA---------PDCV---LSPLG 99
+IF D+ C FY K G C+ G C H R P PD + + P G
Sbjct: 7 SIFGTEKDRVNCPFYFKIGACRHGDRCSRLHNRPTISPTLLLSNMYQRPDMITPGVDPQG 66
Query: 100 LPLRPGE 106
PL P +
Sbjct: 67 QPLDPSK 73
>At5g51730 unknown protein
Length = 412
Score = 32.3 bits (72), Expect = 0.17
Identities = 20/62 (32%), Positives = 30/62 (48%)
Query: 135 YNVSASPLAEAAGRRLLGSSSGTAALSLSSEGLVESGSVKPRRLSLSETRPNPSGDDDID 194
Y VS P + +RLL ++LS S S +P+ L+LSE+ +DDI
Sbjct: 280 YEVSGGPFIKKPCQRLLARQKIPLCMALSVLARSTSNSDQPQALNLSESEAYNRPNDDIK 339
Query: 195 DE 196
D+
Sbjct: 340 DK 341
>At1g29560 hypothetical protein
Length = 521
Score = 32.0 bits (71), Expect = 0.22
Identities = 12/22 (54%), Positives = 15/22 (67%)
Query: 70 GDCKFGAVCRFHHPRERTIPAP 91
G CKFG+VC F H R+R + P
Sbjct: 210 GYCKFGSVCGFQHIRDRDVAEP 231
>At5g12440 unknown protein
Length = 552
Score = 31.6 bits (70), Expect = 0.28
Identities = 16/42 (38%), Positives = 26/42 (61%), Gaps = 3/42 (7%)
Query: 109 CVFYSRYGICKFGPSCKFDHPMGIFTYNVSASPLAEAAGRRL 150
CV++SR G+CK G SCKF H G + N+ + + + R++
Sbjct: 135 CVYFSR-GLCKNGESCKFIH--GGYPDNMDGNGIVADSPRKM 173
>At1g10320 unknown protein
Length = 757
Score = 31.6 bits (70), Expect = 0.28
Identities = 17/52 (32%), Positives = 22/52 (41%), Gaps = 4/52 (7%)
Query: 54 FPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPG 105
F D+ C F++KTG C+FG C R P C L + PG
Sbjct: 237 FGTEQDKAHCPFHLKTGACRFGQRC----SRVHFYPNKSCTLLMKNMYNGPG 284
>At1g27650 U2 snRNP auxiliary factor, putative
Length = 296
Score = 30.4 bits (67), Expect = 0.63
Identities = 14/38 (36%), Positives = 17/38 (43%)
Query: 52 NIFPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIP 89
+IF D+ C FY K G C+ G C H R P
Sbjct: 7 SIFGTEKDRVNCPFYFKIGACRHGDRCSRLHNRPTISP 44
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.137 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,063,512
Number of Sequences: 26719
Number of extensions: 222715
Number of successful extensions: 641
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 26
Number of HSP's successfully gapped in prelim test: 19
Number of HSP's that attempted gapping in prelim test: 508
Number of HSP's gapped (non-prelim): 96
length of query: 197
length of database: 11,318,596
effective HSP length: 94
effective length of query: 103
effective length of database: 8,807,010
effective search space: 907122030
effective search space used: 907122030
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 57 (26.6 bits)
Medicago: description of AC148289.7


