

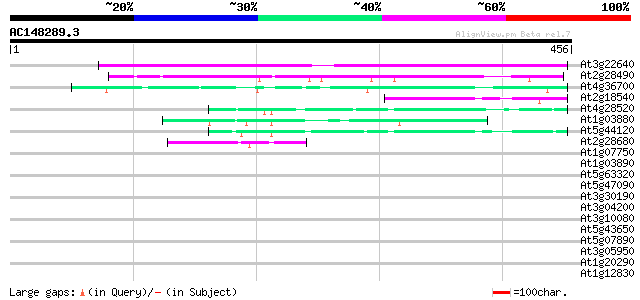
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148289.3 + phase: 0 /pseudo
(456 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g22640 unknown protein 241 6e-64
At2g28490 putative seed storage protein (vicilin-like) 89 4e-18
At4g36700 globulin-like protein 64 1e-10
At2g18540 putative vicilin storage protein (globulin-like) 49 4e-06
At4g28520 12S cruciferin seed storage protein 47 2e-05
At1g03880 putative cruciferin 12S seed storage protein 44 1e-04
At5g44120 legumin-like protein 44 2e-04
At2g28680 legumin-like protein 44 2e-04
At1g07750 globulin like protein 41 0.002
At1g03890 putative cruciferin 12S seed storage protein 37 0.017
At5g63320 putative protein 36 0.050
At5g47090 unknown protein 35 0.066
At3g30190 hypothetical protein 35 0.066
At3g04200 germin-like protein 35 0.066
At3g10080 germin-like protein 35 0.11
At5g43650 putative protein 34 0.15
At5g07890 putative protein 34 0.15
At3g05950 germin-like protein 34 0.15
At1g20290 hypothetical protein 34 0.15
At1g12830 unknown protein 34 0.15
>At3g22640 unknown protein
Length = 486
Score = 241 bits (615), Expect = 6e-64
Identities = 126/385 (32%), Positives = 215/385 (55%), Gaps = 21/385 (5%)
Query: 73 DHDQENPFFFNANRFQTLFENENGHIRLLQRFDKRSK-IFENLQNYRLLEYHSKPHTLFL 131
+ +P+ F F F+++ G +R+L +F K + +F ++NYR +P T F+
Sbjct: 57 EESTNHPYHFRKRSFSDWFQSKEGFVRVLPKFTKHAPALFRGIENYRFSLVEMEPTTFFV 116
Query: 132 PQHNDADFILAVLSGKAILTVLNPNDRNSFNLERGDTIKLPAGSIAYLANRADNEDLRVL 191
P H DAD + VL GK ++ + + SF++ +GD +++P+G ++ N LR+
Sbjct: 117 PHHLDADAVFIVLQGKGVIEFVTDKTKESFHITKGDVVRIPSGVTNFITNTNQTVPLRLA 176
Query: 192 DLAIPVNRPGQFQSFSLSGNQNQQSFFSGFSKNILEAAFNSNYEEIERVLIEEQEQEPRH 251
+ +PVN PG ++ + + +Q QQS+F+GF+K +L +FN E + R++ +E
Sbjct: 177 QITVPVNNPGNYKDYFPAASQFQQSYFNGFTKEVLSTSFNVPEELLGRLVTRSKE----- 231
Query: 252 RRGLRDRRHKQSQEANVIVKVSREQIEELSRHAKS-SSRRSASSESASRRSASSESAPFN 310
+I ++S +QI+EL+ HA S S++ A E + + PFN
Sbjct: 232 ------------IGQGIIRRISPDQIKELAEHATSPSNKHKAKKEKEEDKDLRTLWTPFN 279
Query: 311 LRSREPIYSNEFGNFFEITPEKNPQLQDLDILVNYAEIREGSLLLPHFNSRATVIVAVEE 370
L + +PIYSN+FG+F E P+ QLQDL I +A + +GSL LPHFNS+ T + VE
Sbjct: 280 LFAIDPIYSNDFGHFHEAHPKNYNQLQDLHIAAAWANMTQGSLFLPHFNSKTTFVTFVEN 339
Query: 371 GKGEFEL-VGQRNENQQEQREYEEDEQEEERSQQVQRYRARLTPGDVYVIPAGYP-NVVK 428
G FE+ + + Q+Q + E+EE+ S+ V + +R+ G+V+++PAG+P ++
Sbjct: 340 GCARFEMATPYKFQRGQQQWPGQGQEEEEDMSENVHKVVSRVCKGEVFIVPAGHPFTILS 399
Query: 429 ASSDLSLLGFGINAENNQRSFLAAK 453
D +GFGI A N++R+FLA +
Sbjct: 400 QDQDFIAVGFGIYATNSKRTFLAGE 424
>At2g28490 putative seed storage protein (vicilin-like)
Length = 511
Score = 89.4 bits (220), Expect = 4e-18
Identities = 92/396 (23%), Positives = 171/396 (42%), Gaps = 53/396 (13%)
Query: 81 FFNANRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQHNDADFI 140
+F + + ++E G +R++ R +I E + L +P TLF+PQ+ D+ +
Sbjct: 85 WFMMRESRQVIKSEGGEMRVV--LSPRGRIIEKPMHIGFLTM--EPKTLFVPQYLDSSLL 140
Query: 141 LAVLSGKAILTVLNPNDRNSFNLERGDTIKLPAGSIAYLANRADNEDLRVLDLAIPVNRP 200
+ + G+A L V+ ++ L+ GD +PAGS+ YL N + L V+ P
Sbjct: 141 IFIRQGEATLGVICKDEFGERKLKAGDIYWIPAGSVFYLHNTGLGQRLHVICSIDPTQSL 200
Query: 201 G--QFQSFSLSGNQNQQSFFSGFSKNILEAAFNSNYEEIERVLI-----------EEQEQ 247
G FQ F + G + S +GF + L +AFN + E++++++ E +
Sbjct: 201 GFETFQPFYIGGGPS--SVLAGFDPHTLTSAFNVSLPELQQMMMSQFRGPIVYVTEGPQP 258
Query: 248 EPRHR-----RGLR-DRRHKQSQEANVIVKVSREQIEELSRHAKSSSRRSA--SSESASR 299
+P+ GLR + +HKQ ++ + S + + S + + RS +E ++
Sbjct: 259 QPQSTVWTQFLGLRGEEKHKQLKKLLETKQGSPQDQQYSSGWSWRNIVRSILDLTEEKNK 318
Query: 300 RSASSE-SAPFNL--RSREPIYSNEFGNFFEITPEKNPQLQDLDILVNYAEIREGSLLLP 356
S SSE +N+ + +P + N++G + + L+ I V + G+++ P
Sbjct: 319 GSGSSECEDSYNIYDKKDKPSFDNKYGWSIALDYDDYKPLKHSGIGVYLVNLTAGAMMAP 378
Query: 357 HFNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQVQRYRARLTPGDV 416
H N AT V G GE ++V + R++ GDV
Sbjct: 379 HMNPTATEYGIVLAGSGEIQVVFPNGTSAM---------------------NTRVSVGDV 417
Query: 417 YVIPA--GYPNVVKASSDLSLLGFGINAENNQRSFL 450
+ IP + + + +GF +A N+ FL
Sbjct: 418 FWIPRYFAFCQIASRTGPFEFVGFTTSAHKNRPQFL 453
>At4g36700 globulin-like protein
Length = 499
Score = 64.3 bits (155), Expect = 1e-10
Identities = 83/419 (19%), Positives = 163/419 (38%), Gaps = 68/419 (16%)
Query: 51 PFQLLMLLGIFFLASVCVSSRSDHDQE----------NPFFFNANRFQTLFENENGHIRL 100
P +L+L+ +F S + + +P ++++ +FE + G I
Sbjct: 8 PLSVLLLVLLFLCTESLAKSEESEEYDVAVPSCCGFSSPLLIKKDQWKPIFETKFGQIST 67
Query: 101 LQRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQHNDADFILAVLSGKAILTVLNPNDRNS 160
+Q + + Y++ +P+T+ LP +D + V SG IL ++ + S
Sbjct: 68 VQIGNGCG----GMGPYKIHSITLEPNTILLPLLLHSDMVFFVDSGSGILNWVD-EEAKS 122
Query: 161 FNLERGDTIKLPAGSIAYLANRADNEDLRVLDLAIPVNRP--GQFQSFSLSGNQNQQSFF 218
+ GD +L GS+ YL ++ D P G + S +
Sbjct: 123 TEIRLGDVYRLRPGSVFYLQSKPD---------------PCFGAYSSIT--------DLM 159
Query: 219 SGFSKNILEAAFNSNYEEIERVLIEEQEQEPRHRRGLRDRRHKQSQEANVIVKVSREQIE 278
GF + IL++AF IE L+ + + P ++ + + Q
Sbjct: 160 FGFDETILQSAFGVPEGIIE--LMRNRTKPPLIV--------SETLCTPGVANTWQLQPR 209
Query: 279 ELSRHAKSSSR--RSASSESASRRSASSESAPFNLRSREPIYSNEFGNFFEITPEKNPQL 336
L A S+ E ++ ++ FN+ EP + + +G I + L
Sbjct: 210 LLKLFAGSADLVDNKKKKEKKEKKEKVKKAKTFNVFESEPDFESPYGRTITINRKDLKVL 269
Query: 337 QDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQ 396
+ + V+ + +GS++ PH+N A I V +G G ++
Sbjct: 270 KGSMVGVSMVNLTQGSMMGPHWNPWACEISIVLKGAGMVRVL--------------RSSI 315
Query: 397 EEERSQQVQRYRARLTPGDVYVIPAGYPNVVKASSDLSL--LGFGINAENNQRSFLAAK 453
S + + R ++ GD++ +P +P + ++ SL +GF +A+NN+ FLA +
Sbjct: 316 SSNTSSECKNVRFKVEEGDIFAVPRLHPMAQMSFNNDSLVFVGFTTSAKNNEPQFLAGE 374
>At2g18540 putative vicilin storage protein (globulin-like)
Length = 699
Score = 49.3 bits (116), Expect = 4e-06
Identities = 38/151 (25%), Positives = 64/151 (42%), Gaps = 16/151 (10%)
Query: 305 ESAPFNLRSREPIYSNEFGNFFEITPEKNPQLQDLDILVNYAEIREGSLLLPHFNSRATV 364
+S FN+ +P + N G + + L+ V + +GS++ PH+N A
Sbjct: 235 KSRTFNVFEEDPDFENNNGRSIVVDEKDLDALKGSRFGVFMVNLTKGSMIGPHWNPSACE 294
Query: 365 IVAVEEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQVQRYRARLTPGDVYVIPAGYP 424
I V EG+G +V NQQ + D + E + GDV+V+P +P
Sbjct: 295 ISIVLEGEGMVRVV-----NQQSLSSCKNDRKSES---------FMVEEGDVFVVPKFHP 340
Query: 425 NVVKA--SSDLSLLGFGINAENNQRSFLAAK 453
+ +S +GF +A+ N FL +
Sbjct: 341 MAQMSFENSSFVFMGFSTSAKTNHPQFLVGQ 371
Score = 41.6 bits (96), Expect = 0.001
Identities = 75/377 (19%), Positives = 137/377 (35%), Gaps = 74/377 (19%)
Query: 73 DHDQENPFFFNANRFQTLFENENGHIRLLQRFD------KRSKIFE-NLQNYRLLEYHSK 125
D +++ FN FEN NG ++ D R +F NL ++ H
Sbjct: 230 DTNKKKSRTFNVFEEDPDFENNNGRSIVVDEKDLDALKGSRFGVFMVNLTKGSMIGPHWN 289
Query: 126 PHTLFLPQHNDADFILAVLSGKAILTVLNPNDRNSFNLERGDTIKLPA-GSIAYLANRAD 184
P + + + ++ V++ +++ + N SF +E GD +P +A ++
Sbjct: 290 PSACEISIVLEGEGMVRVVNQQSLSSCKNDRKSESFMVEEGDVFVVPKFHPMAQMSFE-- 347
Query: 185 NEDLRVLDLAIPVNRPGQFQSFSLSGNQNQQSFFSGFS-------KNILEAAFNSNYEEI 237
N F FS S N F G S ++++ +FN + E I
Sbjct: 348 -------------NSSFVFMGFSTSAKTNHPQFLVGQSSVLKVLDRDVVAVSFNLSNETI 394
Query: 238 ERVLIEEQEQ--------------------EPRHRRGLRD--RRHKQSQEANVIVKVSRE 275
+ +L ++E E R RR + RR K+ +EA + R
Sbjct: 395 KGLLKAQKESVIFECASCAEGELSKLMREIEERKRREEEEIERRRKEEEEARKREEAKRR 454
Query: 276 QIEELSRHAKSSSRRSASSESASRRSASSESAPFNLRSREPIYSNEFGNFFEITPEKNPQ 335
+ EE R + + R E +R+ R E E E+ +
Sbjct: 455 EEEEAKRREEEETERKKREEEEARKREEERK-----REEEEAKRRE--------EERKKR 501
Query: 336 LQDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEED- 394
++ + E RE + E + E E V ++ +QE++ EE+
Sbjct: 502 EEEAEQARKREEEREKEEEMAKKREE-------ERQRKEREEVERKRREEQERKRREEEA 554
Query: 395 -EQEEERSQQVQRYRAR 410
++EEER ++ + + R
Sbjct: 555 RKREEERKREEEMAKRR 571
Score = 30.8 bits (68), Expect = 1.6
Identities = 19/76 (25%), Positives = 33/76 (43%), Gaps = 4/76 (5%)
Query: 235 EEIERVLIEEQEQEPRHRRGLRDRRHKQSQEANVIVKVSREQ----IEELSRHAKSSSRR 290
EE+ER + EEQE++ R + +Q +E + + RE+ EE + R+
Sbjct: 581 EEVERKIREEQERKREEEMAKRREQERQKKEREEMERKKREEEARKREEEMAKIREEERQ 640
Query: 291 SASSESASRRSASSES 306
E R+ E+
Sbjct: 641 RKEREDVERKRREEEA 656
>At4g28520 12S cruciferin seed storage protein
Length = 524
Score = 47.4 bits (111), Expect = 2e-05
Identities = 68/309 (22%), Positives = 116/309 (37%), Gaps = 52/309 (16%)
Query: 162 NLERGDTIKLPAGSIAYLANRADNEDLRVLDLAIPVNRPGQFQS----FSLSGN------ 211
++ RGD GS ++ N + + L ++ L N Q F L+GN
Sbjct: 198 HVRRGDVFANTPGSAHWIYNSGE-QPLVIIALLDIANYQNQLDRNPRVFHLAGNNQQGGF 256
Query: 212 ------QNQQSFFSGFSKNILEAAFNSNYEEIERVLIEEQEQEPRHRRGLRDRRHKQSQE 265
Q Q++ +SGF ++ A + V + +Q Q + RG R Q
Sbjct: 257 GGSQQQQEQKNLWSGFDAQVIAQALKID------VQLAQQLQNQQDSRGNIVRVKGPFQV 310
Query: 266 ANVIVKVSREQIEELSRHAKSSSRRSASSESASRRSASSESAPFNLRSREPIYSNEFGNF 325
++ E E RH +S S RS + P +R +Y G
Sbjct: 311 VRPPLRQPYESEEW--RHPRSPQGNGLEETICSMRSHENIDDP----ARADVYKPSLGRV 364
Query: 326 FEITPEKNPQLQDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQRNENQ 385
+ P L+ + + ++ +++LP +N A I+ G+G ++V +N
Sbjct: 365 TSVNSYTLPILEYVRLSATRGVLQGNAMVLPKYNMNANEILYCTGGQGRIQVVNDNGQNV 424
Query: 386 QEQREYEEDEQEEERSQQVQRYRARLTPGDVYVIPAGYPNVVKA-SSDLSLLGFGINAEN 444
+ QQVQ+ G + VIP G+ VV++ + + F N EN
Sbjct: 425 LD--------------QQVQK-------GQLVVIPQGFAYVVQSHGNKFEWISFKTN-EN 462
Query: 445 NQRSFLAAK 453
S LA +
Sbjct: 463 AMISTLAGR 471
>At1g03880 putative cruciferin 12S seed storage protein
Length = 455
Score = 44.3 bits (103), Expect = 1e-04
Identities = 63/315 (20%), Positives = 112/315 (35%), Gaps = 76/315 (24%)
Query: 125 KPHTLFLPQHNDADFILAVLSGKAILTVLNPNDRNSF----------------------- 161
+P LFLP +A + V+ G+ ++ + P +F
Sbjct: 74 EPQGLFLPTFLNAGKLTFVVHGRGLMGRVIPGCAETFMESPVFGEGQGQGQSQGFRDMHQ 133
Query: 162 ---NLERGDTIKLPAGSIAYLANRADNEDLRVL---DLAIPVNRPGQ-FQSFSLSGN--- 211
+L GDTI P+G + N NE L ++ DLA N+ + + F ++GN
Sbjct: 134 KVEHLRCGDTIATPSGVAQWFYNNG-NEPLILVAAADLASNQNQLDRNLRPFLIAGNNPQ 192
Query: 212 ----------QNQQSFFSGFSKNILEAAFNSNYEEIERVLIEEQEQEPRHRRGLRDRRHK 261
Q Q + F+GF+ IL AF N E +++ +++
Sbjct: 193 GQEWLQGRKQQKQNNIFNGFAPEILAQAFKINVETAQQL------------------QNQ 234
Query: 262 QSQEANVIVKVSREQIEELSRHAKSSSRRSASSESASRRSASSESAPFNLRSRE------ 315
Q N++ ++ + RR + + E +R E
Sbjct: 235 QDNRGNIV------KVNGPFGVIRPPLRRGEGGQQPHEIANGLEETLCTMRCTENLDDPS 288
Query: 316 --PIYSNEFGNFFEITPEKNPQLQDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKG 373
+Y G + P L+ L + IR+ +++LP +N A + V GK
Sbjct: 289 DADVYKPSLGYISTLNSYNLPILRLLRLSALRGSIRKNAMVLPQWNVNANAALYVTNGKA 348
Query: 374 EFELVGQRNENQQEQ 388
++V E +Q
Sbjct: 349 HIQMVNDNGERVFDQ 363
>At5g44120 legumin-like protein
Length = 472
Score = 43.5 bits (101), Expect = 2e-04
Identities = 64/311 (20%), Positives = 126/311 (39%), Gaps = 53/311 (17%)
Query: 162 NLERGDTIKLPAGSIAYLANRADNED----LRVLDLAIPVNRPGQF-QSFSLSGN----- 211
++ GDTI G + N D ++ + V DLA N+ + + F L+GN
Sbjct: 144 HIRSGDTIATTPGVAQWFYN--DGQEPLVIVSVFDLASHQNQLDRNPRPFYLAGNNPQGQ 201
Query: 212 --------QNQQSFFSGFSKNILEAAFNSNYEEIERVLIEEQEQEPRHRRGLRDRRHKQS 263
Q Q++ F+GF ++ A + + +++ Q Q+ +R +
Sbjct: 202 VWLQGREQQPQKNIFNGFGPEVIAQALKIDLQTAQQL----QNQDDNRGNIVRVQGPFGV 257
Query: 264 QEANVIVKVSREQIEELSRHAKSSSRRSASSESASRRSASSESAPFNLRSREPIYSNEFG 323
+ + +E+ EE RH + + S R + P SR +Y + G
Sbjct: 258 IRPPLRGQRPQEEEEEEGRHGRHGN--GLEETICSARCTDNLDDP----SRADVYKPQLG 311
Query: 324 NFFEITPEKNPQLQDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQRNE 383
+ P L+ + + IR+ +++LP +N+ A I+ V +G+ + ++V
Sbjct: 312 YISTLNSYDLPILRFIRLSALRGSIRQNAMVLPQWNANANAILYVTDGEAQIQIV----- 366
Query: 384 NQQEQREYEEDEQEEERSQQVQRYRARLTPGDVYVIPAGYPNVVKASSD-LSLLGFGINA 442
N R ++ +++ G + +P G+ V +A+S+ + F NA
Sbjct: 367 NDNGNRVFD----------------GQVSQGQLIAVPQGFSVVKRATSNRFQWVEFKTNA 410
Query: 443 ENNQRSFLAAK 453
N Q + LA +
Sbjct: 411 -NAQINTLAGR 420
Score = 33.1 bits (74), Expect = 0.33
Identities = 40/168 (23%), Positives = 70/168 (40%), Gaps = 15/168 (8%)
Query: 90 LFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQHN-DADFILAVLSGKA 148
+++ + G+I L +D F L L + + + LPQ N +A+ IL V G+A
Sbjct: 305 VYKPQLGYISTLNSYDLPILRFIRLS---ALRGSIRQNAMVLPQWNANANAILYVTDGEA 361
Query: 149 ILTVLNPNDRNSFN--LERGDTIKLPAGSIAYLANRADNEDLRVLDLAIPVNRPGQFQSF 206
+ ++N N F+ + +G I +P G + RA + + ++ N Q
Sbjct: 362 QIQIVNDNGNRVFDGQVSQGQLIAVPQG--FSVVKRATSNRFQWVEFKTNANA----QIN 415
Query: 207 SLSGNQNQQSFFSGFSKNILEAAFNSNYEEIERVLIEEQEQEPRHRRG 254
+L+G + S G ++ F + EE RV E H G
Sbjct: 416 TLAG---RTSVLRGLPLEVITNGFQISPEEARRVKFNTLETTLTHSSG 460
>At2g28680 legumin-like protein
Length = 356
Score = 43.5 bits (101), Expect = 2e-04
Identities = 32/116 (27%), Positives = 60/116 (51%), Gaps = 7/116 (6%)
Query: 129 LFLPQHNDADFILAVLSGKAILTVLNPN-DRNSFNLERGDTIKLPAGSIAYLANRADNED 187
L LP+++D+ + VL G ++ P + +++GD+I LP G + + N D E
Sbjct: 49 LALPRYSDSPKVAYVLQGAGTAGIVLPEKEEKVIAIKKGDSIALPFGVVTWWFNNEDTE- 107
Query: 188 LRVLDL--AIPVNRPGQFQSFSLSGNQNQQSFFSGFSKNILEAAFNSNYEEIERVL 241
L VL L ++ GQF F L+G+ F+GFS + A++ + +++++
Sbjct: 108 LVVLFLGETHKGHKAGQFTDFYLTGS---NGIFTGFSTEFVGRAWDLDETTVKKLV 160
Score = 29.6 bits (65), Expect = 3.6
Identities = 19/74 (25%), Positives = 36/74 (47%), Gaps = 1/74 (1%)
Query: 312 RSREPIYSNEFGNFFEITPEKNPQLQDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEG 371
R + +Y + G++F PE+ P L+D +I + + + L LP + S + + V +G
Sbjct: 8 RLPKKVYGGDGGSYFAWCPEELPMLRDGNIGASKLALEKYGLALPRY-SDSPKVAYVLQG 66
Query: 372 KGEFELVGQRNENQ 385
G +V E +
Sbjct: 67 AGTAGIVLPEKEEK 80
>At1g07750 globulin like protein
Length = 356
Score = 40.8 bits (94), Expect = 0.002
Identities = 27/113 (23%), Positives = 56/113 (48%), Gaps = 5/113 (4%)
Query: 131 LPQHNDADFILAVLSGKAILTVLNPN-DRNSFNLERGDTIKLPAGSIAYLANRADNE-DL 188
+P+++D+ + VL G ++ P + +++GD+I LP G + + N D E +
Sbjct: 51 VPRYSDSSKVAYVLQGSGTAGIVLPEKEEKVIAIKQGDSIALPFGVVTWWFNNEDPELVI 110
Query: 189 RVLDLAIPVNRPGQFQSFSLSGNQNQQSFFSGFSKNILEAAFNSNYEEIERVL 241
L ++ GQF F L+G F+GFS + A++ + +++++
Sbjct: 111 LFLGETHKGHKAGQFTEFYLTGT---NGIFTGFSTEFVGRAWDLDENTVKKLV 160
>At1g03890 putative cruciferin 12S seed storage protein
Length = 451
Score = 37.4 bits (85), Expect = 0.017
Identities = 28/94 (29%), Positives = 43/94 (44%), Gaps = 16/94 (17%)
Query: 162 NLERGDTIKLPAGSIAYLANRADNEDLRVLDLAIPVNRPGQFQS----FSLSGNQNQQ-- 215
N RGD AG + NR D++ + V+ L + NR Q F L+G++ Q+
Sbjct: 144 NFRRGDVFASLAGVSQWWYNRGDSDAVIVIVLDV-TNRENQLDQVPRMFQLAGSRTQEEE 202
Query: 216 ---------SFFSGFSKNILEAAFNSNYEEIERV 240
+ FSGF NI+ AF N E +++
Sbjct: 203 QPLTWPSGNNAFSGFDPNIIAEAFKINIETAKQL 236
>At5g63320 putative protein
Length = 569
Score = 35.8 bits (81), Expect = 0.050
Identities = 40/186 (21%), Positives = 68/186 (36%), Gaps = 24/186 (12%)
Query: 235 EEIERVLIEEQEQEPRHRRGLRDRRHKQSQEANVIVKVSREQIEELSRHAKSSSRRSASS 294
EE E+ L EE+E+ + + R K EA + REQ E +R A ++
Sbjct: 104 EEFEKRLREEKERLQAEAKAAEEARRKAKAEAAEKARREREQEREAARQALQKMEKTVEI 163
Query: 295 ESASR----------RSASSESAPFNLRSREPIYSNEFGNF--FEITPEKNPQLQDLDIL 342
R + P ++ P +S + F++ NP L+ L +
Sbjct: 164 NEGIRFMEDLQMLRATGTEGDQLPTSMEVMSPKFSEDMLGLGSFKMESNSNP-LEHLGLY 222
Query: 343 VNYAEIREGSLLLPHFNSRATVIVAVEEG----------KGEFELVGQRNENQQEQREYE 392
+ E + PHF+ R + +GE +LV NE Q ++
Sbjct: 223 MKMDEDEDEEEDPPHFSQRKVEDNPFDRSEKQEHSPHRVEGEDQLV-SGNEEPVSQEAHD 281
Query: 393 EDEQEE 398
+QE+
Sbjct: 282 NGDQED 287
Score = 31.6 bits (70), Expect = 0.95
Identities = 36/151 (23%), Positives = 61/151 (39%), Gaps = 5/151 (3%)
Query: 270 VKVSREQIEELSRHAKSSSRRSASSESASRRSASSESAPFNLRSREP--IYSNEFGNFFE 327
+++ RE+ E+ R K + A + +RR A +E+A R RE + + E
Sbjct: 99 LRIEREEFEKRLREEKERLQAEAKAAEEARRKAKAEAAEKARREREQEREAARQALQKME 158
Query: 328 ITPEKNPQLQDL-DILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQRN--EN 384
T E N ++ + D+ + A EG L + G G F++ N E+
Sbjct: 159 KTVEINEGIRFMEDLQMLRATGTEGDQLPTSMEVMSPKFSEDMLGLGSFKMESNSNPLEH 218
Query: 385 QQEQREYEEDEQEEERSQQVQRYRARLTPGD 415
+ +EDE EEE + + P D
Sbjct: 219 LGLYMKMDEDEDEEEDPPHFSQRKVEDNPFD 249
>At5g47090 unknown protein
Length = 310
Score = 35.4 bits (80), Expect = 0.066
Identities = 40/154 (25%), Positives = 62/154 (39%), Gaps = 26/154 (16%)
Query: 261 KQSQEANVIVKVSREQIEELSRHAKSSSRRSASSESASRRSASSESAPFN---LRSREPI 317
K E + +K R++I S KS S + A SE F+ +R REP
Sbjct: 84 KHDYEVDWHLKNLRKKISPTSEEIKSRSVAVRNRRLAYLNKLVSEGQYFSEDAMRDREPY 143
Query: 318 YSNEFGNFFEITPEKN---PQLQDLDILVNYAEIREGSLLLPHFNSRATVIVAVEE---- 370
+E+ F+ +N P + + L+ AE A ++ + E
Sbjct: 144 LHHEYVGKFQDVMGRNMARPGERWSETLMRRAE-------------EAVLVTRIREEQQR 190
Query: 371 -GKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
G E + VG NE +E E EE+E EEE ++
Sbjct: 191 LGVSESDWVG--NEKMEESEEEEEEESEEEEEEE 222
>At3g30190 hypothetical protein
Length = 263
Score = 35.4 bits (80), Expect = 0.066
Identities = 20/51 (39%), Positives = 28/51 (54%)
Query: 257 DRRHKQSQEANVIVKVSREQIEELSRHAKSSSRRSASSESASRRSASSESA 307
DRRHK +++ N S S + SSS S+SS S+S S+SS S+
Sbjct: 15 DRRHKMTKQCNGSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSTSSSSSSSS 65
>At3g04200 germin-like protein
Length = 227
Score = 35.4 bits (80), Expect = 0.066
Identities = 33/119 (27%), Positives = 50/119 (41%), Gaps = 22/119 (18%)
Query: 324 NFFEITPEKNPQLQDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQRNE 383
N + K P L L + + + +G PH + RAT I+ V +GK LVG +
Sbjct: 80 NVTTVDVNKIPGLNTLGVSLARLDFAQGGQNPPHIHPRATEILVVTKGK---LLVGFVSS 136
Query: 384 NQQEQREYEEDEQEEERSQQVQRYRARLTPGDVYVIPAG---YPNVVKASSDLSLLGFG 439
NQ R + Y+ L GDV+V P G + V+ + ++ GFG
Sbjct: 137 NQDNNRLF---------------YKV-LKRGDVFVFPIGLIHFQMNVRRTRAVAFAGFG 179
>At3g10080 germin-like protein
Length = 227
Score = 34.7 bits (78), Expect = 0.11
Identities = 35/134 (26%), Positives = 56/134 (41%), Gaps = 9/134 (6%)
Query: 323 GNFFE-------ITPEKNPQLQDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEF 375
GNF E + PE P L L I A+++ GS+ PH++ RAT + + +G+
Sbjct: 74 GNFTETGFATVPVGPENFPGLNTLGISFVRADLKPGSINPPHYHPRATEVAHLVKGRVYS 133
Query: 376 ELVGQRNENQQEQREYEEDEQEEERSQQVQRYRARLTPGDVYVIPAGYPNVVKASSDLSL 435
V N+ + E E + Q +T V + + P + K S +
Sbjct: 134 GFVDSNNKVYAKVMEEGEMMVYPKGLVHFQMNVGDVTATIVGGLNSQNPGIQKIPS--VV 191
Query: 436 LGFGINAENNQRSF 449
G GIN E ++F
Sbjct: 192 FGSGINEELLMKAF 205
>At5g43650 putative protein
Length = 247
Score = 34.3 bits (77), Expect = 0.15
Identities = 28/102 (27%), Positives = 44/102 (42%), Gaps = 17/102 (16%)
Query: 233 NYEEIERVLIEEQEQEPRHRRGLRDRRHKQSQ---------------EANVIVKVSREQI 277
N+EE + + E+E+ RH R RR KQ Q + N IV+ + ++I
Sbjct: 72 NWEEKKNTVAPEKERSRRHMLKERTRREKQKQSYLALHSLLPFATKNDKNSIVEKAVDEI 131
Query: 278 EELSRHAKSSSRRSASSESASRRSASSESAPFNLR--SREPI 317
+L R K RR E S + E + +R +EP+
Sbjct: 132 AKLQRLKKELVRRIKVIEEKSAKDGHDEMSETKVRVNLKEPL 173
>At5g07890 putative protein
Length = 389
Score = 34.3 bits (77), Expect = 0.15
Identities = 37/172 (21%), Positives = 68/172 (39%), Gaps = 17/172 (9%)
Query: 243 EEQEQEPRHRRGLRDRRHKQSQEANVIVKVSREQIEELSRHAKSSSRRSASS--ESASRR 300
EE QE +G+ + QSQ A VK +Q E+L R ++S +S +S R
Sbjct: 187 EESIQEKDQLKGIIEESQFQSQRAKENVKYIEKQNEDL-REKFTASEKSIKDFFQSTKER 245
Query: 301 SASSESAPFNLRSREPIYSNEFGNFFEITPEKNPQLQDLDILVNYAEIREGSLLLPHFNS 360
S + P N F + P N D ++ E+ + L+
Sbjct: 246 LESEDEQPLNAMC-------FFAELSHVLPVSNEVRNCFDAIMKKLELSQNVNLIDKVEG 298
Query: 361 RATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQVQRYRARLT 412
I + E V ++ + + +Q + + E+ E+ +Q++ R ++T
Sbjct: 299 MGKQI-------HQHEDVVKQLKEELKQEKLKAKEEAEDLTQEMAELRYKMT 343
>At3g05950 germin-like protein
Length = 229
Score = 34.3 bits (77), Expect = 0.15
Identities = 29/99 (29%), Positives = 39/99 (39%), Gaps = 19/99 (19%)
Query: 324 NFFEITPEKNPQLQDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQRNE 383
N + +K P L L + + + G PH + RAT I+ V EG LVG
Sbjct: 79 NVTNVNVDKIPGLNTLGVSLVRIDFAPGGQNPPHTHPRATEILVVVEGT---LLVGFVTS 135
Query: 384 NQQEQREYEEDEQEEERSQQVQRYRARLTPGDVYVIPAG 422
NQ R + + L PGDV+V P G
Sbjct: 136 NQDNNRLFSK----------------VLYPGDVFVFPIG 158
>At1g20290 hypothetical protein
Length = 497
Score = 34.3 bits (77), Expect = 0.15
Identities = 15/41 (36%), Positives = 26/41 (62%)
Query: 363 TVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
T++ A EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 359 TIVQAEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 399
Score = 33.1 bits (74), Expect = 0.33
Identities = 14/39 (35%), Positives = 25/39 (63%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQVQRY 407
EE + E E + E ++E+ E EE+E+EEE ++ ++Y
Sbjct: 446 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEKKY 484
Score = 30.8 bits (68), Expect = 1.6
Identities = 15/47 (31%), Positives = 24/47 (50%)
Query: 357 HFNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
H V EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 354 HVGEGTIVQAEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 400
Score = 30.8 bits (68), Expect = 1.6
Identities = 13/38 (34%), Positives = 24/38 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQVQR 406
EE + E E + E ++E+ E EE+E+EEE ++ ++
Sbjct: 445 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEK 482
Score = 30.4 bits (67), Expect = 2.1
Identities = 13/42 (30%), Positives = 25/42 (58%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQVQRYRAR 410
EE + E E + E ++E+ E EE+E+EEE+ + + + +
Sbjct: 451 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEKKYMIVKKKKK 492
Score = 30.4 bits (67), Expect = 2.1
Identities = 12/36 (33%), Positives = 21/36 (58%)
Query: 368 VEEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
V G+G + E ++E+ E EE+E+EEE ++
Sbjct: 353 VHVGEGTIVQAEEEEEEEEEEEEEEEEEEEEEEEEE 388
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 386 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 420
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 414 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 448
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 380 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 414
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 369 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 403
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 370 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 404
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 393 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 427
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 406 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 440
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 439 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 473
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 381 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 415
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 438 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 472
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 404 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 438
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 379 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 413
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 417 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 451
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 373 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 407
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 410 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 444
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 421 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 455
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 429 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 463
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 415 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 449
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 405 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 439
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 394 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 428
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 401 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 435
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 409 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 443
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 391 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 425
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 416 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 450
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 375 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 409
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 402 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 436
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 435 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 469
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 397 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 431
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 371 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 405
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 368 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 402
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 384 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 418
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 400 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 434
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 418 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 452
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 395 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 429
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 392 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 426
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 377 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 411
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 440 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 474
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 425 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 459
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 407 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 441
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 382 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 416
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 396 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 430
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 423 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 457
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 399 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 433
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 389 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 423
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 398 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 432
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 434 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 468
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 367 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 401
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 428 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 462
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 419 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 453
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 390 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 424
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 426 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 460
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 388 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 422
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 432 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 466
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 443 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 477
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 420 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 454
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 437 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 471
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 413 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 447
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 376 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 410
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 442 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 476
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 436 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 470
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 433 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 467
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 430 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 464
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 424 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 458
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 383 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 417
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 403 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 437
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 387 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 421
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 444 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 478
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 412 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 446
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 372 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 406
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 431 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 465
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 427 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 461
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 408 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 442
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 411 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 445
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 378 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 412
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 385 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 419
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 422 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 456
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 441 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 475
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQ 403
EE + E E + E ++E+ E EE+E+EEE ++
Sbjct: 374 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 408
Score = 28.9 bits (63), Expect = 6.1
Identities = 13/42 (30%), Positives = 26/42 (60%)
Query: 369 EEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQVQRYRAR 410
EE + E E + E ++E+ E EE+E+EE++ V++ + +
Sbjct: 452 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEKKYMIVKKKKKK 493
>At1g12830 unknown protein
Length = 213
Score = 34.3 bits (77), Expect = 0.15
Identities = 26/115 (22%), Positives = 44/115 (37%), Gaps = 12/115 (10%)
Query: 303 SSESAPFNLRSREPIYSNEFGNFFEITPEKNPQLQDLDILVNYAEIREGSLLLPHFNSRA 362
+ + + F ++ + ++ + N + NP L D E GS + + NS
Sbjct: 6 NQQDSSFPVKRKSDLFCQDQDNVTNKAQKLNPSLNSADSESKDGETN-GSGQIENLNSST 64
Query: 363 TVIVAVEEG-----------KGEFELVGQRNENQQEQREYEEDEQEEERSQQVQR 406
+ E G VG E + E+ E E+DE+EEE + V R
Sbjct: 65 EETIGAESSVLSEKAAEEVESGVIGDVGAEEEQEDEEDEEEDDEEEEEEEEVVDR 119
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.133 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,751,658
Number of Sequences: 26719
Number of extensions: 420208
Number of successful extensions: 3586
Number of sequences better than 10.0: 117
Number of HSP's better than 10.0 without gapping: 48
Number of HSP's successfully gapped in prelim test: 72
Number of HSP's that attempted gapping in prelim test: 2903
Number of HSP's gapped (non-prelim): 459
length of query: 456
length of database: 11,318,596
effective HSP length: 103
effective length of query: 353
effective length of database: 8,566,539
effective search space: 3023988267
effective search space used: 3023988267
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Medicago: description of AC148289.3


