

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148289.2 + phase: 0 /pseudo
(440 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
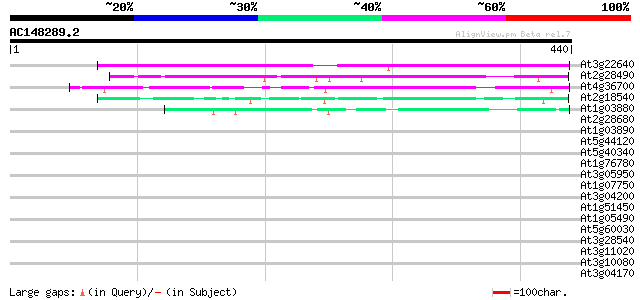
Score E
Sequences producing significant alignments: (bits) Value
At3g22640 unknown protein 228 5e-60
At2g28490 putative seed storage protein (vicilin-like) 92 4e-19
At4g36700 globulin-like protein 71 1e-12
At2g18540 putative vicilin storage protein (globulin-like) 54 2e-07
At1g03880 putative cruciferin 12S seed storage protein 52 7e-07
At2g28680 legumin-like protein 39 0.006
At1g03890 putative cruciferin 12S seed storage protein 39 0.006
At5g44120 legumin-like protein 39 0.007
At5g40340 unknown protein 36 0.037
At1g76780 putative heat shock protein 35 0.082
At3g05950 germin-like protein 35 0.11
At1g07750 globulin like protein 35 0.11
At3g04200 germin-like protein 34 0.14
At1g51450 unknown protein 34 0.18
At1g05490 hypothetical protein 34 0.18
At5g60030 KED - like protein 33 0.24
At3g28540 hypothetical protein 33 0.24
At3g11020 DREB2B transcription factor 33 0.24
At3g10080 germin-like protein 33 0.24
At3g04170 germin-like protein 33 0.24
>At3g22640 unknown protein
Length = 486
Score = 228 bits (581), Expect = 5e-60
Identities = 118/384 (30%), Positives = 212/384 (54%), Gaps = 32/384 (8%)
Query: 70 DQDQENPFIFNSNRFQTLFENENGHIRLLQRFDKRSK-IFENLQNYRLLEYHSKPHTLFL 128
++ +P+ F F F+++ G +R+L +F K + +F ++NYR +P T F+
Sbjct: 57 EESTNHPYHFRKRSFSDWFQSKEGFVRVLPKFTKHAPALFRGIENYRFSLVEMEPTTFFV 116
Query: 129 PQHNDADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKDLRVL 188
P H DAD + +V+ GK ++ + + SF++ +GD +++P+G ++ N + LR+
Sbjct: 117 PHHLDADAVFIVLQGKGVIEFVTDKTKESFHITKGDVVRIPSGVTNFITNTNQTVPLRLA 176
Query: 189 DLAIPVNRPGQFQSFSLSESENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENEQEPQH 248
+ +PVN PG ++ + + S+ QQS+ +GF+K +L +FN E + R++
Sbjct: 177 QITVPVNNPGNYKDYFPAASQFQQSYFNGFTKEVLSTSFNVPEELLGRLV---------- 226
Query: 249 RRGLRKDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESSESE-----------PI 297
R + +I ++S +QI+EL+++A S S + ++ + + P
Sbjct: 227 --------TRSKEIGQGIIRRISPDQIKELAEHATSPSNKHKAKKEKEEDKDLRTLWTPF 278
Query: 298 NLRNQKPIYSNKFGNFFEITPEKNPQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVE 357
NL PIYSN FG+F E P+ QL+DL I +A + +GSL LPHFNS+ T + VE
Sbjct: 279 NLFAIDPIYSNDFGHFHEAHPKNYNQLQDLHIAAAWANMTQGSLFLPHFNSKTTFVTFVE 338
Query: 358 EGKGEFEL-VGQRNENQQEQREYEEDEQQQERSQQVQRYRARLSPGDVYVIPAGHPI-VV 415
G FE+ + + Q+Q + E++++ S+ V + +R+ G+V+++PAGHP ++
Sbjct: 339 NGCARFEMATPYKFQRGQQQWPGQGQEEEEDMSENVHKVVSRVCKGEVFIVPAGHPFTIL 398
Query: 416 TASSDLSLLGFGINAENNQRNFLA 439
+ D +GFGI A N++R FLA
Sbjct: 399 SQDQDFIAVGFGIYATNSKRTFLA 422
>At2g28490 putative seed storage protein (vicilin-like)
Length = 511
Score = 92.4 bits (228), Expect = 4e-19
Identities = 93/395 (23%), Positives = 163/395 (40%), Gaps = 62/395 (15%)
Query: 79 FNSNRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQHNDADFIL 138
F + + ++E G +R++ R +I E + L +P TLF+PQ+ D+ ++
Sbjct: 86 FMMRESRQVIKSEGGEMRVV--LSPRGRIIEKPMHIGFLTM--EPKTLFVPQYLDSSLLI 141
Query: 139 VVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKDLRVLDLAIPVNRPG 198
+ G+A L V+ + L+ GD +PAG++ YL N + L V+ P G
Sbjct: 142 FIRQGEATLGVICKDEFGERKLKAGDIYWIPAGSVFYLHNTGLGQRLHVICSIDPTQSLG 201
Query: 199 --QFQSFSLSESENQQSFLSGFSKNILEAAFNSNYEEIERVLI-----------EENEQE 245
FQ F + + S L+GF + L +AFN + E++++++ E + +
Sbjct: 202 FETFQPFYIGGGPS--SVLAGFDPHTLTSAFNVSLPELQQMMMSQFRGPIVYVTEGPQPQ 259
Query: 246 PQHR-----RGLRKDERRQQSQEANVIVKVSREQ---------------IEELSKNAKSS 285
PQ GLR +E+ +Q ++ + S + I +L++
Sbjct: 260 PQSTVWTQFLGLRGEEKHKQLKKLLETKQGSPQDQQYSSGWSWRNIVRSILDLTEEKNKG 319
Query: 286 SRRSESSESEPINLRNQKPIYSNKFGNFFEITPEKNPQLKDLDILVNYAEIREGSLLLPH 345
S SE +S I + KP + NK+G + + LK I V + G+++ PH
Sbjct: 320 SGSSECEDSYNIYDKKDKPSFDNKYGWSIALDYDDYKPLKHSGIGVYLVNLTAGAMMAPH 379
Query: 346 FNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQQQERSQQVQRYRARLSPGDVY 405
N AT V G GE ++V + R+S GDV+
Sbjct: 380 MNPTATEYGIVLAGSGEIQVVFPNGTSAM---------------------NTRVSVGDVF 418
Query: 406 VIPAGHPI--VVTASSDLSLLGFGINAENNQRNFL 438
IP + + + +GF +A N+ FL
Sbjct: 419 WIPRYFAFCQIASRTGPFEFVGFTTSAHKNRPQFL 453
>At4g36700 globulin-like protein
Length = 499
Score = 70.9 bits (172), Expect = 1e-12
Identities = 79/409 (19%), Positives = 169/409 (41%), Gaps = 61/409 (14%)
Query: 48 PCSLLMLLGIVFLASICVSSRSDQDQ-----------ENPFIFNSNRFQTLFENENGHIR 96
P S+L+L+ ++FL + ++ + ++ +P + ++++ +FE + G I
Sbjct: 8 PLSVLLLV-LLFLCTESLAKSEESEEYDVAVPSCCGFSSPLLIKKDQWKPIFETKFGQIS 66
Query: 97 LLQRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQHNDADFILVVVSGKAILTVLNPNNRN 156
+Q + + Y++ +P+T+ LP +D + V SG IL ++ ++
Sbjct: 67 TVQIGNGCG----GMGPYKIHSITLEPNTILLPLLLHSDMVFFVDSGSGILNWVDEEAKS 122
Query: 157 SFNLERGDTIKLPAGTLGYLANRDDNKDLRVLDLAIPVNRPGQFQSFSLSESENQQSFLS 216
+ + GD +L G++ YL ++ D G + S + +
Sbjct: 123 T-EIRLGDVYRLRPGSVFYLQSKPDPCF-------------GAYSSIT--------DLMF 160
Query: 217 GFSKNILEAAFNSNYEEIERVLIEENEQEP----QHRRGLRKDERRQQSQEANVIVKVSR 272
GF + IL++AF IE + N +P Q Q + +
Sbjct: 161 GFDETILQSAFGVPEGIIE---LMRNRTKPPLIVSETLCTPGVANTWQLQPRLLKLFAGS 217
Query: 273 EQIEELSKNAKSSSRRSESSESEPINLRNQKPIYSNKFGNFFEITPEKNPQLKDLDILVN 332
+ + K + ++ + +++ N+ +P + + +G I + LK + V+
Sbjct: 218 ADLVDNKKKKEKKEKKEKVKKAKTFNVFESEPDFESPYGRTITINRKDLKVLKGSMVGVS 277
Query: 333 YAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQQQERSQQV 392
+ +GS++ PH+N A I V +G G ++ S +
Sbjct: 278 MVNLTQGSMMGPHWNPWACEISIVLKGAGMVRVL--------------RSSISSNTSSEC 323
Query: 393 QRYRARLSPGDVYVIPAGHPIVVTASSDLSL--LGFGINAENNQRNFLA 439
+ R ++ GD++ +P HP+ + ++ SL +GF +A+NN+ FLA
Sbjct: 324 KNVRFKVEEGDIFAVPRLHPMAQMSFNNDSLVFVGFTTSAKNNEPQFLA 372
Score = 31.2 bits (69), Expect = 1.2
Identities = 42/183 (22%), Positives = 70/183 (37%), Gaps = 53/183 (28%)
Query: 137 ILVVVSGKAILTVLNPN---NRNS------FNLERGDTIKLPAGTLGYLANRDDNKDLRV 187
I +V+ G ++ VL + N +S F +E GD +P L +A N D V
Sbjct: 298 ISIVLKGAGMVRVLRSSISSNTSSECKNVRFKVEEGDIFAVPR--LHPMAQMSFNNDSLV 355
Query: 188 LDLAIPVNRPGQFQSFSLSESENQQSFLSG-------FSKNILEAAFNSNYE-------- 232
F F+ S N+ FL+G + +L A+ N +
Sbjct: 356 ------------FVGFTTSAKNNEPQFLAGEDSALRMLDRQVLAASLNVSSVTIDGLLGA 403
Query: 233 ---------------EIERVLIEENEQEPQHRRGLRKDERRQQSQEANVIVKVSREQIEE 277
EIE++ +E ++ R R DER+++ +EA + R++ EE
Sbjct: 404 QKEAVILECHSCAEGEIEKLKVEIERKKIDDERKRRHDERKKEEEEAKREEEERRKREEE 463
Query: 278 LSK 280
K
Sbjct: 464 EEK 466
>At2g18540 putative vicilin storage protein (globulin-like)
Length = 699
Score = 53.9 bits (128), Expect = 2e-07
Identities = 84/381 (22%), Positives = 151/381 (39%), Gaps = 64/381 (16%)
Query: 70 DQDQENPFIFNSNRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFLP 129
D +P + ++ ++ E G+I +Q D Y + +P+ L LP
Sbjct: 40 DPSYSSPLLVKKDQRTSVVATEFGNISAVQIGD----------GYHIQFITLEPNALLLP 89
Query: 130 QHNDADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKDLRV-- 187
+D + V +G + + ++F + KL G N N+ LRV
Sbjct: 90 LLLHSDMVFFVHTGTS-------KHSSTFVFSK----KLLIGP----KNLKPNEKLRVYA 134
Query: 188 -LDLAIPVNRPGQFQSFSLSESENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENEQE- 245
++ +N P L + + L GF L +AF + E+I R + + +
Sbjct: 135 IFNVGKCLNDP------CLGAYSSVRDLLLGFDDRTLRSAF-AVPEDILRKIRDATKPPL 187
Query: 246 -----PQHR-RGLRKDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESSESEPINL 299
P++R +GL +D + QS+ + V V K ++++ +S N+
Sbjct: 188 IVNALPRNRTQGLEED--KWQSRLVRLFVSVEDVTDHLAMKPIVDTNKK----KSRTFNV 241
Query: 300 RNQKPIYSNKFGNFFEITPEKNPQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEG 359
+ P + N G + + LK V + +GS++ PH+N A I V EG
Sbjct: 242 FEEDPDFENNNGRSIVVDEKDLDALKGSRFGVFMVNLTKGSMIGPHWNPSACEISIVLEG 301
Query: 360 KGEFELVGQRNENQQEQREYEEDEQQQERSQQVQRYRARLSPGDVYVIPAGHPIVVTA-- 417
+G +V NQQ + D + + + GDV+V+P HP+ +
Sbjct: 302 EGMVRVV-----NQQSLSSCKNDRKSES---------FMVEEGDVFVVPKFHPMAQMSFE 347
Query: 418 SSDLSLLGFGINAENNQRNFL 438
+S +GF +A+ N FL
Sbjct: 348 NSSFVFMGFSTSAKTNHPQFL 368
Score = 41.2 bits (95), Expect = 0.001
Identities = 71/360 (19%), Positives = 131/360 (35%), Gaps = 75/360 (20%)
Query: 70 DQDQENPFIFNSNRFQTLFENENGHIRLLQRFD------KRSKIFE-NLQNYRLLEYHSK 122
D +++ FN FEN NG ++ D R +F NL ++ H
Sbjct: 230 DTNKKKSRTFNVFEEDPDFENNNGRSIVVDEKDLDALKGSRFGVFMVNLTKGSMIGPHWN 289
Query: 123 PHTLFLPQHNDADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDN 182
P + + + ++ VV+ +++ + N SF +E GD +P
Sbjct: 290 PSACEISIVLEGEGMVRVVNQQSLSSCKNDRKSESFMVEEGDVFVVP------------- 336
Query: 183 KDLRVLDLAIPVNRPGQFQSFSLSESENQQSFLSGFS-------KNILEAAFNSNYEEIE 235
K + ++ N F FS S N FL G S ++++ +FN + E I+
Sbjct: 337 KFHPMAQMSFE-NSSFVFMGFSTSAKTNHPQFLVGQSSVLKVLDRDVVAVSFNLSNETIK 395
Query: 236 RVLIEENE---------------------QEPQHRRGLRKDERRQQSQEANVIVKVSREQ 274
+L + E +E + R + RR++ +EA + R +
Sbjct: 396 GLLKAQKESVIFECASCAEGELSKLMREIEERKRREEEEIERRRKEEEEARKREEAKRRE 455
Query: 275 IEELSKNAKSSSRRSESSESEPINLRNQKPIYSNKFGNFFEITPEKNPQLKDLDILVNYA 334
EE + + + R + E E ++ E + + K + A
Sbjct: 456 EEEAKRREEEETERKKREEEEARKREEERKREE-------EEAKRREEERKKREEEAEQA 508
Query: 335 EIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQQQERSQQVQR 394
RE EE + E E+ +R E +Q ++E EE E+++ Q+ +R
Sbjct: 509 RKRE------------------EEREKEEEMAKKREEERQ-RKEREEVERKRREEQERKR 549
Score = 33.1 bits (74), Expect = 0.31
Identities = 18/68 (26%), Positives = 37/68 (53%), Gaps = 4/68 (5%)
Query: 232 EEIERVLIEENEQEPQHRRGLRKDERRQQSQEANVIVK----VSREQIEELSKNAKSSSR 287
EE+ER + EE E++ + R+++ RQ+ + + K +R++ EE++K + +
Sbjct: 581 EEVERKIREEQERKREEEMAKRREQERQKKEREEMERKKREEEARKREEEMAKIREEERQ 640
Query: 288 RSESSESE 295
R E + E
Sbjct: 641 RKEREDVE 648
Score = 32.3 bits (72), Expect = 0.53
Identities = 33/169 (19%), Positives = 72/169 (42%), Gaps = 38/169 (22%)
Query: 232 EEIERVLIEENEQEPQHRRGLRKDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSES 291
EE E+ E E+E + +++E RQ+ + RE++E + + RR E
Sbjct: 503 EEAEQARKREEEREKEEEMAKKREEERQRKE---------REEVERKRREEQERKRREEE 553
Query: 292 SESEPINLRNQKPIYSNKFGNFFEITPEKNPQLKDLDILVNYAEIREGSLLLPHFNSRAT 351
+ + ++ + + E+ Q K+ + + +IRE
Sbjct: 554 ARKREEERKREEEMAKRR---------EQERQRKEREEVER--KIRE------------- 589
Query: 352 VIVAVEEGKGEFELVGQR-NENQQEQREYEEDEQQQERSQQVQRYRARL 399
+E K E E+ +R E Q+++RE E ++++E +++ + A++
Sbjct: 590 ----EQERKREEEMAKRREQERQKKEREEMERKKREEEARKREEEMAKI 634
Score = 29.6 bits (65), Expect = 3.5
Identities = 20/74 (27%), Positives = 36/74 (48%), Gaps = 10/74 (13%)
Query: 232 EEIERVLIEENE----------QEPQHRRGLRKDERRQQSQEANVIVKVSREQIEELSKN 281
EE+ER EE +E + +R R+D R++ +E + + R++ EE +K
Sbjct: 613 EEMERKKREEEARKREEEMAKIREEERQRKEREDVERKRREEEAMRREEERKREEEAAKR 672
Query: 282 AKSSSRRSESSESE 295
A+ R+ E E +
Sbjct: 673 AEEERRKKEEEEEK 686
Score = 29.3 bits (64), Expect = 4.5
Identities = 23/93 (24%), Positives = 40/93 (42%), Gaps = 6/93 (6%)
Query: 207 ESENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENEQEPQHRRGLRKDERRQQSQEANV 266
E E Q+ + E EE R EE ++E + + R+++ RQ+ + V
Sbjct: 526 EEERQRKEREEVERKRREEQERKRREEEARKREEERKREEEMAK--RREQERQRKEREEV 583
Query: 267 IVKVSREQI----EELSKNAKSSSRRSESSESE 295
K+ EQ EE++K + ++ E E E
Sbjct: 584 ERKIREEQERKREEEMAKRREQERQKKEREEME 616
>At1g03880 putative cruciferin 12S seed storage protein
Length = 455
Score = 52.0 bits (123), Expect = 7e-07
Identities = 76/373 (20%), Positives = 130/373 (34%), Gaps = 96/373 (25%)
Query: 122 KPHTLFLPQHNDADFILVVVSGKAILTVLNPNNRNSF----------------------- 158
+P LFLP +A + VV G+ ++ + P +F
Sbjct: 74 EPQGLFLPTFLNAGKLTFVVHGRGLMGRVIPGCAETFMESPVFGEGQGQGQSQGFRDMHQ 133
Query: 159 ---NLERGDTIKLPAGTLGY----------------LANRDDNKDLRVLDLAIPVNRPGQ 199
+L GDTI P+G + LA+ + D + I N P
Sbjct: 134 KVEHLRCGDTIATPSGVAQWFYNNGNEPLILVAAADLASNQNQLDRNLRPFLIAGNNPQG 193
Query: 200 FQSFSLSESENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENEQEPQH----------- 248
+ + + Q + +GF+ IL AF N E +++ +N+Q+ +
Sbjct: 194 QEWLQGRKQQKQNNIFNGFAPEILAQAFKINVETAQQL---QNQQDNRGNIVKVNGPFGV 250
Query: 249 -RRGLRKDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESSESEPINLRNQKPIYS 307
R LR+ E QQ E +EE + + + S+++ +Y
Sbjct: 251 IRPPLRRGEGGQQPHEI-------ANGLEETLCTMRCTENLDDPSDAD---------VYK 294
Query: 308 NKFGNFFEITPEKNPQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVG 367
G + P L+ L + IR+ +++LP +N A + V GK ++V
Sbjct: 295 PSLGYISTLNSYNLPILRLLRLSALRGSIRKNAMVLPQWNVNANAALYVTNGKAHIQMVN 354
Query: 368 QRNENQQEQREYEEDEQQQERSQQVQRYRARLSPGDVYVIPAGHPIVVTA-SSDLSLLGF 426
E +Q +S G + V+P G ++ A + F
Sbjct: 355 DNGERVFDQ---------------------EISSGQLLVVPQGFSVMKHAIGEQFEWIEF 393
Query: 427 GINAENNQRNFLA 439
N EN Q N LA
Sbjct: 394 KTN-ENAQVNTLA 405
>At2g28680 legumin-like protein
Length = 356
Score = 38.9 bits (89), Expect = 0.006
Identities = 34/142 (23%), Positives = 68/142 (46%), Gaps = 17/142 (11%)
Query: 126 LFLPQHNDADFILVVVSGKAILTVLNPNNRNS-FNLERGDTIKLPAGTLGYLANRDDNKD 184
L LP+++D+ + V+ G ++ P +++GD+I LP G + + N +D +
Sbjct: 49 LALPRYSDSPKVAYVLQGAGTAGIVLPEKEEKVIAIKKGDSIALPFGVVTWWFNNEDT-E 107
Query: 185 LRVLDL--AIPVNRPGQFQSFSLSESENQQSFLSGFSKNILEAAFNSNYEEIER------ 236
L VL L ++ GQF F L+ S +GFS + A++ + +++
Sbjct: 108 LVVLFLGETHKGHKAGQFTDFYLTGS---NGIFTGFSTEFVGRAWDLDETTVKKLVGSQT 164
Query: 237 ----VLIEENEQEPQHRRGLRK 254
V ++ + + P+ ++G RK
Sbjct: 165 GNGIVKVDASLKMPEPKKGDRK 186
Score = 30.4 bits (67), Expect = 2.0
Identities = 20/74 (27%), Positives = 35/74 (47%), Gaps = 1/74 (1%)
Query: 300 RNQKPIYSNKFGNFFEITPEKNPQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEG 359
R K +Y G++F PE+ P L+D +I + + + L LP + S + + V +G
Sbjct: 8 RLPKKVYGGDGGSYFAWCPEELPMLRDGNIGASKLALEKYGLALPRY-SDSPKVAYVLQG 66
Query: 360 KGEFELVGQRNENQ 373
G +V E +
Sbjct: 67 AGTAGIVLPEKEEK 80
>At1g03890 putative cruciferin 12S seed storage protein
Length = 451
Score = 38.9 bits (89), Expect = 0.006
Identities = 65/338 (19%), Positives = 123/338 (36%), Gaps = 75/338 (22%)
Query: 122 KPHTLFLPQHNDADFILVVVSGKAILTVLNPNNRNSF----------------------- 158
+P+++FLP + VV G+ ++ + +F
Sbjct: 80 QPNSIFLPAFFSPPALAYVVQGEGVMGTIASGCPETFAEVEGSSGRGGGGDPGRRFEDMH 139
Query: 159 ----NLERGDTIKLPAGTLGYLANRDDNKDLRVLDLAIPVNRPGQFQS----FSLSESEN 210
N RGD AG + NR D+ + V+ L + NR Q F L+ S
Sbjct: 140 QKLENFRRGDVFASLAGVSQWWYNRGDSDAVIVIVLDV-TNRENQLDQVPRMFQLAGSRT 198
Query: 211 QQ-----------SFLSGFSKNILEAAFNSNYEEIERVLIEENEQEPQHRRGLRKDERRQ 259
Q+ + SGF NI+ AF N E +++ Q+++ R + R
Sbjct: 199 QEEEQPLTWPSGNNAFSGFDPNIIAEAFKINIETAKQL---------QNQKDNRGNIIRA 249
Query: 260 QSQEANVIVKVSREQIEELSKNAKSSSRRSESSESEPINLRNQKPIYSNKFGNFFEITPE 319
VI Q + ++ + + ++ E+ I+ + +S + G +
Sbjct: 250 NGPLHFVIPPPREWQQDGIANGIEETYCTAKIHEN--IDDPERSDHFSTRAGRISTLNSL 307
Query: 320 KNPQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQRNENQQEQREY 379
P L+ + + + G ++LP + + A ++ V G+ + ++V
Sbjct: 308 NLPVLRLVRLNALRGYLYSGGMVLPQWTANAHTVLYVTGGQAKIQVV------------- 354
Query: 380 EEDEQQQERSQQVQRYRARLSPGDVYVIPAGHPIVVTA 417
+D Q ++QV G + VIP G + TA
Sbjct: 355 -DDNGQSVFNEQV-------GQGQIIVIPQGFAVSKTA 384
>At5g44120 legumin-like protein
Length = 472
Score = 38.5 bits (88), Expect = 0.007
Identities = 45/242 (18%), Positives = 99/242 (40%), Gaps = 40/242 (16%)
Query: 207 ESENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENEQEPQHR--------RGLRKDERR 258
E + Q++ +GF ++ A + + +++ +++ + R R + +R
Sbjct: 208 EQQPQKNIFNGFGPEVIAQALKIDLQTAQQLQNQDDNRGNIVRVQGPFGVIRPPLRGQRP 267
Query: 259 QQSQEANVIVKVSREQIEELSKNAKSSSRRSESSESEPINLRNQKPIYSNKFGNFFEITP 318
Q+ +E +EE +A+ + + S ++ +Y + G +
Sbjct: 268 QEEEEEEGRHGRHGNGLEETICSARCTDNLDDPSRAD---------VYKPQLGYISTLNS 318
Query: 319 EKNPQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQRNENQQEQRE 378
P L+ + + IR+ +++LP +N+ A I+ V +G+ + ++V N R
Sbjct: 319 YDLPILRFIRLSALRGSIRQNAMVLPQWNANANAILYVTDGEAQIQIV-----NDNGNRV 373
Query: 379 YEEDEQQQERSQQVQRYRARLSPGDVYVIPAGHPIVVTASSD-LSLLGFGINAENNQRNF 437
++ ++S G + +P G +V A+S+ + F NA N Q N
Sbjct: 374 FD----------------GQVSQGQLIAVPQGFSVVKRATSNRFQWVEFKTNA-NAQINT 416
Query: 438 LA 439
LA
Sbjct: 417 LA 418
Score = 32.3 bits (72), Expect = 0.53
Identities = 37/168 (22%), Positives = 67/168 (39%), Gaps = 15/168 (8%)
Query: 87 LFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQHN-DADFILVVVSGKA 145
+++ + G+I L +D F L L + + + LPQ N +A+ IL V G+A
Sbjct: 305 VYKPQLGYISTLNSYDLPILRFIRLS---ALRGSIRQNAMVLPQWNANANAILYVTDGEA 361
Query: 146 ILTVLNPNNRNSFN--LERGDTIKLPAGTLGYLANRDDNKDLRVLDLAIPVNRPGQFQSF 203
+ ++N N F+ + +G I +P G + R + + ++ N
Sbjct: 362 QIQIVNDNGNRVFDGQVSQGQLIAVPQGF--SVVKRATSNRFQWVEFKTNANA------- 412
Query: 204 SLSESENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENEQEPQHRRG 251
++ + S L G ++ F + EE RV E H G
Sbjct: 413 QINTLAGRTSVLRGLPLEVITNGFQISPEEARRVKFNTLETTLTHSSG 460
>At5g40340 unknown protein
Length = 1008
Score = 36.2 bits (82), Expect = 0.037
Identities = 42/188 (22%), Positives = 74/188 (39%), Gaps = 28/188 (14%)
Query: 208 SENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENEQEP----QHRRGLRKDERRQQSQE 263
SE + G K + EEIE EENE + + R +K E +++ E
Sbjct: 686 SEMGKPVTKGKEKKDKKGKAKQKAEEIEVTGKEENETDKHGKMKKERKRKKSESKKEGGE 745
Query: 264 ANVIVKVSREQIEELSKNAKSSSRRSESSESEPINLRNQKPIYSNKFGNFFEITPEKNPQ 323
K + E ++ K KS S++ E E ++P S K + KNP+
Sbjct: 746 GEETQKEANESTKKERKRKKSESKKQSDGEEE----TQKEPSESTK-----KERKRKNPE 796
Query: 324 LKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDE 383
K V E R+ S VE K E + +++ ++ E E+ E
Sbjct: 797 SKKKAEAVEEEETRKES---------------VESTKKERKRKKPKHDEEEVPNETEKPE 841
Query: 384 QQQERSQQ 391
+++++ ++
Sbjct: 842 KKKKKKRE 849
>At1g76780 putative heat shock protein
Length = 1871
Score = 35.0 bits (79), Expect = 0.082
Identities = 44/219 (20%), Positives = 87/219 (39%), Gaps = 27/219 (12%)
Query: 204 SLSESENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENEQEPQHRRGLRKDERRQQSQE 263
+ E N+Q G+++ I+E N + +V QE ++ L + R ++
Sbjct: 509 TFQEETNKQP--EGYNEKIMETGKKINEDGTRKV------QEMIRQQELDEPARSEKENR 560
Query: 264 ANVIVKVSREQIEELSKNAKSSSRRSESSESEPINLRNQKPIYS-----NKFGNFFEITP 318
+ +VK S+ EE + + + R E P LR Q+ KF + E+
Sbjct: 561 SRELVK-SKTNDEEKKEKEIAGTERKEKESDRPKILREQEVADEVAEDKTKFSIYGEVKE 619
Query: 319 EKNPQLK--------DLDILVNYAEIREGSLLLPHFNSRATVIVAVEE-----GKGEFEL 365
E+ K D+ +V E + + + + +EE GKG +
Sbjct: 620 EEEIAGKEKEFGSDDDIARIVRDTEQLDSNAMKGQEEKDMIQELVLEEKVCDGGKGIIAV 679
Query: 366 VGQRNENQQEQREYEEDEQQQERSQQVQRYRARLSPGDV 404
+ EN + +R E +EQ+ ++ ++ +L G++
Sbjct: 680 AETKAENNKSKRVQETEEQKLDKEDTCGKHFQKLIEGEI 718
Score = 30.8 bits (68), Expect = 1.6
Identities = 24/89 (26%), Positives = 43/89 (47%), Gaps = 10/89 (11%)
Query: 215 LSGFSKNILEAAFNSNYEEIERVLIEENEQEPQHRRGLRKDERRQQSQEANVIVKVSREQ 274
+SGF +E EE ERV + E E + + +G KD+ ++S + + E+
Sbjct: 55 ISGFK---IEEEEEEEEEEEERVDVSEAEHKEETEKGELKDDYLEKSHQID-------ER 104
Query: 275 IEELSKNAKSSSRRSESSESEPINLRNQK 303
IEE A S+ +SS +P ++ ++
Sbjct: 105 IEEEKGLADSNKESVDSSLRKPPDIEGRE 133
>At3g05950 germin-like protein
Length = 229
Score = 34.7 bits (78), Expect = 0.11
Identities = 31/104 (29%), Positives = 42/104 (39%), Gaps = 20/104 (19%)
Query: 308 NKFG-NFFEITPEKNPQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELV 366
N+ G N + +K P L L + + + G PH + RAT I+ V EG LV
Sbjct: 74 NRVGSNVTNVNVDKIPGLNTLGVSLVRIDFAPGGQNPPHTHPRATEILVVVEGT---LLV 130
Query: 367 GQRNENQQEQREYEEDEQQQERSQQVQRYRARLSPGDVYVIPAG 410
G NQ R + + L PGDV+V P G
Sbjct: 131 GFVTSNQDNNRLFSK----------------VLYPGDVFVFPIG 158
>At1g07750 globulin like protein
Length = 356
Score = 34.7 bits (78), Expect = 0.11
Identities = 25/114 (21%), Positives = 56/114 (48%), Gaps = 7/114 (6%)
Query: 128 LPQHNDADFILVVVSGKAILTVLNPNNRNS-FNLERGDTIKLPAGTLGYLANRDDNKDLR 186
+P+++D+ + V+ G ++ P +++GD+I LP G + + N +D +L
Sbjct: 51 VPRYSDSSKVAYVLQGSGTAGIVLPEKEEKVIAIKQGDSIALPFGVVTWWFNNED-PELV 109
Query: 187 VLDL--AIPVNRPGQFQSFSLSESENQQSFLSGFSKNILEAAFNSNYEEIERVL 238
+L L ++ GQF F L+ +GFS + A++ + +++++
Sbjct: 110 ILFLGETHKGHKAGQFTEFYLT---GTNGIFTGFSTEFVGRAWDLDENTVKKLV 160
>At3g04200 germin-like protein
Length = 227
Score = 34.3 bits (77), Expect = 0.14
Identities = 33/119 (27%), Positives = 49/119 (40%), Gaps = 22/119 (18%)
Query: 312 NFFEITPEKNPQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQRNE 371
N + K P L L + + + +G PH + RAT I+ V +GK LVG +
Sbjct: 80 NVTTVDVNKIPGLNTLGVSLARLDFAQGGQNPPHIHPRATEILVVTKGK---LLVGFVSS 136
Query: 372 NQQEQREYEEDEQQQERSQQVQRYRARLSPGDVYVIPAG---HPIVVTASSDLSLLGFG 427
NQ R + Y+ L GDV+V P G + V + ++ GFG
Sbjct: 137 NQDNNRLF---------------YKV-LKRGDVFVFPIGLIHFQMNVRRTRAVAFAGFG 179
>At1g51450 unknown protein
Length = 509
Score = 33.9 bits (76), Expect = 0.18
Identities = 38/171 (22%), Positives = 74/171 (43%), Gaps = 20/171 (11%)
Query: 132 NDADFILVVVSGKAILTVLNPNNRNSFN-LERGDTIKLPAGTLGYLANRDDNKDLRVLDL 190
++A+ + ++ G T L P+ N LE I+ P T DL LD+
Sbjct: 62 SEAEVSISLLEGTTTGTALLPSEENDLAPLESSGIIEEPIDT-----------DLEKLDV 110
Query: 191 -AIPVNRPG---QFQSFSLSESENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENEQEP 246
A+ V++PG + +S S SE S L++ N N +E +E + +P
Sbjct: 111 VAMDVDQPGSDLKIESDSFSEEAPTTSSSDNPKSPKLDSVANQNGSAMEEDEGDEEQDDP 170
Query: 247 QHRRGLRKD----ERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESSE 293
H++ + D ++ +E ++ +EE + S++++S+S +
Sbjct: 171 PHKKLKQLDCLTSVAVKEEEEPEQVLPSEAMVVEEAATLVASAAKKSKSKK 221
>At1g05490 hypothetical protein
Length = 731
Score = 33.9 bits (76), Expect = 0.18
Identities = 28/105 (26%), Positives = 46/105 (43%), Gaps = 11/105 (10%)
Query: 190 LAIPVNRPGQFQSFSLSESENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENEQEPQHR 249
L + + PG+ + S SESE + + K + +EE R++ ++ E
Sbjct: 201 LVLEKSVPGEIEILSDSESETEARRRASAKKKL--------FEESSRIVESISDGEDSSS 252
Query: 250 RGLRKDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESSES 294
++E Q S++ N V+ +E LS SSS S SS S
Sbjct: 253 ETDEEEEENQDSEDNNTKDNVT---VESLSSEDPSSSSSSSSSSS 294
>At5g60030 KED - like protein
Length = 292
Score = 33.5 bits (75), Expect = 0.24
Identities = 36/156 (23%), Positives = 76/156 (48%), Gaps = 27/156 (17%)
Query: 241 ENEQEPQHRRGLRKDERRQQS-QEANVIVKVSREQIEELSKNAKSSSRRSESSESEPINL 299
E EQ + RR +K+++++++ ++ +V+ + +E++E+ K+A R+ + S
Sbjct: 138 EAEQRSEERRERKKEKKKKKNNKDEDVVDEKVKEKLEDEQKSADRKERKKKKS------- 190
Query: 300 RNQKPIYSNKFGNFFEITPEKNPQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEG 359
K N ++ EK +L+D AEI+E N V+ E+
Sbjct: 191 ---------KKNNDEDVVDEKE-KLEDEQ---KSAEIKEKKK-----NKDEDVVDEKEKE 232
Query: 360 KGEFEL-VGQRNENQQEQREYEEDEQQQERSQQVQR 394
K E E G+R + ++++R+ +E+ +ER + +R
Sbjct: 233 KLEDEQRSGERKKEKKKKRKSDEEIVSEERKSKKKR 268
>At3g28540 hypothetical protein
Length = 510
Score = 33.5 bits (75), Expect = 0.24
Identities = 14/50 (28%), Positives = 31/50 (62%)
Query: 347 NSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQQQERSQQVQRYR 396
+++ IV +E+ +L GQR + ++E + EE+E+++E + ++R R
Sbjct: 287 DTKGKSIVVIEDIDCSLDLTGQRKKKKEEDEDEEEEEKKKEAEKLLKRER 336
>At3g11020 DREB2B transcription factor
Length = 330
Score = 33.5 bits (75), Expect = 0.24
Identities = 33/128 (25%), Positives = 57/128 (43%), Gaps = 5/128 (3%)
Query: 289 SESSESEPINLRNQKPIYSNKFGNFFEITPEKNPQLKDLDILVNYAEIREGSLLLPH--F 346
S SS+SE + N+ + + + E NP + LD+ R S + H
Sbjct: 143 STSSQSEVCTVENKAVVCGDVCVKHEDTDCESNPFSQILDVREESCGTRPDSCTVGHQDM 202
Query: 347 NSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQQQERSQQVQRYRARLSPGDVYV 406
NS + +E E + GQ + +++ ++ EE+ QQQ++ QQ Q+ + L Y
Sbjct: 203 NSSLNYDLLLEF---EQQYWGQVLQEKEKPKQEEEEIQQQQQEQQQQQLQPDLLTVADYG 259
Query: 407 IPAGHPIV 414
P + IV
Sbjct: 260 WPWSNDIV 267
>At3g10080 germin-like protein
Length = 227
Score = 33.5 bits (75), Expect = 0.24
Identities = 21/68 (30%), Positives = 33/68 (47%), Gaps = 7/68 (10%)
Query: 311 GNFFE-------ITPEKNPQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEF 363
GNF E + PE P L L I A+++ GS+ PH++ RAT + + +G+
Sbjct: 74 GNFTETGFATVPVGPENFPGLNTLGISFVRADLKPGSINPPHYHPRATEVAHLVKGRVYS 133
Query: 364 ELVGQRNE 371
V N+
Sbjct: 134 GFVDSNNK 141
>At3g04170 germin-like protein
Length = 227
Score = 33.5 bits (75), Expect = 0.24
Identities = 28/104 (26%), Positives = 44/104 (41%), Gaps = 21/104 (20%)
Query: 307 SNKFGNFFEITPEKNPQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELV 366
+ + G+F + P P L L + + + G L+ PH + RA+ I+ V +GK LV
Sbjct: 73 NKRLGSF--VNPANIPGLNTLGVGIARIDFAPGGLIPPHIHPRASEIILVIKGK---LLV 127
Query: 367 GQRNENQQEQREYEEDEQQQERSQQVQRYRARLSPGDVYVIPAG 410
G + N + + L PGDV+V P G
Sbjct: 128 GFVSSNDYNYTLFSK----------------ILYPGDVFVHPIG 155
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.133 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,441,694
Number of Sequences: 26719
Number of extensions: 412469
Number of successful extensions: 2782
Number of sequences better than 10.0: 132
Number of HSP's better than 10.0 without gapping: 39
Number of HSP's successfully gapped in prelim test: 93
Number of HSP's that attempted gapping in prelim test: 2475
Number of HSP's gapped (non-prelim): 305
length of query: 440
length of database: 11,318,596
effective HSP length: 102
effective length of query: 338
effective length of database: 8,593,258
effective search space: 2904521204
effective search space used: 2904521204
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Medicago: description of AC148289.2


