

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148289.12 - phase: 0
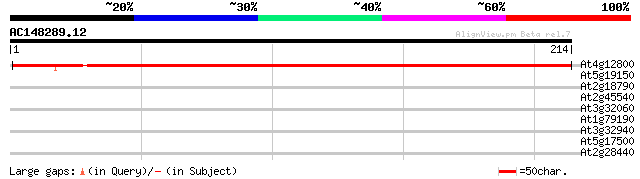
(214 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g12800 PSI-L protein (PsaL, PSI subunit XI) 355 8e-99
At5g19150 unknown protein 33 0.15
At2g18790 phytochrome B 29 2.1
At2g45540 unknown protein 28 2.8
At3g32060 hypothetical protein 28 3.6
At1g79190 hypothetical protein 28 3.6
At3g32940 hypothetical protein 28 4.7
At5g17500 putative protein 27 6.2
At2g28440 En/Spm-like transposon protein 27 8.1
>At4g12800 PSI-L protein (PsaL, PSI subunit XI)
Length = 219
Score = 355 bits (912), Expect = 8e-99
Identities = 174/218 (79%), Positives = 197/218 (89%), Gaps = 6/218 (2%)
Query: 2 AAASPMASQLKSSFT-----RPLVTPRGLCGSSPLHQLPSRRQFNFTVKAIQSEKPNYQV 56
A+ASPMASQL+SSF+ + L P+G+ G+ P P++R +FTV+A++S+K +QV
Sbjct: 3 ASASPMASQLRSSFSSASLSQRLAVPKGISGA-PFGVSPTKRVSSFTVRAVKSDKTTFQV 61
Query: 57 IQPINGDPFIGSLETPVTSSPLVAWYLSNLPGYRTAVNPLLRGIEVGLAHGFFLVGPFVK 116
+QPINGDPFIGSLETPVTSSPL+AWYLSNLPGYRTAVNPLLRG+EVGLAHGFFLVGPFVK
Sbjct: 62 VQPINGDPFIGSLETPVTSSPLIAWYLSNLPGYRTAVNPLLRGVEVGLAHGFFLVGPFVK 121
Query: 117 AGPLRNTEYAGSAGSLAAAGLIVILSICLTIYGVSSFNEGDPSIAPSLTLTGRKKQPDPL 176
AGPLRNT YAGSAGSLAAAGL+VILS+CLTIYG+SSF EG+PSIAPSLTLTGRKKQPD L
Sbjct: 122 AGPLRNTAYAGSAGSLAAAGLVVILSMCLTIYGISSFKEGEPSIAPSLTLTGRKKQPDQL 181
Query: 177 QTADGWSKFTGGFFFGGISGVTWAYFLLYVLDLPYFVK 214
QTADGW+KFTGGFFFGGISGVTWAYFLLYVLDLPYFVK
Sbjct: 182 QTADGWAKFTGGFFFGGISGVTWAYFLLYVLDLPYFVK 219
>At5g19150 unknown protein
Length = 365
Score = 32.7 bits (73), Expect = 0.15
Identities = 26/96 (27%), Positives = 47/96 (48%), Gaps = 8/96 (8%)
Query: 110 LVGPFVKAGPLRNTEYA-GSAGSLAAAGLIVILSICLT--IYGVSSFNEGDP-----SIA 161
LV P + +G +R T ++ S+ S+ ++ ++C + I +SS +E D ++
Sbjct: 2 LVKPSIISGLVRLTSHSPSSSSSVLRRQEFLVRTLCGSPIIRAMSSTSEADAESVLRTVT 61
Query: 162 PSLTLTGRKKQPDPLQTADGWSKFTGGFFFGGISGV 197
PSL L K Q + G ++TG +F IS +
Sbjct: 62 PSLDLKRHKGQAGKIAVIGGCREYTGAPYFAAISAL 97
>At2g18790 phytochrome B
Length = 1172
Score = 28.9 bits (63), Expect = 2.1
Identities = 23/84 (27%), Positives = 41/84 (48%), Gaps = 5/84 (5%)
Query: 63 DPFIGSLETPVTSSPLVAWYLSNLPGYRTAVN----PLLRGIEVGLAHGFFLVGPFVKAG 118
+P+IG L P T P + +L R V+ P+L + L LVG ++A
Sbjct: 296 EPYIG-LHYPATDIPQASRFLFKQNRVRMIVDCNATPVLVVQDDRLTQSMCLVGSTLRAP 354
Query: 119 PLRNTEYAGSAGSLAAAGLIVILS 142
+++Y + GS+A+ + VI++
Sbjct: 355 HGCHSQYMANMGSIASLAMAVIIN 378
>At2g45540 unknown protein
Length = 2946
Score = 28.5 bits (62), Expect = 2.8
Identities = 16/46 (34%), Positives = 22/46 (47%)
Query: 124 EYAGSAGSLAAAGLIVILSICLTIYGVSSFNEGDPSIAPSLTLTGR 169
E G A +V+ S+ T+ GV SF E + S PS+ L R
Sbjct: 189 ENFGCGEESEATAFLVVDSLIATMGGVESFEEDEDSNPPSVMLNSR 234
>At3g32060 hypothetical protein
Length = 487
Score = 28.1 bits (61), Expect = 3.6
Identities = 18/80 (22%), Positives = 36/80 (44%), Gaps = 9/80 (11%)
Query: 27 GSSPLHQLPSRRQFNFTVKAIQSE-------KPNYQVIQPINGDPFIGSLETPVTSSPLV 79
G + + + F + +K + E K N Q++ + D F+G +ETPV ++
Sbjct: 124 GGTVVDRAAFANSFGYVIKKLDKERYVLKKRKGNPQLVAAVLHDHFLGQVETPVPR--II 181
Query: 80 AWYLSNLPGYRTAVNPLLRG 99
+ G + + + +LRG
Sbjct: 182 MELVQTKLGVKVSYSTVLRG 201
>At1g79190 hypothetical protein
Length = 1280
Score = 28.1 bits (61), Expect = 3.6
Identities = 13/36 (36%), Positives = 20/36 (55%)
Query: 143 ICLTIYGVSSFNEGDPSIAPSLTLTGRKKQPDPLQT 178
+CL + +S E DP+ P+L L R+ P LQ+
Sbjct: 32 LCLELLNLSQNPEKDPTTIPALLLLLRRTPPSSLQS 67
>At3g32940 hypothetical protein
Length = 590
Score = 27.7 bits (60), Expect = 4.7
Identities = 23/81 (28%), Positives = 37/81 (45%), Gaps = 3/81 (3%)
Query: 10 QLKSSFTRPLVTPRGLCGSSPLHQLPSRRQFNFTVKAIQSEKPNYQVIQPIN--GDPFIG 67
Q + T P ++PR S+ L LP + +F +S+ + Q+ QP++ PF G
Sbjct: 462 QQSNIITTPHISPRPNSQSAHLPHLPFQASQHFGQNFTRSQHFDRQMDQPLSHPSAPFHG 521
Query: 68 SLETPVTSSPLVAWYLSNLPG 88
+ + V PL N PG
Sbjct: 522 NARSNV-GPPLSQMMPRNFPG 541
>At5g17500 putative protein
Length = 526
Score = 27.3 bits (59), Expect = 6.2
Identities = 24/78 (30%), Positives = 33/78 (41%), Gaps = 10/78 (12%)
Query: 103 GLAH---GFFLVGPFVKAGPLRNTEYAGSAGSLAAAGLIVILSICLTIYGVSSFNE---- 155
GL H G + P++ PL N + SL ++VIL T+ G N+
Sbjct: 110 GLDHELQGIYTHNPYIVNTPLINV-FQAVVYSLGRHDVMVILDNHKTVPGWCCSNDDPDA 168
Query: 156 --GDPSIAPSLTLTGRKK 171
GDP P L + G KK
Sbjct: 169 FFGDPKFNPDLWMLGLKK 186
>At2g28440 En/Spm-like transposon protein
Length = 268
Score = 26.9 bits (58), Expect = 8.1
Identities = 15/37 (40%), Positives = 20/37 (53%), Gaps = 2/37 (5%)
Query: 138 IVILSICLTIYGVSSFNEGDPSIAPSLTLTGRKKQPD 174
IV+LSICL I+ + E PS P+ GR+ D
Sbjct: 8 IVMLSICLLIFDFAGAQEESPS--PAAVSPGREPSTD 42
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.139 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,154,799
Number of Sequences: 26719
Number of extensions: 232109
Number of successful extensions: 442
Number of sequences better than 10.0: 9
Number of HSP's better than 10.0 without gapping: 3
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 438
Number of HSP's gapped (non-prelim): 9
length of query: 214
length of database: 11,318,596
effective HSP length: 95
effective length of query: 119
effective length of database: 8,780,291
effective search space: 1044854629
effective search space used: 1044854629
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Medicago: description of AC148289.12


