

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148289.1 + phase: 0
(340 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
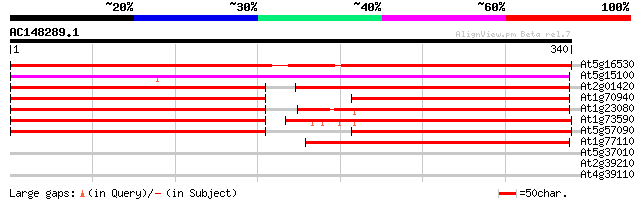
At5g16530 auxin efflux carrier -like protein 465 e-131
At5g15100 auxin transport protein - like 256 2e-68
At2g01420 auxin transporter splice variant b (PIN4) 178 3e-45
At1g70940 auxin transport like protein 177 7e-45
At1g23080 auxin transport protein (PIN7) 175 4e-44
At1g73590 auxin transporter splice variant b, putative 171 4e-43
At5g57090 root gravitropism control protein (PIN2) 171 5e-43
At1g77110 auxin transport protein (PIN6) 146 2e-35
At5g37010 serine-rich protein - like 29 4.2
At2g39210 nodulin-like protein 28 5.5
At4g39110 putative receptor-like protein kinase 28 9.4
>At5g16530 auxin efflux carrier -like protein
Length = 351
Score = 465 bits (1197), Expect = e-131
Identities = 235/340 (69%), Positives = 274/340 (80%), Gaps = 12/340 (3%)
Query: 1 MIGWEDVYKVIVAVVPLYFALILGYGSVRWWKIFTREQCDAINKLVCYFTLPLFAFEFTA 60
MI DVYKVI A+VPLY ALILGYGSV+WW IFTR+QCDAIN+LVCYFTLPLF EFTA
Sbjct: 1 MINCGDVYKVIEAMVPLYVALILGYGSVKWWHIFTRDQCDAINRLVCYFTLPLFTIEFTA 60
Query: 61 HIDPFKMNFLFIGADTLSKLIIVAVIALWAKCSSKVSYSWSITSFSLCTLTNSLVVGIPM 120
H+DPF MN+ FI AD LSK+IIV V+ALWAK S+K SY WSITSFSLCTLTNSLVVG+P+
Sbjct: 61 HVDPFNMNYRFIAADVLSKVIIVTVLALWAKYSNKGSYCWSITSFSLCTLTNSLVVGVPL 120
Query: 121 VKPMYGPMGVDLVVQASVVQAIIWLTLLLFVLEFRRTGIEGTITTLKPKASSISNVTCEG 180
K MYG VDLVVQ+SV QAI+WLTLLLFVLEFR+ G +++IS+V +
Sbjct: 121 AKAMYGQQAVDLVVQSSVFQAIVWLTLLLFVLEFRKAGF---------SSNNISDVQVDN 171
Query: 181 EESKDVEANNIVEYTSSRLPFLQLMKRVWLKLIANPNSYGCVIGISWAFISNRWNLELPS 240
+ + +V S FL++M VWLKL NPN Y C++GI+WAFISNRW+LELP
Sbjct: 172 INIESGKRETVVVGEKS---FLEVMSLVWLKLATNPNCYSCILGIAWAFISNRWHLELPG 228
Query: 241 MVEGSILIMSKAGTGTAMFSMGIFMALQEKVISCGPSLTVFGLVLKFIAGPAAMAISAFI 300
++EGSILIMSKAGTGTAMF+MGIFMALQEK+I CG SLTV G+VLKFIAGPAAMAI + +
Sbjct: 229 ILEGSILIMSKAGTGTAMFNMGIFMALQEKLIVCGTSLTVMGMVLKFIAGPAAMAIGSIV 288
Query: 301 VGLRGDVLRIAIIQAAVPQSITSFIFAKEYELHAEVLSTA 340
+GL GDVLR+AIIQAA+PQSITSFIFAKEY LHA+VLSTA
Sbjct: 289 LGLHGDVLRVAIIQAALPQSITSFIFAKEYGLHADVLSTA 328
>At5g15100 auxin transport protein - like
Length = 367
Score = 256 bits (653), Expect = 2e-68
Identities = 126/345 (36%), Positives = 205/345 (58%), Gaps = 6/345 (1%)
Query: 1 MIGWEDVYKVIVAVVPLYFALILGYGSVRWWKIFTREQCDAINKLVCYFTLPLFAFEFTA 60
MI W D+Y V+ A VPLY ++ LG+ S R K+F+ EQC INK V F++PL +F+ +
Sbjct: 1 MISWLDIYHVVSATVPLYVSMTLGFLSARHLKLFSPEQCAGINKFVAKFSIPLLSFQIIS 60
Query: 61 HIDPFKMNFLFIGADTLSKLIIVAVIAL----WAKCSSKVS-YSWSITSFSLCTLTNSLV 115
+PFKM+ I +D L K ++V V+A+ W + W IT S+ L N+L+
Sbjct: 61 ENNPFKMSPKLILSDILQKFLVVVVLAMVLRFWHPTGGRGGKLGWVITGLSISVLPNTLI 120
Query: 116 VGIPMVKPMYGPMGVDLVVQASVVQAIIWLTLLLFVLEFRRT-GIEGTITTLKPKASSIS 174
+G+P++ +YG ++ Q V+Q++IW T+LLF+ E + + +L+ +
Sbjct: 121 LGMPILSAIYGDEAASILEQIVVLQSLIWYTILLFLFELNAARALPSSGASLEHTGNDQE 180
Query: 175 NVTCEGEESKDVEANNIVEYTSSRLPFLQLMKRVWLKLIANPNSYGCVIGISWAFISNRW 234
E E ++ + + + + ++++ + W KLI NPN+Y +IGI WA + R
Sbjct: 181 EANIEDEPKEEEDEEEVAIVRTRSVGTMKILLKAWRKLIINPNTYATLIGIIWATLHFRL 240
Query: 235 NLELPSMVEGSILIMSKAGTGTAMFSMGIFMALQEKVISCGPSLTVFGLVLKFIAGPAAM 294
LP M++ SI ++S G G AMFS+G+FMA Q +I+CG + + ++LKF+ GPA M
Sbjct: 241 GWNLPEMIDKSIHLLSDGGLGMAMFSLGLFMASQSSIIACGTKMAIITMLLKFVLGPALM 300
Query: 295 AISAFIVGLRGDVLRIAIIQAAVPQSITSFIFAKEYELHAEVLST 339
SA+ + L+ + ++AI+QAA+PQ + F+FAKEY LH E++ST
Sbjct: 301 IASAYCIRLKSTLFKVAILQAALPQGVVPFVFAKEYNLHPEIIST 345
>At2g01420 auxin transporter splice variant b (PIN4)
Length = 612
Score = 178 bits (452), Expect = 3e-45
Identities = 83/155 (53%), Positives = 109/155 (69%)
Query: 1 MIGWEDVYKVIVAVVPLYFALILGYGSVRWWKIFTREQCDAINKLVCYFTLPLFAFEFTA 60
MI W D+Y V+ AVVPLY A+IL YGSV+WWKIF+ +QC IN+ V F +PL +F F +
Sbjct: 1 MITWHDLYTVLTAVVPLYVAMILAYGSVQWWKIFSPDQCSGINRFVAIFAVPLLSFHFIS 60
Query: 61 HIDPFKMNFLFIGADTLSKLIIVAVIALWAKCSSKVSYSWSITSFSLCTLTNSLVVGIPM 120
DP+ MNF F+ ADTL K+I++ ++ALWA + S W IT FSL TL N+LV+GIP+
Sbjct: 61 TNDPYAMNFRFVAADTLQKIIMLVLLALWANLTKNGSLEWMITIFSLSTLPNTLVMGIPL 120
Query: 121 VKPMYGPMGVDLVVQASVVQAIIWLTLLLFVLEFR 155
+ MYG L+VQ V+Q IIW TLLLF+ E+R
Sbjct: 121 LIAMYGTYAGSLMVQVVVLQCIIWYTLLLFLFEYR 155
Score = 162 bits (410), Expect = 2e-40
Identities = 79/167 (47%), Positives = 113/167 (67%), Gaps = 1/167 (0%)
Query: 174 SNVTCEGEESK-DVEANNIVEYTSSRLPFLQLMKRVWLKLIANPNSYGCVIGISWAFISN 232
SN T E E + D NN + + ++ VW KLI NPN+Y +IG+ WA ++
Sbjct: 424 SNSTAELEAAGGDGGGNNGTHMPPTSVMTRLILIMVWRKLIRNPNTYSSLIGLIWALVAY 483
Query: 233 RWNLELPSMVEGSILIMSKAGTGTAMFSMGIFMALQEKVISCGPSLTVFGLVLKFIAGPA 292
RW++ +P +++ SI I+S AG G AMFS+G+FMALQ K+I+CG S+ F + ++FI GPA
Sbjct: 484 RWHVAMPKILQQSISILSDAGLGMAMFSLGLFMALQPKIIACGNSVATFAMAVRFITGPA 543
Query: 293 AMAISAFIVGLRGDVLRIAIIQAAVPQSITSFIFAKEYELHAEVLST 339
MA++ +GL GD+LRIAI+QAA+PQ I F+FAKEY +H +LST
Sbjct: 544 IMAVAGIAIGLHGDLLRIAIVQAALPQGIVPFVFAKEYNVHPTILST 590
>At1g70940 auxin transport like protein
Length = 640
Score = 177 bits (449), Expect = 7e-45
Identities = 83/155 (53%), Positives = 109/155 (69%)
Query: 1 MIGWEDVYKVIVAVVPLYFALILGYGSVRWWKIFTREQCDAINKLVCYFTLPLFAFEFTA 60
MI W D+Y V+ AV+PLY A+IL YGSVRWWKIF+ +QC IN+ V F +PL +F F +
Sbjct: 1 MISWHDLYTVLTAVIPLYVAMILAYGSVRWWKIFSPDQCSGINRFVAIFAVPLLSFHFIS 60
Query: 61 HIDPFKMNFLFIGADTLSKLIIVAVIALWAKCSSKVSYSWSITSFSLCTLTNSLVVGIPM 120
+P+ MN FI ADTL K+I+++++ LWA + S WSIT FSL TL N+LV+GIP+
Sbjct: 61 TNNPYAMNLRFIAADTLQKIIMLSLLVLWANFTRSGSLEWSITIFSLSTLPNTLVMGIPL 120
Query: 121 VKPMYGPMGVDLVVQASVVQAIIWLTLLLFVLEFR 155
+ MYG L+VQ V+Q IIW TLLLF+ EFR
Sbjct: 121 LIAMYGEYSGSLMVQIVVLQCIIWYTLLLFLFEFR 155
Score = 163 bits (412), Expect = 1e-40
Identities = 71/132 (53%), Positives = 102/132 (76%)
Query: 208 VWLKLIANPNSYGCVIGISWAFISNRWNLELPSMVEGSILIMSKAGTGTAMFSMGIFMAL 267
VW KLI NPN+Y +IG+ WA ++ RW++ +P +++ SI I+S AG G AMFS+G+FMAL
Sbjct: 487 VWRKLIRNPNTYSSLIGLIWALVAFRWHVAMPKIIQQSISILSDAGLGMAMFSLGLFMAL 546
Query: 268 QEKVISCGPSLTVFGLVLKFIAGPAAMAISAFIVGLRGDVLRIAIIQAAVPQSITSFIFA 327
Q K+I+CG S+ F + ++F+ GPA MA++A +GLRGD+LR+AI+QAA+PQ I F+FA
Sbjct: 547 QPKLIACGNSVATFAMAVRFLTGPAVMAVAAIAIGLRGDLLRVAIVQAALPQGIVPFVFA 606
Query: 328 KEYELHAEVLST 339
KEY +H +LST
Sbjct: 607 KEYNVHPAILST 618
>At1g23080 auxin transport protein (PIN7)
Length = 619
Score = 175 bits (443), Expect = 4e-44
Identities = 82/155 (52%), Positives = 108/155 (68%)
Query: 1 MIGWEDVYKVIVAVVPLYFALILGYGSVRWWKIFTREQCDAINKLVCYFTLPLFAFEFTA 60
MI W D+Y V+ AV+PLY A+IL YGSVRWWKIF+ +QC IN+ V F +PL +F F +
Sbjct: 1 MITWHDLYTVLTAVIPLYVAMILAYGSVRWWKIFSPDQCSGINRFVAIFAVPLLSFHFIS 60
Query: 61 HIDPFKMNFLFIGADTLSKLIIVAVIALWAKCSSKVSYSWSITSFSLCTLTNSLVVGIPM 120
+P+ MN FI ADTL KLI++ ++ +WA + S WSIT FSL TL N+LV+GIP+
Sbjct: 61 SNNPYAMNLRFIAADTLQKLIMLTLLIIWANFTRSGSLEWSITIFSLSTLPNTLVMGIPL 120
Query: 121 VKPMYGPMGVDLVVQASVVQAIIWLTLLLFVLEFR 155
+ MYG L+VQ V+Q IIW TLLLF+ E+R
Sbjct: 121 LIAMYGEYSGSLMVQIVVLQCIIWYTLLLFLFEYR 155
Score = 162 bits (410), Expect = 2e-40
Identities = 79/170 (46%), Positives = 112/170 (65%), Gaps = 7/170 (4%)
Query: 175 NVTCEGEESKDVEANNIVEYTSSRLPFLQLMKR-----VWLKLIANPNSYGCVIGISWAF 229
N T E + +E V +P +M R VW KLI NPN+Y +IG+ WA
Sbjct: 430 NSTAELNPKEAIETGETVPV--KHMPPASVMTRLILIMVWRKLIRNPNTYSSLIGLIWAL 487
Query: 230 ISNRWNLELPSMVEGSILIMSKAGTGTAMFSMGIFMALQEKVISCGPSLTVFGLVLKFIA 289
++ RW++ +P +++ SI I+S AG G AMFS+G+FMALQ K+I+CG S F + ++F
Sbjct: 488 VAFRWDVAMPKIIQQSISILSDAGLGMAMFSLGLFMALQPKLIACGNSTATFAMAVRFFT 547
Query: 290 GPAAMAISAFIVGLRGDVLRIAIIQAAVPQSITSFIFAKEYELHAEVLST 339
GPA MA++A +GLRGD+LR+AI+QAA+PQ I F+FAKEY +H +LST
Sbjct: 548 GPAVMAVAAMAIGLRGDLLRVAIVQAALPQGIVPFVFAKEYNVHPAILST 597
>At1g73590 auxin transporter splice variant b, putative
Length = 622
Score = 171 bits (434), Expect = 4e-43
Identities = 81/155 (52%), Positives = 109/155 (70%)
Query: 1 MIGWEDVYKVIVAVVPLYFALILGYGSVRWWKIFTREQCDAINKLVCYFTLPLFAFEFTA 60
MI D Y V+ A+VPLY A+IL YGSV+WWKIFT +QC IN+ V F +PL +F F A
Sbjct: 1 MITAADFYHVMTAMVPLYVAMILAYGSVKWWKIFTPDQCSGINRFVALFAVPLLSFHFIA 60
Query: 61 HIDPFKMNFLFIGADTLSKLIIVAVIALWAKCSSKVSYSWSITSFSLCTLTNSLVVGIPM 120
+P+ MN F+ AD+L K+I+++++ LW K S S W+IT FSL TL N+LV+GIP+
Sbjct: 61 ANNPYAMNLRFLAADSLQKVIVLSLLFLWCKLSRNGSLDWTITLFSLSTLPNTLVMGIPL 120
Query: 121 VKPMYGPMGVDLVVQASVVQAIIWLTLLLFVLEFR 155
+K MYG DL+VQ V+Q IIW TL+LF+ E+R
Sbjct: 121 LKGMYGNFSGDLMVQIVVLQCIIWYTLMLFLFEYR 155
Score = 160 bits (404), Expect = 1e-39
Identities = 85/192 (44%), Positives = 122/192 (63%), Gaps = 19/192 (9%)
Query: 168 PKASSISNVTCEGEE----SKDVEA--------NNIVEYTSSR--LPFLQLMKR-----V 208
P+ +S N E EE +KD ++ NNI T+ +P +M R V
Sbjct: 410 PQGNSNDNQYVEREEFSFGNKDDDSKVLATDGGNNISNKTTQAKVMPPTSVMTRLILIMV 469
Query: 209 WLKLIANPNSYGCVIGISWAFISNRWNLELPSMVEGSILIMSKAGTGTAMFSMGIFMALQ 268
W KLI NPNSY + GI+W+ IS +WN+E+P+++ SI I+S AG G AMFS+G+FMAL
Sbjct: 470 WRKLIRNPNSYSSLFGITWSLISFKWNIEMPALIAKSISILSDAGLGMAMFSLGLFMALN 529
Query: 269 EKVISCGPSLTVFGLVLKFIAGPAAMAISAFIVGLRGDVLRIAIIQAAVPQSITSFIFAK 328
++I+CG F ++F+ GPA M ++++ VGLRG +L +AIIQAA+PQ I F+FAK
Sbjct: 530 PRIIACGNRRAAFAAAMRFVVGPAVMLVASYAVGLRGVLLHVAIIQAALPQGIVPFVFAK 589
Query: 329 EYELHAEVLSTA 340
EY +H ++LSTA
Sbjct: 590 EYNVHPDILSTA 601
>At5g57090 root gravitropism control protein (PIN2)
Length = 647
Score = 171 bits (433), Expect = 5e-43
Identities = 80/155 (51%), Positives = 110/155 (70%)
Query: 1 MIGWEDVYKVIVAVVPLYFALILGYGSVRWWKIFTREQCDAINKLVCYFTLPLFAFEFTA 60
MI +D+Y V+ A+VPLY A+IL YGSVRWW IFT +QC IN+ V F +PL +F F +
Sbjct: 1 MITGKDMYDVLAAMVPLYVAMILAYGSVRWWGIFTPDQCSGINRFVAVFAVPLLSFHFIS 60
Query: 61 HIDPFKMNFLFIGADTLSKLIIVAVIALWAKCSSKVSYSWSITSFSLCTLTNSLVVGIPM 120
DP+ MN+ F+ AD+L K++I+A + LW S + S W IT FSL TL N+LV+GIP+
Sbjct: 61 SNDPYAMNYHFLAADSLQKVVILAALFLWQAFSRRGSLEWMITLFSLSTLPNTLVMGIPL 120
Query: 121 VKPMYGPMGVDLVVQASVVQAIIWLTLLLFVLEFR 155
++ MYG +L+VQ V+Q+IIW TL+LF+ EFR
Sbjct: 121 LRAMYGDFSGNLMVQIVVLQSIIWYTLMLFLFEFR 155
Score = 162 bits (411), Expect = 2e-40
Identities = 69/133 (51%), Positives = 104/133 (77%)
Query: 208 VWLKLIANPNSYGCVIGISWAFISNRWNLELPSMVEGSILIMSKAGTGTAMFSMGIFMAL 267
VW KLI NPN+Y + G++W+ +S +WN+++P+++ GSI I+S AG G AMFS+G+FMAL
Sbjct: 494 VWRKLIRNPNTYSSLFGLAWSLVSFKWNIKMPTIMSGSISILSDAGLGMAMFSLGLFMAL 553
Query: 268 QEKVISCGPSLTVFGLVLKFIAGPAAMAISAFIVGLRGDVLRIAIIQAAVPQSITSFIFA 327
Q K+I+CG S+ F + ++F+ GPA +A ++ +G+RGD+L IAI+QAA+PQ I F+FA
Sbjct: 554 QPKIIACGKSVAGFAMAVRFLTGPAVIAATSIAIGIRGDLLHIAIVQAALPQGIVPFVFA 613
Query: 328 KEYELHAEVLSTA 340
KEY +H ++LSTA
Sbjct: 614 KEYNVHPDILSTA 626
>At1g77110 auxin transport protein (PIN6)
Length = 429
Score = 146 bits (368), Expect = 2e-35
Identities = 72/160 (45%), Positives = 108/160 (67%)
Query: 180 GEESKDVEANNIVEYTSSRLPFLQLMKRVWLKLIANPNSYGCVIGISWAFISNRWNLELP 239
G+++ V A E S+ + ++ V KL NPN+Y ++G+ W+ IS +WN+ +P
Sbjct: 248 GKDTTPVAAIGKQEMPSAIVMMRLILTVVGRKLSRNPNTYSSLLGLVWSLISFKWNIPMP 307
Query: 240 SMVEGSILIMSKAGTGTAMFSMGIFMALQEKVISCGPSLTVFGLVLKFIAGPAAMAISAF 299
++V+ SI I+S AG G AMFS+G+FMALQ K+I CG G++++FI+GP MA ++
Sbjct: 308 NIVDFSIKIISDAGLGMAMFSLGLFMALQPKMIPCGAKKATMGMLIRFISGPLFMAGASL 367
Query: 300 IVGLRGDVLRIAIIQAAVPQSITSFIFAKEYELHAEVLST 339
+VGLRG L AI+QAA+PQ I F+FA+EY LH ++LST
Sbjct: 368 LVGLRGSRLHAAIVQAALPQGIVPFVFAREYNLHPDLLST 407
>At5g37010 serine-rich protein - like
Length = 637
Score = 28.9 bits (63), Expect = 4.2
Identities = 14/44 (31%), Positives = 26/44 (58%)
Query: 149 LFVLEFRRTGIEGTITTLKPKASSISNVTCEGEESKDVEANNIV 192
+FV++ RR+ + + TT P+ SS+S+V G V+ + I+
Sbjct: 73 VFVIKHRRSHEKSSKTTTDPEDSSVSDVKSTGSNHNPVDVDAIL 116
>At2g39210 nodulin-like protein
Length = 601
Score = 28.5 bits (62), Expect = 5.5
Identities = 25/102 (24%), Positives = 45/102 (43%), Gaps = 14/102 (13%)
Query: 95 KVSYSWSITSFSLCTLTNSLVVGIPMVKPMYGPMGVDLVVQASVVQAIIWLTLLLFVLEF 154
KV Y++ S L T +++ + + G + A+VV ++ L +++ +LE
Sbjct: 214 KVFYNFLYISLGLATFLMVVII----INKLSGFTQSEFGGSAAVVIVLLLLPIIVVILEE 269
Query: 155 RRTGIEG----------TITTLKPKASSISNVTCEGEESKDV 186
++ E + T KPK S +GEESK+V
Sbjct: 270 KKLWKEKQVALNDPAPINVVTEKPKLDSSEFKDDDGEESKEV 311
>At4g39110 putative receptor-like protein kinase
Length = 878
Score = 27.7 bits (60), Expect = 9.4
Identities = 12/30 (40%), Positives = 20/30 (66%)
Query: 273 SCGPSLTVFGLVLKFIAGPAAMAISAFIVG 302
S PS+ + +L F++GP+A A++A VG
Sbjct: 18 SSKPSMALLLAILLFLSGPSASAVAAAAVG 47
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.326 0.139 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,953,534
Number of Sequences: 26719
Number of extensions: 267569
Number of successful extensions: 861
Number of sequences better than 10.0: 11
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 1
Number of HSP's that attempted gapping in prelim test: 841
Number of HSP's gapped (non-prelim): 17
length of query: 340
length of database: 11,318,596
effective HSP length: 100
effective length of query: 240
effective length of database: 8,646,696
effective search space: 2075207040
effective search space used: 2075207040
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC148289.1


