

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148236.5 - phase: 0 /pseudo
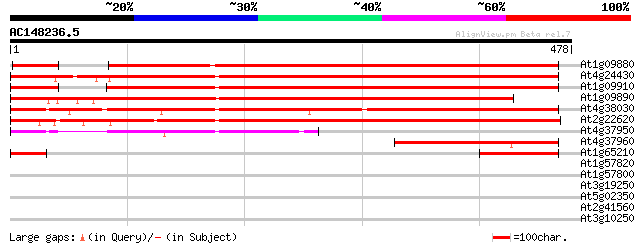
(478 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g09880 hypothetical protein 561 e-160
At4g24430 LG27/30-like gene 551 e-157
At1g09910 hypothetical protein 527 e-150
At1g09890 hypothetical protein 503 e-143
At4g38030 LG127/30 like gene 489 e-138
At2g22620 unknown protein 475 e-134
At4g37950 hypothetical protein 211 7e-55
At4g37960 putative protein 204 7e-53
At1g65210 hypothetical protein 103 3e-22
At1g57820 putative transcription factor protein (At1g57820) 33 0.45
At1g57800 hypothetical protein 31 1.7
At3g19250 hypothetical protein 30 2.2
At5g02350 putative protein 29 5.0
At2g41560 putative Ca2+-ATPase 29 5.0
At3g10250 unknown protein 29 6.5
>At1g09880 hypothetical protein
Length = 631
Score = 561 bits (1447), Expect = e-160
Identities = 258/383 (67%), Positives = 311/383 (80%), Gaps = 3/383 (0%)
Query: 85 WPAFDLDNTRVAFKLRKDKFNYMAIADNRQRFMPLPDDRLPPRGQVLAYPEAVRLVNPVE 144
WPA +LDN R+ FKL K KF+YMAI+D+RQR+MP+PDDR+PPRGQ LAYPEAV+L++P+E
Sbjct: 123 WPAVELDNMRLVFKLNKKKFHYMAISDDRQRYMPVPDDRVPPRGQPLAYPEAVQLLDPIE 182
Query: 145 PEFKGEVDDKYEYACESRSNQVHGWISIDSSSDSTGCWLITPSYEFRSAGPLKQYLGSHL 204
PEFKGEVDDKYEY+ ES+ +VHGWIS ++DS G W ITPS EFRSAGPLKQ+LGSH+
Sbjct: 183 PEFKGEVDDKYEYSMESKDIKVHGWIS---TNDSVGFWQITPSNEFRSAGPLKQFLGSHV 239
Query: 205 GPTMLSVFHSTHYSGADLIMKFGENEPWKKVFGPIFIYLNSNSDGFSPIKLWEDAKQQMV 264
GPT L+VFHSTHY GADLIM F E WKKVFGP+FIYLNS G P+ LW +AK Q
Sbjct: 240 GPTNLAVFHSTHYVGADLIMSFKNGEAWKKVFGPVFIYLNSFPKGVDPLLLWHEAKNQTK 299
Query: 265 NEVESWPYTFPASEDFLSSAQRGKFEGRLLVRDRYIRDAFVPVSGAYVGLAAPGDVGSWQ 324
E E WPY F AS+DF +S QRG GRLLVRDR+I +P +G+YVGLAAPGDVGSWQ
Sbjct: 300 IEEEKWPYNFTASDDFPASDQRGSVSGRLLVRDRFISSEDIPANGSYVGLAAPGDVGSWQ 359
Query: 325 REYKGYQFWTITDDKGYFSIINVLPGDYNLYSWVNGFIGDYQCNNIINITSGSEINVGEL 384
RE KGYQFW+ D+ G FSI NV G YNLY++ GFIGDY + + +I+ GS+I++G+L
Sbjct: 360 RECKGYQFWSKADENGSFSINNVRSGRYNLYAFAPGFIGDYHNDTVFDISPGSKISLGDL 419
Query: 385 VYEPPRHGPTLWEIGIPDRSAAEFYVPDPNPLYVNRLYVNHSDRFRQYGLWERYADLYPN 444
VYEPPR G TLWEIG+PDRSAAEFY+PDPNP +VN+LY+NHSD++RQYGLWERY++LYP+
Sbjct: 420 VYEPPRDGSTLWEIGVPDRSAAEFYIPDPNPSFVNKLYLNHSDKYRQYGLWERYSELYPD 479
Query: 445 EDLVYTVGISDYKKDWFFAQVTR 467
ED+VY V I DY K+WFF QVTR
Sbjct: 480 EDMVYNVDIDDYSKNWFFMQVTR 502
Score = 48.9 bits (115), Expect = 6e-06
Identities = 23/40 (57%), Positives = 30/40 (74%), Gaps = 1/40 (2%)
Query: 3 IDNDIVQVNISNPEGCVTGIQYNGLDNLLE-INNHESDRG 41
++N +Q+ +SNPEG VTGIQYNG+DN+L N E DRG
Sbjct: 1 MENRFLQLTLSNPEGFVTGIQYNGIDNVLAYYTNKEYDRG 40
>At4g24430 LG27/30-like gene
Length = 646
Score = 551 bits (1421), Expect = e-157
Identities = 284/509 (55%), Positives = 343/509 (66%), Gaps = 48/509 (9%)
Query: 1 VVIDNDIVQVNISNPEGCVTGIQYNGLDNLLEINNHE----------SDRG*KAGNDKYE 50
VV+ N V+V IS P+G VTGI Y G+DNLLE +N + SD G K E
Sbjct: 16 VVMGNGKVKVTISKPDGFVTGISYQGVDNLLETHNEDFNRGYWDLVWSDEGTPGTTGKSE 75
Query: 51 GDNGK*RTSRAIFH*NMEFIS*-------------------------RKACATQY**K-- 83
G TS + N E + RK Y
Sbjct: 76 RIKG---TSFEVVVENEELVEISFSRKWDSSLQDSIAPINVDKRFIMRKDVTGFYSYAIF 132
Query: 84 ----EWPAFDLDNTRVAFKLRKDKFNYMAIADNRQRFMPLPDDRLPPRGQVLAYPEAVRL 139
EWPAF+L TR+ +KLRKDKF YMAIADNRQR MPLP+DRL RG+ LAYPEAV L
Sbjct: 133 EHLAEWPAFNLPQTRIVYKLRKDKFKYMAIADNRQRKMPLPEDRLGKRGRPLAYPEAVLL 192
Query: 140 VNPVEPEFKGEVDDKYEYACESRSNQVHGWISIDSSSDSTGCWLITPSYEFRSAGPLKQY 199
V+PVE EFKGEVDDKYEY+ E++ +VHGWIS + GCW I PS EFRS G KQ
Sbjct: 193 VHPVEDEFKGEVDDKYEYSSENKDLKVHGWISHNLD---LGCWQIIPSNEFRSGGLSKQN 249
Query: 200 LGSHLGPTMLSVFHSTHYSGADLIMKFGENEPWKKVFGPIFIYLNSNSDGFS-PIKLWED 258
L SH+GP L++F S HY+G D++MK + WKKVFGP+F YLN D S P+ LW+D
Sbjct: 250 LTSHVGPISLAMFLSAHYAGEDMVMKVKAGDSWKKVFGPVFTYLNCLPDKTSDPLSLWQD 309
Query: 259 AKQQMVNEVESWPYTFPASEDFLSSAQRGKFEGRLLVRDRYIRDAFVPVSGAYVGLAAPG 318
AK QM+ EV+SWPY FPASEDF S +RG GRLLV D+++ D F+P +GA+VGLA PG
Sbjct: 310 AKNQMLTEVQSWPYDFPASEDFPVSDKRGCISGRLLVCDKFLSDDFLPANGAFVGLAPPG 369
Query: 319 DVGSWQREYKGYQFWTITDDKGYFSIINVLPGDYNLYSWVNGFIGDYQCNNIINITSGSE 378
+VGSWQ E KGYQFWT D GYF+I ++ G+YNL +V G+IGDYQ +INIT+G +
Sbjct: 370 EVGSWQLESKGYQFWTEADSDGYFAINDIREGEYNLNGYVTGWIGDYQYEQLINITAGCD 429
Query: 379 INVGELVYEPPRHGPTLWEIGIPDRSAAEFYVPDPNPLYVNRLYVNHSDRFRQYGLWERY 438
I+VG +VYEPPR GPT+WEIGIPDRSAAEF+VPDPNP Y+N+LY+ H DRFRQYGLWERY
Sbjct: 430 IDVGNIVYEPPRDGPTVWEIGIPDRSAAEFFVPDPNPKYINKLYIGHPDRFRQYGLWERY 489
Query: 439 ADLYPNEDLVYTVGISDYKKDWFFAQVTR 467
+LYP EDLV+T+G+SDYKKDWFFA VTR
Sbjct: 490 TELYPKEDLVFTIGVSDYKKDWFFAHVTR 518
>At1g09910 hypothetical protein
Length = 675
Score = 527 bits (1358), Expect = e-150
Identities = 238/385 (61%), Positives = 295/385 (75%), Gaps = 3/385 (0%)
Query: 83 KEWPAFDLDNTRVAFKLRKDKFNYMAIADNRQRFMPLPDDRLPPRGQVLAYPEAVRLVNP 142
K+WP F+L TR+AFKLRKDKF+YMA+AD+R+R MP PDD R Q L Y EA L P
Sbjct: 175 KDWPGFELGETRIAFKLRKDKFHYMAVADDRKRIMPFPDDLCKGRCQTLDYQEASLLTAP 234
Query: 143 VEPEFKGEVDDKYEYACESRSNQVHGWISIDSSSDSTGCWLITPSYEFRSAGPLKQYLGS 202
+P +GEVDDKY+Y+CE++ +VHGWIS D G W ITPS EFRS GPLKQ L S
Sbjct: 235 CDPRLQGEVDDKYQYSCENKDLRVHGWISFDPP---VGFWQITPSNEFRSGGPLKQNLTS 291
Query: 203 HLGPTMLSVFHSTHYSGADLIMKFGENEPWKKVFGPIFIYLNSNSDGFSPIKLWEDAKQQ 262
H+GPT L+VFHSTHY+G ++ +F EPWKKV+GP+FIYLNS ++G P+ LW+DAK +
Sbjct: 292 HVGPTTLAVFHSTHYAGKTMMPRFEHGEPWKKVYGPVFIYLNSTANGDDPLCLWDDAKIK 351
Query: 263 MVNEVESWPYTFPASEDFLSSAQRGKFEGRLLVRDRYIRDAFVPVSGAYVGLAAPGDVGS 322
M+ EVE WPY+F AS+D+ S +RG GRLL+RDR+I + + GAYVGLA PGD GS
Sbjct: 352 MMAEVERWPYSFVASDDYPKSEERGTARGRLLIRDRFINNDLISARGAYVGLAPPGDSGS 411
Query: 323 WQREYKGYQFWTITDDKGYFSIINVLPGDYNLYSWVNGFIGDYQCNNIINITSGSEINVG 382
WQ E KGYQFW I D+ GYFSI NV PG+YNLY+WV FIGDY I+ +TSG I +G
Sbjct: 412 WQIECKGYQFWAIADEAGYFSIGNVRPGEYNLYAWVPSFIGDYHNGTIVRVTSGCMIEMG 471
Query: 383 ELVYEPPRHGPTLWEIGIPDRSAAEFYVPDPNPLYVNRLYVNHSDRFRQYGLWERYADLY 442
++VYEPPR GPTLWEIGIPDR A+EF++PDP+P VNR+ V+H DRFRQYGLW++Y D+Y
Sbjct: 472 DIVYEPPRDGPTLWEIGIPDRKASEFFIPDPDPTLVNRVLVHHQDRFRQYGLWKKYTDMY 531
Query: 443 PNEDLVYTVGISDYKKDWFFAQVTR 467
PN+DLVYTVG+SDY++DWFFA V R
Sbjct: 532 PNDDLVYTVGVSDYRRDWFFAHVPR 556
Score = 58.2 bits (139), Expect = 1e-08
Identities = 24/41 (58%), Positives = 34/41 (82%)
Query: 1 VVIDNDIVQVNISNPEGCVTGIQYNGLDNLLEINNHESDRG 41
VV+DN I+QV +S P G +TGI+YNG+DN+LE+ N E++RG
Sbjct: 58 VVMDNGILQVTLSKPGGIITGIEYNGIDNVLEVRNKETNRG 98
>At1g09890 hypothetical protein
Length = 477
Score = 503 bits (1296), Expect = e-143
Identities = 256/465 (55%), Positives = 320/465 (68%), Gaps = 38/465 (8%)
Query: 1 VVIDNDIVQVNISNPEGCVTGIQYNGLDNLL-----EINNHESD-----RG*KAGNDKYE 50
VV+DN I +V +S P+G VTGI+YNG+DNLL E+N D G G D +
Sbjct: 15 VVMDNGIARVTLSKPDGIVTGIEYNGIDNLLEVLNEEVNRGYWDLVWGGSGTAGGFDVIK 74
Query: 51 GDNGK*-------------------RTSRAI-FH*NMEFI-----S*RKACATQY**KEW 85
G N + + +A+ + + F+ S A KEW
Sbjct: 75 GSNFEVIVKNEEQIELSFTRKWDPSQEGKAVPLNIDKRFVMLSGSSGFYTYAIYEHLKEW 134
Query: 86 PAFDLDNTRVAFKLRKDKFNYMAIADNRQRFMPLPDDRLPPRGQVLAYPEAVRLVNPVEP 145
PAF L TR+AFKLRK+KF+YMA+ D+RQRFMPLPDDRLP RGQ LAYPEAV LVNP+E
Sbjct: 135 PAFSLAETRIAFKLRKEKFHYMAVTDDRQRFMPLPDDRLPDRGQALAYPEAVLLVNPLES 194
Query: 146 EFKGEVDDKYEYACESRSNQVHGWISIDSSSDSTGCWLITPSYEFRSAGPLKQYLGSHLG 205
+FKGEVDDKY+Y+CE++ VHGWI + S G WLITPS+E+R+ GP KQ L SH+G
Sbjct: 195 QFKGEVDDKYQYSCENKDITVHGWICTEQPS--VGFWLITPSHEYRTGGPQKQNLTSHVG 252
Query: 206 PTMLSVFHSTHYSGADLIMKFGENEPWKKVFGPIFIYLNSNSDGFS-PIKLWEDAKQQMV 264
PT L+VF S HY+G DL+ KF E E WKKVFGP+F+YLNS++D + P+ LW+DAK QM
Sbjct: 253 PTALAVFISAHYTGEDLVPKFSEGEAWKKVFGPVFVYLNSSTDDDNDPLWLWQDAKSQMN 312
Query: 265 NEVESWPYTFPASEDFLSSAQRGKFEGRLLVRDRYIRDAFVPVSGAYVGLAAPGDVGSWQ 324
E ESWPY+FPAS+D++ + QRG GRLLV+DRY+ F+ + YVGLA PG GSWQ
Sbjct: 313 VEAESWPYSFPASDDYVKTEQRGNVVGRLLVQDRYVDKDFIAANRGYVGLAVPGAAGSWQ 372
Query: 325 REYKGYQFWTITDDKGYFSIINVLPGDYNLYSWVNGFIGDYQCNNIINITSGSEINVGEL 384
RE K YQFWT TD++G+F I + PG YNLY+W+ GFIGDY+ +++I ITSG I V +L
Sbjct: 373 RECKEYQFWTRTDEEGFFYISGIRPGQYNLYAWIPGFIGDYKYDDVITITSGCYIYVEDL 432
Query: 385 VYEPPRHGPTLWEIGIPDRSAAEFYVPDPNPLYVNRLYVNHSDRF 429
VY+PPR+G TLWEIG PDRSAAEFYVPDPNP Y+N LY NH DRF
Sbjct: 433 VYQPPRNGATLWEIGFPDRSAAEFYVPDPNPKYINNLYQNHPDRF 477
>At4g38030 LG127/30 like gene
Length = 649
Score = 489 bits (1259), Expect = e-138
Identities = 246/488 (50%), Positives = 323/488 (65%), Gaps = 33/488 (6%)
Query: 1 VVIDNDIVQVNISNPEGCVTGIQYNGLDNLLEINNHESDRG*KAGNDKY----------- 49
V++DN I++V+ SNP+G +TGI+Y G+DN+L + H DRG + N +
Sbjct: 54 VIVDNGIIRVSFSNPQGLITGIKYKGIDNVL--HPHLRDRGIEGTNFRIITQTQEQVEIS 111
Query: 50 ------EGDNGK*RTSRAIFH*NMEFIS*RKACATQY**KEWPAFDLDNTRVAFKLRKDK 103
+G R I N I EWP D+ R+AFKL ++
Sbjct: 112 FSRTWEDGHIPLNVDKRYIIRRNTSGIYMYGIFERL---PEWPELDMGLIRIAFKLNPER 168
Query: 104 FNYMAIADNRQRFMPLPDDRLPPRG--QVLAYPEAVRLVNPVEPEFKGEVDDKYEYACES 161
F+YMA+ADNRQR MP DDR RG + L Y EAV+L +P FK +VDDKY+Y CE
Sbjct: 169 FHYMAVADNRQREMPTEDDRDIKRGHAKALGYKEAVQLTHPHNSMFKNQVDDKYQYTCEI 228
Query: 162 RSNQVHGWISIDSSSDSTGCWLITPSYEFRSAGPLKQYLGSHLGPTMLSVFHSTHYSGAD 221
+ N+VHGWIS S G W+I+PS E+RS GP+KQ L SH+GPT ++ F S HY GAD
Sbjct: 229 KDNKVHGWISTKSR---VGFWIISPSGEYRSGGPIKQELTSHVGPTAITTFISGHYVGAD 285
Query: 222 LIMKFGENEPWKKVFGPIFIYLNSNSDGFSPIK--LWEDAKQQMVNEVESWPYTFPASED 279
+ + E WKKV GP+FIYLNS+S + + LWEDAKQQ EV++WPY F AS D
Sbjct: 286 MEAHYRPGEAWKKVLGPVFIYLNSDSTSNNKPQDLLWEDAKQQSEKEVKAWPYDFVASSD 345
Query: 280 FLSSAQRGKFEGRLLVRDRYIRDAFVPVSGAYVGLAAPGDVGSWQREYKGYQFWTITDDK 339
+LS +RG GRLLV DR++ P AYVGLA PG+ GSWQ KGYQFWT T++
Sbjct: 346 YLSRRERGSVTGRLLVNDRFL----TPGKSAYVGLAPPGEAGSWQTNTKGYQFWTKTNET 401
Query: 340 GYFSIINVLPGDYNLYSWVNGFIGDYQCNNIINITSGSEINVGELVYEPPRHGPTLWEIG 399
GYF+I NV PG YNLY WV GFIGD++ N++N+ +GS I++G +VY+PPR+GPTLWEIG
Sbjct: 402 GYFTIENVRPGTYNLYGWVPGFIGDFRYQNLVNVAAGSVISLGRVVYKPPRNGPTLWEIG 461
Query: 400 IPDRSAAEFYVPDPNPLYVNRLYVNHSDRFRQYGLWERYADLYPNEDLVYTVGISDYKKD 459
+PDR+A E+++P+P +N LY+NH+D+FRQYGLW+RY +LYP DLVYTVG+S+Y +D
Sbjct: 462 VPDRTAREYFIPEPYKDTMNPLYLNHTDKFRQYGLWQRYTELYPTHDLVYTVGVSNYSQD 521
Query: 460 WFFAQVTR 467
WF+AQVTR
Sbjct: 522 WFYAQVTR 529
>At2g22620 unknown protein
Length = 677
Score = 475 bits (1222), Expect = e-134
Identities = 257/505 (50%), Positives = 314/505 (61%), Gaps = 43/505 (8%)
Query: 1 VVIDNDIVQVNISNPEGCVTGIQY----NGLDNLLEINNH------ESDRG*KAGNDKYE 50
VV+DN IVQV SNPEG +TGI+Y N LD+ ++ + E ++ K DK E
Sbjct: 62 VVVDNGIVQVTFSNPEGLITGIKYHGIDNVLDDKIDDRGYWDVVWYEPEK--KQKTDKLE 119
Query: 51 GDNGK*RTSRA----IFH*NMEFIS*RKACATQY**KE---------------------W 85
G + T I IS R + K W
Sbjct: 120 GTKFEIITQNEEQIEISFTRTWTISRRGSLVPLNVDKRYIIRSGVSGLYMYGILERLEGW 179
Query: 86 PAFDLDNTRVAFKLRKDKFNYMAIADNRQRFMPLPDDRLPPRGQVLAYPEAVRLVNPVEP 145
P D+D R+ FKL KF++MAI+D+RQR MP DR + LAY EAV L NP P
Sbjct: 180 PDVDMDQIRIVFKLNPKKFDFMAISDDRQRSMPSMADR--ENSKSLAYKEAVLLTNPSNP 237
Query: 146 EFKGEVDDKYEYACESRSNQVHGWISIDSSSDSTGCWLITPSYEFRSAGPLKQYLGSHLG 205
FKGEVDDKY Y+ E + N VHGWIS D G W+ITPS EFR GP+KQ L SH G
Sbjct: 238 MFKGEVDDKYMYSMEDKDNNVHGWISSDPP---VGFWMITPSDEFRLGGPIKQDLTSHAG 294
Query: 206 PTMLSVFHSTHYSGADLIMKFGENEPWKKVFGPIFIYLNSNSDGFSPIKLWEDAKQQMVN 265
P LS+F STHY+G ++ M + EPWKKVFGP+ YLNS S S ++LW DAK+QM
Sbjct: 295 PITLSMFTSTHYAGKEMRMDYRNGEPWKKVFGPVLAYLNSVSPKDSTLRLWRDAKRQMAA 354
Query: 266 EVESWPYTFPASEDFLSSAQRGKFEGRLLVRDRYIRDAFVPVSGAYVGLAAPGDVGSWQR 325
EV+SWPY F SED+ QRG EG+ L++D Y+ + A+VGLA G+ GSWQ
Sbjct: 355 EVKSWPYDFITSEDYPLRHQRGTLEGQFLIKDSYVSRLKIYGKFAFVGLAPIGEAGSWQT 414
Query: 326 EYKGYQFWTITDDKGYFSIINVLPGDYNLYSWVNGFIGDYQCNNIINITSGSEINVGELV 385
E KGYQFWT D +G F I NV G+Y+LY+W +GFIGDY+ I IT GSE+NVG LV
Sbjct: 415 ESKGYQFWTKADRRGRFIIENVRAGNYSLYAWGSGFIGDYKYEQNITITPGSEMNVGPLV 474
Query: 386 YEPPRHGPTLWEIGIPDRSAAEFYVPDPNPLYVNRLYVNH-SDRFRQYGLWERYADLYPN 444
YEPPR+GPTLWEIG+PDR+A EFY+PDP P +N+LYVN DRFRQYGLW+RYADLYP
Sbjct: 475 YEPPRNGPTLWEIGVPDRTAGEFYIPDPYPTLMNKLYVNPLQDRFRQYGLWDRYADLYPQ 534
Query: 445 EDLVYTVGISDYKKDWFFAQVTRFV 469
DLVYT+G+SDY+ DWFFA V R V
Sbjct: 535 NDLVYTIGVSDYRSDWFFAHVARNV 559
>At4g37950 hypothetical protein
Length = 296
Score = 211 bits (537), Expect = 7e-55
Identities = 115/265 (43%), Positives = 152/265 (56%), Gaps = 51/265 (19%)
Query: 1 VVIDNDIVQVNISNPEGCVTGIQYNGLDNLLEINNHESDRG*KAGNDKYEGDNGK*RTSR 60
VV+DN I+ V S+P+G +T I+YNGL+N+L N+ +RG
Sbjct: 69 VVVDNGIIDVTFSSPQGLITRIKYNGLNNVL--NDQIENRG------------------- 107
Query: 61 AIFH*NMEFIS*RKACATQY**KEWPAFDLDNTRVAFKLRKDKFNYMAIADNRQRFMPLP 120
WP D+D R+ FKL KF++MA++DNRQ+ MP
Sbjct: 108 ----------------------YSWPDVDMDQIRIVFKLNTTKFDFMAVSDNRQKIMPFD 145
Query: 121 DDRLPPRGQV--LAYPEAVRLVNPVEPEFKGEVDDKYEYACESRSNQVHGWISIDSSSDS 178
DR +G+ LAY EAV L+NP KG+VDDKY Y+ E++ N+VHGWIS D
Sbjct: 146 TDRDITKGRASPLAYKEAVHLINPQNHMLKGQVDDKYMYSVENKDNKVHGWISSDQR--- 202
Query: 179 TGCWLITPSYEFRSAGPLKQYLGSHLGPTMLSVFHSTHYSGADLIMKFGENEPWKKVFGP 238
G W+ITPS EF + GP+KQ L SH+GPT LS+F S HY+G D+ + EPWKKVFGP
Sbjct: 203 IGFWMITPSDEFHACGPIKQDLTSHVGPTTLSMFTSVHYAGKDMNTNYKSKEPWKKVFGP 262
Query: 239 IFIYLNSNSDGFSPIKLWEDAKQQM 263
+F+YLNS S S LW DAK+Q+
Sbjct: 263 VFVYLNSAS---SRNLLWTDAKRQV 284
>At4g37960 putative protein
Length = 289
Score = 204 bits (520), Expect = 7e-53
Identities = 92/142 (64%), Positives = 111/142 (77%), Gaps = 3/142 (2%)
Query: 329 GYQFWTITDDKGYFSIINVLPGDYNLYSWVNGFIGDYQCNNIINITSGSEINVGELVYEP 388
GYQFWT D G F+I NV PG Y+LY+WV+GFIGDY+ I IT G EI+VG +VY P
Sbjct: 28 GYQFWTRADKMGMFTIANVRPGTYSLYAWVSGFIGDYKYVRDITITPGREIDVGHIVYVP 87
Query: 389 PRHGPTLWEIGIPDRSAAEFYVPDPNPLYVNRLYVNHS---DRFRQYGLWERYADLYPNE 445
PR+GPTLWEIG PDR+AAEFY+PDP+P +LY+N+S DRFRQYGLW+RY+ LYP
Sbjct: 88 PRNGPTLWEIGQPDRTAAEFYIPDPDPTLFTKLYLNYSNPQDRFRQYGLWDRYSVLYPRN 147
Query: 446 DLVYTVGISDYKKDWFFAQVTR 467
DLV+T G+SDYKKDWF+A V R
Sbjct: 148 DLVFTAGVSDYKKDWFYAHVNR 169
>At1g65210 hypothetical protein
Length = 248
Score = 103 bits (256), Expect = 3e-22
Identities = 40/67 (59%), Positives = 56/67 (82%)
Query: 401 PDRSAAEFYVPDPNPLYVNRLYVNHSDRFRQYGLWERYADLYPNEDLVYTVGISDYKKDW 460
P + A E++VP+P +N LY+NH+D+FRQYGLW+RY +LYPN DL+YT+G+S+Y KDW
Sbjct: 62 PHQRAREYFVPEPYKNTMNPLYLNHTDKFRQYGLWQRYTELYPNHDLIYTIGVSNYSKDW 121
Query: 461 FFAQVTR 467
F++QVTR
Sbjct: 122 FYSQVTR 128
Score = 42.0 bits (97), Expect = 7e-04
Identities = 15/31 (48%), Positives = 26/31 (83%)
Query: 1 VVIDNDIVQVNISNPEGCVTGIQYNGLDNLL 31
V++DN I+ V+ S+P+G +TGI+Y G++N+L
Sbjct: 30 VIVDNGIISVSFSSPQGLITGIKYKGVNNVL 60
>At1g57820 putative transcription factor protein (At1g57820)
Length = 645
Score = 32.7 bits (73), Expect = 0.45
Identities = 15/45 (33%), Positives = 26/45 (57%)
Query: 254 KLWEDAKQQMVNEVESWPYTFPASEDFLSSAQRGKFEGRLLVRDR 298
+L ++ K Q+ +V + P T P + +F + KF G+ LVR+R
Sbjct: 511 RLLKEFKCQICQQVLTLPVTTPCAHNFCKACLEAKFAGKTLVRER 555
>At1g57800 hypothetical protein
Length = 650
Score = 30.8 bits (68), Expect = 1.7
Identities = 15/45 (33%), Positives = 25/45 (55%)
Query: 254 KLWEDAKQQMVNEVESWPYTFPASEDFLSSAQRGKFEGRLLVRDR 298
+L ++ K Q+ +V + P T P + +F + KF G LVR+R
Sbjct: 511 RLLKEFKCQICQKVMTNPVTTPCAHNFCKACLESKFAGTALVRER 555
>At3g19250 hypothetical protein
Length = 398
Score = 30.4 bits (67), Expect = 2.2
Identities = 15/41 (36%), Positives = 21/41 (50%), Gaps = 1/41 (2%)
Query: 147 FKGEVDDKYEYACESRSNQVHGWISIDSSSDSTGCWLITPS 187
F+GE + EY C +H +SID SS + C + PS
Sbjct: 7 FQGEYEPSSEYLCPGLIQHIH-TLSIDESSSAFRCRTVLPS 46
>At5g02350 putative protein
Length = 651
Score = 29.3 bits (64), Expect = 5.0
Identities = 31/103 (30%), Positives = 45/103 (43%), Gaps = 8/103 (7%)
Query: 120 PDDRLPPRGQVLAYPEAVRL-VNPVEPEFKGEVDDKYEYACESRSNQVHGWISIDSSSDS 178
P RL G+ +Y E + +P P G VD + ++ +S S QVH + D+S D
Sbjct: 57 PSLRLRVYGENNSYTEFLPYNTSPHFPSITG-VDQQVQHLLDSDSQQVHDLLHSDNSHDD 115
Query: 179 TG---CWL-ITPSYEFRSAGPLKQYLGSHLGPTMLSVFHSTHY 217
G C L + P Y S +YL G LS + +Y
Sbjct: 116 DGDEICKLPVVPIYWCNS--KKDEYLQFTCGACQLSKIGTDYY 156
>At2g41560 putative Ca2+-ATPase
Length = 1030
Score = 29.3 bits (64), Expect = 5.0
Identities = 27/103 (26%), Positives = 45/103 (43%), Gaps = 6/103 (5%)
Query: 254 KLWEDAKQQMVNEVESWPYTFPASEDFLSSAQRGKFE--GRLLVRDRYIRDAFV--PVSG 309
K+W K Q E + SE+ S+ +G F+ G +V+D+ + P
Sbjct: 465 KVWICDKVQERQEGSKESFELELSEEVQSTLLQGIFQNTGSEVVKDKDGNTQILGSPTER 524
Query: 310 AYV--GLAAPGDVGSWQREYKGYQFWTITDDKGYFSIINVLPG 350
A + GL GD + ++E+K + DK S++ LPG
Sbjct: 525 AILEFGLLLGGDFNTQRKEHKILKIEPFNSDKKKMSVLIALPG 567
>At3g10250 unknown protein
Length = 324
Score = 28.9 bits (63), Expect = 6.5
Identities = 17/65 (26%), Positives = 31/65 (47%), Gaps = 2/65 (3%)
Query: 229 NEPWKKVFGPIFIYLNSNSDGFSPIKLWEDAKQ--QMVNEVESWPYTFPASEDFLSSAQR 286
++P + F +L FS L D Q +++ + P+ P +E+FL S R
Sbjct: 240 SDPLLEAEASTFGFLGQIPRNFSLSDLTADFSQSSEILESYDRSPFLVPNAENFLDSRDR 299
Query: 287 GKFEG 291
G+++G
Sbjct: 300 GEYQG 304
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.326 0.144 0.466
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,754,609
Number of Sequences: 26719
Number of extensions: 549701
Number of successful extensions: 1395
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 11
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 1359
Number of HSP's gapped (non-prelim): 24
length of query: 478
length of database: 11,318,596
effective HSP length: 103
effective length of query: 375
effective length of database: 8,566,539
effective search space: 3212452125
effective search space used: 3212452125
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 62 (28.5 bits)
Medicago: description of AC148236.5


