

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148176.1 + phase: 0
(700 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
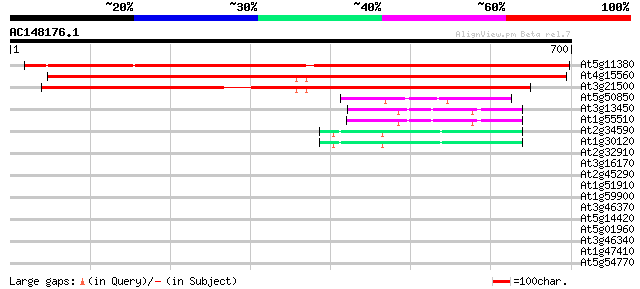
Score E
Sequences producing significant alignments: (bits) Value
At5g11380 1-D-deoxyxylulose 5-phosphate synthase - like protein 922 0.0
At4g15560 1-D-deoxyxylulose 5-phosphate synthase, putative 766 0.0
At3g21500 1-D-deoxyxylulose 5-phosphate synthase, putative 642 0.0
At5g50850 pyruvate dehydrogenase E1 component beta subunit, mito... 70 5e-12
At3g13450 branched chain alpha-keto acid dehydrogenase E1 beta s... 55 2e-07
At1g55510 branched-chain alpha-keto acid decarboxylase E1 beta s... 51 3e-06
At2g34590 putative pyruvate dehydrogenase E1 beta subunit 45 1e-04
At1g30120 unknown protein 45 2e-04
At2g32910 unknown protein 32 1.6
At3g16170 long-chain acyl-CoA like synthetase 31 2.7
At2g45290 putative transketolase precursor 31 2.7
At1g51910 Putative protein kinase 31 2.7
At1g59900 pyruvate dehydrogenase E1 alpha subunit, putative 30 3.5
At3g46370 receptor-like protein kinase homolog 30 4.6
At5g14420 unknown protein 30 6.0
At5g01960 unknown protein 30 6.0
At3g46340 receptor-like protein kinase homolog 30 6.0
At1g47410 unknown protein 30 6.0
At5g54770 thiazole biosynthetic enzyme precursor (ARA6) (sp|Q38814) 29 7.8
>At5g11380 1-D-deoxyxylulose 5-phosphate synthase - like protein
Length = 700
Score = 922 bits (2382), Expect = 0.0
Identities = 472/682 (69%), Positives = 558/682 (81%), Gaps = 14/682 (2%)
Query: 19 TLTRRLDCSI-SQFPLSRITYSSSSRILIHRVCSRPDIDDFYWEKVPTPILDTVQNPLCL 77
+L +LD SI S F T+ S RVCS P+ D + EK TPILD+++ PL L
Sbjct: 30 SLPCKLDVSIKSLFSAPSSTHKEYSNRA--RVCSLPNTDGYCDEKFETPILDSIETPLQL 87
Query: 78 KNLSQQELKQLAAEIRLELSSIL-SGTQILLNPSMAVVDLTVAIHHVFHAPVDKILWDVG 136
KNLS +ELK LA EIR EL S+L TQ +NPS A ++LT+A+H+VF APVD ILWD
Sbjct: 88 KNLSVKELKLLADEIRTELHSVLWKKTQKSMNPSFAAIELTLALHYVFRAPVDNILWDAV 147
Query: 137 DQTYAHKILTGRRSLMKTIRKKNGLSGFTSRFESEYDAFGAGHGCNSISAGLGMAVARDI 196
+QTYAHK+LT R S + + R+ +G+SG TSR ESEYD+FG GHGCNSISAGLG+AVARD+
Sbjct: 148 EQTYAHKVLTRRWSAIPS-RQNSGISGVTSRLESEYDSFGTGHGCNSISAGLGLAVARDM 206
Query: 197 KGRRERVVAVISNWTTMSGQVYEAMSNAGYLDSNLVVILNDSRHSLLPKIEDGSKTSVNA 256
KG+R+RVVAVI N T +GQ YEAMSNAGYLDSN++VILNDSRHSL P +E+GSK S++A
Sbjct: 207 KGKRDRVVAVIDNVTITAGQAYEAMSNAGYLDSNMIVILNDSRHSLHPNMEEGSKASISA 266
Query: 257 LSSTLSRLQSSKSFRKFREAAKGVTKRIGRGMHELAAKVDEYARGMMGPPGSTLFEELGL 316
LSS +S++QSSK FRKFRE AK +TKRIG+GM+E AAKVDEYARGM+GP GSTLFEELGL
Sbjct: 267 LSSIMSKIQSSKVFRKFRELAKAMTKRIGKGMYEWAAKVDEYARGMVGPTGSTLFEELGL 326
Query: 317 YYIGPVDGHNIEDLISVLQEVASLDSMGPVLIHVITNENQVEEHNKKSYMTDKQQDDNAG 376
YYIGPVDGHNIEDL+ VL+EV+SLDSMGPVL+HVIT N+ E K + D++
Sbjct: 327 YYIGPVDGHNIEDLVCVLREVSSLDSMGPVLVHVITEGNRDAETVKNIMVKDRR------ 380
Query: 377 RLQTYGDCFVESLVAEAEKDKDIVVVHAGITTEPSLKLFMEKFPDRIFNVGIAEQHAVTF 436
TY DCFVE+LV EAEKD+DIVVVHAG+ +PSL F E+FPDR FNVG+AEQHAVTF
Sbjct: 381 ---TYSDCFVEALVMEAEKDRDIVVVHAGMEMDPSLLTFQERFPDRFFNVGMAEQHAVTF 437
Query: 437 ASGLSCGGLKPFCIIPSSFLQRAYDQVVHDVDQQKVPVRFVITSAGLVGSDGPLQCGAFD 496
++GLS GGLKPFCIIPS+FLQRAYDQVVHDVD+Q+ VRFVITSAGLVGSDGP+QCGAFD
Sbjct: 438 SAGLSSGGLKPFCIIPSAFLQRAYDQVVHDVDRQRKAVRFVITSAGLVGSDGPVQCGAFD 497
Query: 497 ITFMSCLPNMIVMAPSDEAELVHMVATAAHINDQPVCFRYPRGALVGKDEAILDGIPIEI 556
I FMS LPNMI MAP+DE ELV+MVATAA++ D+PVCFR+PRG++V + + G+PIEI
Sbjct: 498 IAFMSSLPNMIAMAPADEDELVNMVATAAYVTDRPVCFRFPRGSIVNMNYLVPTGLPIEI 557
Query: 557 GKGRILVEGKDVALLGYGSMVQNCLKAYSLLANLGIEVTVADARFCKPLDIELLRQLCKH 616
G+GR+LVEG+DVALLGYG+MVQNCL A+SLL+ LG+ VTVADARFCKPLDI+L+R LC++
Sbjct: 558 GRGRVLVEGQDVALLGYGAMVQNCLHAHSLLSKLGLNVTVADARFCKPLDIKLVRDLCQN 617
Query: 617 HSFLITVEEGSIGGFGSHVAQFIALDGLLDRRIKWRPIVLPDSYIEHASPNQQLNQAGLT 676
H FLITVEEG +GGFGSHVAQFIALDG LD IKWRPIVLPD YIE ASP +QL AGLT
Sbjct: 618 HKFLITVEEGCVGGFGSHVAQFIALDGQLDGNIKWRPIVLPDGYIEEASPREQLALAGLT 677
Query: 677 GHHIAATALSLLGRTREALSFM 698
GHHIAATALSLLGRTREAL M
Sbjct: 678 GHHIAATALSLLGRTREALLLM 699
>At4g15560 1-D-deoxyxylulose 5-phosphate synthase, putative
Length = 717
Score = 766 bits (1977), Expect = 0.0
Identities = 399/661 (60%), Positives = 491/661 (73%), Gaps = 13/661 (1%)
Query: 48 RVC-SRPDIDDFYWEKVPTPILDTVQNPLCLKNLSQQELKQLAAEIRLELSSILSGTQIL 106
+VC S + ++Y + PTP+LDT+ P+ +KNLS +ELKQL+ E+R ++ +S T
Sbjct: 56 KVCASLAEKGEYYSNRPPTPLLDTINYPIHMKNLSVKELKQLSDELRSDVIFNVSKTGGH 115
Query: 107 LNPSMAVVDLTVAIHHVFHAPVDKILWDVGDQTYAHKILTGRRSLMKTIRKKNGLSGFTS 166
L S+ VV+LTVA+H++F+ P DKILWDVG Q+Y HKILTGRR M T+R+ NGLSGFT
Sbjct: 116 LGSSLGVVELTVALHYIFNTPQDKILWDVGHQSYPHKILTGRRGKMPTMRQTNGLSGFTK 175
Query: 167 RFESEYDAFGAGHGCNSISAGLGMAVARDIKGRRERVVAVISNWTTMSGQVYEAMSNAGY 226
R ESE+D FG GH +ISAGLGMAV RD+KG+ VVAVI + +GQ YEAM+NAGY
Sbjct: 176 RGESEHDCFGTGHSSTTISAGLGMAVGRDLKGKNNNVVAVIGDGAMTAGQAYEAMNNAGY 235
Query: 227 LDSNLVVILNDSRHSLLPKIE-DGSKTSVNALSSTLSRLQSSKSFRKFREAAKGVTKRIG 285
LDS+++VILND++ LP DG V ALSS LSRLQS+ + R+ RE AKG+TK+IG
Sbjct: 236 LDSDMIVILNDNKQVSLPTATLDGPSPPVGALSSALSRLQSNPALRELREVAKGMTKQIG 295
Query: 286 RGMHELAAKVDEYARGMMGPPGSTLFEELGLYYIGPVDGHNIEDLISVLQEVASLDSMGP 345
MH+LAAKVDEYARGM+ GS+LFEELGLYYIGPVDGHNI+DL+++L+EV S + GP
Sbjct: 296 GPMHQLAAKVDEYARGMISGTGSSLFEELGLYYIGPVDGHNIDDLVAILKEVKSTRTTGP 355
Query: 346 VLIHVITNENQ----VEEHNKKSYMTDK------QQDDNAGRLQTYGDCFVESLVAEAEK 395
VLIHV+T + + E + K + K +Q + Q+Y F E+LVAEAE
Sbjct: 356 VLIHVVTEKGRGYPYAERADDKYHGVVKFDPATGRQFKTTNKTQSYTTYFAEALVAEAEV 415
Query: 396 DKDIVVVHAGITTEPSLKLFMEKFPDRIFNVGIAEQHAVTFASGLSCGGLKPFCIIPSSF 455
DKD+V +HA + L LF +FP R F+VGIAEQHAVTFA+GL+C GLKPFC I SSF
Sbjct: 416 DKDVVAIHAAMGGGTGLNLFQRRFPTRCFDVGIAEQHAVTFAAGLACEGLKPFCAIYSSF 475
Query: 456 LQRAYDQVVHDVDQQKVPVRFVITSAGLVGSDGPLQCGAFDITFMSCLPNMIVMAPSDEA 515
+QRAYDQVVHDVD QK+PVRF + AGLVG+DGP CGAFD+TFM+CLPNMIVMAPSDEA
Sbjct: 476 MQRAYDQVVHDVDLQKLPVRFAMDRAGLVGADGPTHCGAFDVTFMACLPNMIVMAPSDEA 535
Query: 516 ELVHMVATAAHINDQPVCFRYPRGALVGKD-EAILDGIPIEIGKGRILVEGKDVALLGYG 574
+L +MVATA I+D+P CFRYPRG +G G+PIEIGKGRIL EG+ VALLGYG
Sbjct: 536 DLFNMVATAVAIDDRPSCFRYPRGNGIGVALPPGNKGVPIEIGKGRILKEGERVALLGYG 595
Query: 575 SMVQNCLKAYSLLANLGIEVTVADARFCKPLDIELLRQLCKHHSFLITVEEGSIGGFGSH 634
S VQ+CL A +L G+ VTVADARFCKPLD L+R L K H LITVEEGSIGGFGSH
Sbjct: 596 SAVQSCLGAAVMLEERGLNVTVADARFCKPLDRALIRSLAKSHEVLITVEEGSIGGFGSH 655
Query: 635 VAQFIALDGLLDRRIKWRPIVLPDSYIEHASPNQQLNQAGLTGHHIAATALSLLGRTREA 694
V QF+ALDGLLD ++KWRP+VLPD YI+H +P QL +AGL HIAATAL+L+G REA
Sbjct: 656 VVQFLALDGLLDGKLKWRPMVLPDRYIDHGAPADQLAEAGLMPSHIAATALNLIGAPREA 715
Query: 695 L 695
L
Sbjct: 716 L 716
>At3g21500 1-D-deoxyxylulose 5-phosphate synthase, putative
Length = 628
Score = 642 bits (1655), Expect = 0.0
Identities = 339/622 (54%), Positives = 435/622 (69%), Gaps = 43/622 (6%)
Query: 40 SSSRILIHRVCSRPDIDDFYWEKVPTPILDTVQNPLCLKNLSQQELKQLAAEIRLELSSI 99
SS++ R + + ++Y + PTP+LDT+ +P+ +KNLS +ELK L+ E+R ++
Sbjct: 27 SSTKYSKVRATTFSEKGEYYSNRPPTPLLDTINHPMHMKNLSIKELKVLSDELRSDVIFN 86
Query: 100 LSGTQILLNPSMAVVDLTVAIHHVFHAPVDKILWDVGDQTYAHKILTGRRSLMKTIRKKN 159
+S T L ++ VV+LTVA+H++F+ P DKILWDVG Q+Y HKILTGRR MKTIR+ N
Sbjct: 87 VSKTGGHLGSNLGVVELTVALHYIFNTPHDKILWDVGHQSYPHKILTGRRGKMKTIRQTN 146
Query: 160 GLSGFTSRFESEYDAFGAGHGCNSISAGLGMAVARDIKGRRERVVAVISNWTTMSGQVYE 219
GLSG+T R ESE+D+FG GH ++SAGLGMAV RD+KG VV+VI + +GQ YE
Sbjct: 147 GLSGYTKRRESEHDSFGTGHSSTTLSAGLGMAVGRDLKGMNNSVVSVIGDGAMTAGQAYE 206
Query: 220 AMSNAGYLDSNLVVILNDSRHSLLPKIE-DGSKTSVNALSSTLSRLQSSKSFRKFREAAK 278
AM+NAGYL SN++VILND++ LP DG V ALS LSRLQS+
Sbjct: 207 AMNNAGYLHSNMIVILNDNKQVSLPTANLDGPTQPVGALSCALSRLQSNC---------- 256
Query: 279 GVTKRIGRGMHELAAKVDEYARGMMGPPGSTLFEELGLYYIGPVDGHNIEDLISVLQEVA 338
GM+ STLFEELG +Y+GPVDGHNI+DL+S+L+ +
Sbjct: 257 ----------------------GMIRETSSTLFEELGFHYVGPVDGHNIDDLVSILETLK 294
Query: 339 SLDSMGPVLIHVITNENQ----VEEHNKKSYMTD-----KQQDDNAGRLQTYGDCFVESL 389
S ++GPVLIHV+T + + E + K ++ +Q N + Q+Y CFVE+L
Sbjct: 295 STKTIGPVLIHVVTEKGRGYPYAERADDKYHVLKFDPETGKQFKNISKTQSYTSCFVEAL 354
Query: 390 VAEAEKDKDIVVVHAGITTEPSLKLFMEKFPDRIFNVGIAEQHAVTFASGLSCGGLKPFC 449
+AEAE DKDIV +HA + L LF +FP R F+VGIAEQHAVTFA+GL+C GLKPFC
Sbjct: 355 IAEAEADKDIVAIHAAMGGGTMLNLFESRFPTRCFDVGIAEQHAVTFAAGLACEGLKPFC 414
Query: 450 IIPSSFLQRAYDQVVHDVDQQKVPVRFVITSAGLVGSDGPLQCGAFDITFMSCLPNMIVM 509
I SSF+QRAYDQVVHDVD QK+PVRF I AGL+G+DGP CGAFD+TFM+CLPNMIVM
Sbjct: 415 TIYSSFMQRAYDQVVHDVDLQKLPVRFAIDRAGLMGADGPTHCGAFDVTFMACLPNMIVM 474
Query: 510 APSDEAELVHMVATAAHINDQPVCFRYPRGALVGKD-EAILDGIPIEIGKGRILVEGKDV 568
APSDEAEL +MVATAA I+D+P CFRY RG +G G+P++IG+GRIL +G+ V
Sbjct: 475 APSDEAELFNMVATAAAIDDRPSCFRYHRGNGIGVSLPPGNKGVPLQIGRGRILRDGERV 534
Query: 569 ALLGYGSMVQNCLKAYSLLANLGIEVTVADARFCKPLDIELLRQLCKHHSFLITVEEGSI 628
ALLGYGS VQ CL+A S+L+ G+++TVADARFCKPLD+ L+R L K H LITVEEGSI
Sbjct: 535 ALLGYGSAVQRCLEAASMLSERGLKITVADARFCKPLDVALIRSLAKSHEVLITVEEGSI 594
Query: 629 GGFGSHVAQFIALDGLLDRRIK 650
GGFGSHV QF+ALDGLLD ++K
Sbjct: 595 GGFGSHVVQFLALDGLLDGKLK 616
>At5g50850 pyruvate dehydrogenase E1 component beta subunit,
mitochondrial precursor (PDHE1-B) (sp|Q38799)
Length = 363
Score = 69.7 bits (169), Expect = 5e-12
Identities = 68/230 (29%), Positives = 103/230 (44%), Gaps = 21/230 (9%)
Query: 413 KLFMEKF-PDRIFNVGIAEQHAVTFASGLSCGGLKPFC-IIPSSFLQRAYDQVVHDV--- 467
K +EK+ P+R+++ I E G + GLKP + +F +A D +++
Sbjct: 74 KGLLEKYGPERVYDTPITEAGFTGIGVGAAYAGLKPVVEFMTFNFSMQAIDHIINSAAKS 133
Query: 468 -----DQQKVPVRFVITSAGLVGSDGP-LQCGAFDITFMSCLPNMIVMAPSDEAELVHMV 521
Q VP+ F + G QC A + + +P + V+AP AE +
Sbjct: 134 NYMSAGQINVPIVFRGPNGAAAGVGAQHSQCYA---AWYASVPGLKVLAPYS-AEDARGL 189
Query: 522 ATAAHINDQPVCFRYPRGALVGK-----DEAILDGIPIEIGKGRILVEGKDVALLGYGSM 576
AA + PV F L G+ +EA+ + IGK +I EGKDV ++ + M
Sbjct: 190 LKAAIRDPDPVVF-LENELLYGESFPISEEALDSSFCLPIGKAKIEREGKDVTIVTFSKM 248
Query: 577 VQNCLKAYSLLANLGIEVTVADARFCKPLDIELLRQLCKHHSFLITVEEG 626
V LKA LA GI V + R +PLD + + S L+TVEEG
Sbjct: 249 VGFALKAAEKLAEEGISAEVINLRSIRPLDRATINASVRKTSRLVTVEEG 298
>At3g13450 branched chain alpha-keto acid dehydrogenase E1 beta
subunit
Length = 358
Score = 54.7 bits (130), Expect = 2e-07
Identities = 60/233 (25%), Positives = 101/233 (42%), Gaps = 21/233 (9%)
Query: 422 RIFNVGIAEQHAVTFASGLSCGGLKPFCIIP-SSFLQRAYDQVVHDVDQQKVPVRFVITS 480
R+FN + EQ V F GL+ G + I + ++ A+DQ+V++ + +
Sbjct: 85 RVFNTPLCEQGIVGFGIGLAAMGNRVIAEIQFADYIFPAFDQIVNEAAKFRYRSGNQFNC 144
Query: 481 AGL--------VGSDGPLQCGAFDITFMSCLPNMIVMAPSDEAELVHMVATAAHINDQPV 532
GL VG G + + F +P + V+ P E ++ ++ + PV
Sbjct: 145 GGLTIRAPYGAVGHGGHYHSQSPE-AFFCHVPGIKVVIPRSPREAKGLLLSSIR-DPNPV 202
Query: 533 CFRYPRGALVGKDEAIL-DGIPIEIGKGRILVEGKDVALLGYGS----MVQNCLKAYSLL 587
F P+ E + D I + + ++ EG D+ L+G+G+ M Q CL A
Sbjct: 203 VFFEPKWLYRQAVEDVPEDDYMIPLSEAEVMREGSDITLVGWGAQLTIMEQACLDA---- 258
Query: 588 ANLGIEVTVADARFCKPLDIELLR-QLCKHHSFLITVEEGSIGGFGSHVAQFI 639
N GI + D + P D E++ + K LI+ E GGFG+ +A I
Sbjct: 259 ENEGISCELIDLKTLIPWDKEIVETSVRKTGRLLISHEAPVTGGFGAEIAATI 311
>At1g55510 branched-chain alpha-keto acid decarboxylase E1 beta
subunit
Length = 352
Score = 50.8 bits (120), Expect = 3e-06
Identities = 56/234 (23%), Positives = 98/234 (40%), Gaps = 21/234 (8%)
Query: 421 DRIFNVGIAEQHAVTFASGLSCGGLKPFCIIP-SSFLQRAYDQVVHDVDQQKVPVRFVIT 479
+R+FN + EQ V F GL+ G + I + ++ A+DQ+V++ + +
Sbjct: 78 NRVFNTPLCEQGIVGFGIGLAAMGNRAIVEIQFADYIYPAFDQIVNEAAKFRYRSGNQFN 137
Query: 480 SAGL--------VGSDGPLQCGAFDITFMSCLPNMIVMAPSDEAELVHMVATAAHINDQP 531
GL VG G + + F +P + V+ P E ++ + + P
Sbjct: 138 CGGLTIRAPYGAVGHGGHYHSQSPE-AFFCHVPGIKVVIPRSPREAKGLLLSCIR-DPNP 195
Query: 532 VCFRYPRGALVGKDEAILD-GIPIEIGKGRILVEGKDVALLGYGS----MVQNCLKAYSL 586
V F P+ E + + I + + ++ EG D+ L+G+G+ M Q CL A
Sbjct: 196 VVFFEPKWLYRQAVEEVPEHDYMIPLSEAEVIREGNDITLVGWGAQLTVMEQACLDA--- 252
Query: 587 LANLGIEVTVADARFCKPLDIELLRQLCKHHSFLITVEEGSI-GGFGSHVAQFI 639
GI + D + P D E + K L+ E + GGFG+ ++ I
Sbjct: 253 -EKEGISCELIDLKTLLPWDKETVEASVKKTGRLLISHEAPVTGGFGAEISATI 305
>At2g34590 putative pyruvate dehydrogenase E1 beta subunit
Length = 406
Score = 45.1 bits (105), Expect = 1e-04
Identities = 61/268 (22%), Positives = 109/268 (39%), Gaps = 18/268 (6%)
Query: 387 ESLVAEAEKDKDIVVV-----HAGITTEPSLKLFMEKFPD-RIFNVGIAEQHAVTFASGL 440
E L E ++D + V+ H G + + + K +KF D R+ + I E G
Sbjct: 94 EGLEEEMDRDPHVCVMGEDVGHYGGSYKVT-KGLADKFGDLRVLDTPICENAFTGMGIGA 152
Query: 441 SCGGLKPFCI-IPSSFLQRAYDQV------VHDVDQQKVPVRFVITSAGLVGSDGPLQCG 493
+ GL+P + FL A++Q+ +H + + VI G VG +
Sbjct: 153 AMTGLRPVIEGMNMGFLLLAFNQISNNCGMLHYTSGGQFTIPVVIRGPGGVGRQLGAEHS 212
Query: 494 AFDITFMSCLPNMIVMAPSDEAELVHMVATAAHINDQPVCFRYPRGALVGKDEAILDGIP 553
++ +P + ++A S ++ A + + F + L E+I D
Sbjct: 213 QRLESYFQSIPGIQMVACSTPYNAKGLMKAAIRSENPVILFEHV--LLYNLKESIPDEEY 270
Query: 554 I-EIGKGRILVEGKDVALLGYGSMVQNCLKAYSLLANLGIEVTVADARFCKPLDIELL-R 611
I + + ++ G+ + +L Y M + ++A L N G + V D R KP D+ +
Sbjct: 271 ICNLEEAEMVRPGEHITILTYSRMRYHVMQAAKTLVNKGYDPEVIDIRSLKPFDLYTIGN 330
Query: 612 QLCKHHSFLITVEEGSIGGFGSHVAQFI 639
+ K H LI E GG G+ + I
Sbjct: 331 SVKKTHRVLIVEECMRTGGIGASLTAAI 358
>At1g30120 unknown protein
Length = 406
Score = 44.7 bits (104), Expect = 2e-04
Identities = 60/268 (22%), Positives = 108/268 (39%), Gaps = 18/268 (6%)
Query: 387 ESLVAEAEKDKDIVVV-----HAGITTEPSLKLFMEKFPD-RIFNVGIAEQHAVTFASGL 440
E L E ++D + V+ H G + + + K +KF D R+ + I E G
Sbjct: 94 EGLEEEMDRDPHVCVMGEDVGHYGGSYKVT-KGLADKFGDLRVLDTPICENAFTGMGIGA 152
Query: 441 SCGGLKPFCI-IPSSFLQRAYDQV------VHDVDQQKVPVRFVITSAGLVGSDGPLQCG 493
+ GL+P + FL A++Q+ +H + + VI G VG +
Sbjct: 153 AMTGLRPVIEGMNMGFLLLAFNQISNNCGMLHYTSGGQFTIPVVIRGPGGVGRQLGAEHS 212
Query: 494 AFDITFMSCLPNMIVMAPSDEAELVHMVATAAHINDQPVCFRYPRGALVGKDEAILDGIP 553
++ +P + ++A S ++ A + + F + L E I D
Sbjct: 213 QRLESYFQSIPGIQMVACSTPYNAKGLMKAAIRSENPVILFEHV--LLYNLKEKIPDEDY 270
Query: 554 I-EIGKGRILVEGKDVALLGYGSMVQNCLKAYSLLANLGIEVTVADARFCKPLDIELL-R 611
+ + + ++ G+ + +L Y M + ++A L N G + V D R KP D+ +
Sbjct: 271 VCNLEEAEMVRPGEHITILTYSRMRYHVMQAAKTLVNKGYDPEVIDIRSLKPFDLHTIGN 330
Query: 612 QLCKHHSFLITVEEGSIGGFGSHVAQFI 639
+ K H LI E GG G+ + I
Sbjct: 331 SVKKTHRVLIVEECMRTGGIGASLTAAI 358
>At2g32910 unknown protein
Length = 691
Score = 31.6 bits (70), Expect = 1.6
Identities = 31/106 (29%), Positives = 48/106 (45%), Gaps = 25/106 (23%)
Query: 352 TNENQVEEHNKKSYMTDKQQDDNAGRLQTYG-----------DCFVESLVAEAEKDKDIV 400
T E +EE ++ +K++ D G+++ G DCF S++ EK KD V
Sbjct: 291 TTEQGMEERKEQPVDPEKREMDGPGKVKIGGLIFMCNTKTRPDCFRFSVMGVQEKRKDFV 350
Query: 401 VVHAGITTEPSLKLFMEKFPDRIFNVGIAEQHAVTFASGLSCGGLK 446
GI +P LKLF+ + ++ GI E S GG+K
Sbjct: 351 ---KGI--KPGLKLFLYDYDLKLL-YGIFE--------ASSAGGMK 382
>At3g16170 long-chain acyl-CoA like synthetase
Length = 544
Score = 30.8 bits (68), Expect = 2.7
Identities = 34/125 (27%), Positives = 57/125 (45%), Gaps = 15/125 (12%)
Query: 129 DKILWDVGDQTYAHKILTGRR-SLMKTIRKKNGLSGFTSRFESEYDAFGAGHGCNSISAG 187
D+I ++Y++ LT + K K + +G + +Y+ FG+ G A
Sbjct: 16 DRIAIKADGKSYSYGQLTSSALRISKLFLKDDTTNG--GQETKKYEGFGSLKG-----AR 68
Query: 188 LGMAVARDIKGRRERVVAVISNWTTMSGQVYEAMSNAGYLDSNLVVILNDSRHSLLPKIE 247
+G+ K E V V+ W + V A+S Y ++ L+ ++NDS SLL E
Sbjct: 69 IGIVA----KPSAEFVAGVLGTWFSGGVAVPLALS---YPEAELLHVMNDSDISLLLSTE 121
Query: 248 DGSKT 252
D S+T
Sbjct: 122 DHSET 126
>At2g45290 putative transketolase precursor
Length = 741
Score = 30.8 bits (68), Expect = 2.7
Identities = 52/230 (22%), Positives = 85/230 (36%), Gaps = 22/230 (9%)
Query: 421 DRIFNVGIAEQHAVTFASGLSCG--GLKPFCIIPSSFLQRAYDQVVHDVDQ-QKVPVRFV 477
+R G+ E +G++ G P+C + F+ Y + + + V +V
Sbjct: 479 ERNLRFGVREHGMGAICNGIALHSPGFIPYCA--TFFVFTDYMRAAMRISALSEAGVIYV 536
Query: 478 ITSAGL-VGSDGPLQCGAFDITFMSCLPNMIVMAPSDEAELVHMVATAAHINDQPVCFRY 536
+T + +G DGP ++ +PN+++ P+D E A P
Sbjct: 537 MTHDSIGLGEDGPTHQPIEHLSSFRAMPNIMMFRPADGNETAGAYKIAVTKRKTPSVLAL 596
Query: 537 PRGALVGKDEAILDGIPIE-IGKGRILVEGK------DVALLGYGSMVQNCLKAYSLLAN 589
R L L G IE + KG + DV L+G GS ++ +A L
Sbjct: 597 SRQKL-----PQLPGTSIESVEKGGYTISDNSTGNKPDVILIGTGSELEIAAQAAEKLRE 651
Query: 590 LGIEVTVAD----ARFCKPLDIELLRQLCKHHSFLITVEEGSIGGFGSHV 635
G V V F + D L S +++E GS G+G V
Sbjct: 652 QGKSVRVVSFVCWELFDEQSDAYKESVLPSDVSARVSIEAGSTFGWGKIV 701
>At1g51910 Putative protein kinase
Length = 879
Score = 30.8 bits (68), Expect = 2.7
Identities = 19/71 (26%), Positives = 33/71 (45%), Gaps = 15/71 (21%)
Query: 313 ELGLYYIGPVDGHNIEDLI---------------SVLQEVASLDSMGPVLIHVITNENQV 357
+ GL PVDG + I ++L E + S G VL+ +ITN+ +
Sbjct: 716 DFGLSRSSPVDGESYVSTIVAGTPGYLDPEYYRTNLLSEKTDVYSFGVVLLEIITNQPVI 775
Query: 358 EEHNKKSYMTD 368
+ +K+++TD
Sbjct: 776 DTTREKAHITD 786
>At1g59900 pyruvate dehydrogenase E1 alpha subunit, putative
Length = 389
Score = 30.4 bits (67), Expect = 3.5
Identities = 18/82 (21%), Positives = 37/82 (44%), Gaps = 2/82 (2%)
Query: 163 GFTSRFESEYDAFGAGHGCNSISAGLGMAVARDIKGRRERVV--AVISNWTTMSGQVYEA 220
G + F + +F GHG LG +A K +E V A+ + GQ++EA
Sbjct: 148 GGSMHFYKKESSFYGGHGIVGAQVPLGCGIAFAQKYNKEEAVTFALYGDGAANQGQLFEA 207
Query: 221 MSNAGYLDSNLVVILNDSRHSL 242
++ + D +++ ++ + +
Sbjct: 208 LNISALWDLPAILVCENNHYGM 229
>At3g46370 receptor-like protein kinase homolog
Length = 793
Score = 30.0 bits (66), Expect = 4.6
Identities = 11/35 (31%), Positives = 25/35 (71%)
Query: 334 LQEVASLDSMGPVLIHVITNENQVEEHNKKSYMTD 368
L E++ + S G +L+ +ITN+N ++ +K+++T+
Sbjct: 666 LAEMSDVYSFGILLLEIITNQNVIDHAREKAHITE 700
>At5g14420 unknown protein
Length = 468
Score = 29.6 bits (65), Expect = 6.0
Identities = 20/51 (39%), Positives = 27/51 (52%)
Query: 183 SISAGLGMAVARDIKGRRERVVAVISNWTTMSGQVYEAMSNAGYLDSNLVV 233
S SA + D K R ER + IS+ + QV EA++ AG SNL+V
Sbjct: 75 SYSAPPSQSYGSDNKKRLERKYSKISDDYSSLEQVTEALARAGLESSNLIV 125
>At5g01960 unknown protein
Length = 426
Score = 29.6 bits (65), Expect = 6.0
Identities = 22/61 (36%), Positives = 29/61 (47%), Gaps = 3/61 (4%)
Query: 299 ARGMMGPPGSTLFEELGLYYIGPVDGHNIEDLISVLQEVASLDSMGPVLIHVITNENQVE 358
A ++ P STL E GL+ P H E L+S + +S S G V I +I N Q
Sbjct: 23 ASNIIQAPISTLLEYSGLFRARPSPSHEAETLVS---DDSSGLSNGEVAIRIIGNTEQDA 79
Query: 359 E 359
E
Sbjct: 80 E 80
>At3g46340 receptor-like protein kinase homolog
Length = 889
Score = 29.6 bits (65), Expect = 6.0
Identities = 12/35 (34%), Positives = 24/35 (68%)
Query: 334 LQEVASLDSMGPVLIHVITNENQVEEHNKKSYMTD 368
L E++ + S G VL+ +ITN+ ++ +KS++T+
Sbjct: 765 LAEMSDVYSFGIVLLEIITNQRVIDPAREKSHITE 799
>At1g47410 unknown protein
Length = 189
Score = 29.6 bits (65), Expect = 6.0
Identities = 16/43 (37%), Positives = 24/43 (55%), Gaps = 4/43 (9%)
Query: 135 VGDQTYAHKILTG--RRSLMKTIRKKN--GLSGFTSRFESEYD 173
+GD Y H+++ R M+ +RKK G+ G T +F EYD
Sbjct: 41 IGDFDYDHEVVVFEIRSRAMEKLRKKRRTGMEGLTYKFVKEYD 83
>At5g54770 thiazole biosynthetic enzyme precursor (ARA6) (sp|Q38814)
Length = 349
Score = 29.3 bits (64), Expect = 7.8
Identities = 51/220 (23%), Positives = 92/220 (41%), Gaps = 22/220 (10%)
Query: 390 VAEAEKDKDIVVVHAGITTEPSLKLFMEKFPDRIFNVGIAEQHAVTFASGLSCGGLKPFC 449
VA E+D +VV HA + T + + + ++FN AE V G GG
Sbjct: 148 VAYDEQDTYVVVKHAALFTSTIMSKLLARPNVKLFNAVAAEDLIV---KGNRVGG----- 199
Query: 450 IIPSSFLQRAYDQVVHDVDQQKVPVRFVITSAGLVGSDGPLQCGAFDITFMSCLPNMIVM 509
++ + L +D + + V++S G DGP GA + + + MI
Sbjct: 200 VVTNWALVAQNHHTQSCMDPNVMEAKIVVSSC---GHDGPF--GATGVKRLKSI-GMIDH 253
Query: 510 APSDEAELVHMVATAAHINDQPVCFRYPRGALVGKDEAILDGIPIEIGK--GRILVEGKD 567
P +A ++ A + V P + G + A +DG P +G G +++ G+
Sbjct: 254 VPGMKALDMNTAEDAIVRLTREV---VPGMIVTGMEVAEIDGAP-RMGPTFGAMMISGQK 309
Query: 568 VALLGYGSM-VQNCLKAYSLLANLGIEVTVADARFCKPLD 606
L ++ + N + +L+ NL E+ +A A + +D
Sbjct: 310 AGQLALKALGLPNAIDG-TLVGNLSPELVLAAADSAETVD 348
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.137 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,164,448
Number of Sequences: 26719
Number of extensions: 641189
Number of successful extensions: 1627
Number of sequences better than 10.0: 19
Number of HSP's better than 10.0 without gapping: 11
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 1601
Number of HSP's gapped (non-prelim): 22
length of query: 700
length of database: 11,318,596
effective HSP length: 106
effective length of query: 594
effective length of database: 8,486,382
effective search space: 5040910908
effective search space used: 5040910908
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 64 (29.3 bits)
Medicago: description of AC148176.1


