

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147964.8 + phase: 0
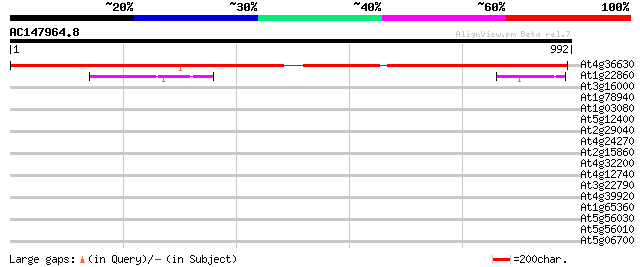
(992 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g36630 unknown protein 1200 0.0
At1g22860 unknown protein 49 1e-05
At3g16000 myosin heavy chain like protein 32 1.8
At1g78940 unknown protein 32 1.8
At1g03080 unknown protein 32 1.8
At5g12400 putative protein 32 2.3
At2g29040 hypothetical protein 32 2.3
At4g24270 unknown protein 31 3.0
At2g15860 unknown protein 31 3.0
At4g32200 putative protein 30 5.2
At4g12740 adenine DNA glycosylase like protein 30 5.2
At3g22790 unknown protein 30 5.2
At4g39920 unknown protein 30 6.8
At1g65360 30 6.8
At5g56030 HEAT SHOCK PROTEIN 81-2 (HSP81-2) (sp|P55737) 30 8.8
At5g56010 heat shock protein 90 30 8.8
At5g06700 unknown protein 30 8.8
>At4g36630 unknown protein
Length = 1003
Score = 1200 bits (3105), Expect = 0.0
Identities = 631/1038 (60%), Positives = 778/1038 (74%), Gaps = 92/1038 (8%)
Query: 1 MVHTAYDSFELLTNSTSKIESIESYGSKLLLGCSNGSLLVYAP-EHSVPAPEEMRKEAYV 59
MVH AYDSF+LL + ++I+++ESYGSKL GC +GSL +Y+P E S P E+ +E YV
Sbjct: 1 MVHNAYDSFQLLKDCPARIDAVESYGSKLFAGCYDGSLRIYSPPESSASDPSELHQETYV 60
Query: 60 LERNVNGFAKKAVVSLQVVESREFLLSLSESIAFHKLPTFETIAVITKAKGANAFCWDEH 119
LE+ V GF+KK +V+++V+ SRE LLSLSESIAFH LP ET+AVITKAKGANA+ WD+
Sbjct: 61 LEKTVAGFSKKPIVAMEVLASRELLLSLSESIAFHGLPNLETVAVITKAKGANAYSWDDR 120
Query: 120 RGFLCFARQKRVCIFRRDGGRGFVEVKDFGVLDVVKSMSWCGENICLGIRKAYVILNATS 179
RGFLCF+RQKRVC+F+ DGG GFVEV+D+GV D VKS+SWCGENICLGI+K YVILN +
Sbjct: 121 RGFLCFSRQKRVCVFKHDGGGGFVEVRDYGVPDTVKSISWCGENICLGIKKEYVILNTAN 180
Query: 180 GSISEVFTSGRLAPPLVVSLPSGELLLGKDNIGVIVDQNGKLRPEGRICWSEAPTEVVIQ 239
G++SEVF SGR+APPLV+SLPSGEL+LGK+NIGV VDQNGKL RICWSEAPT +VIQ
Sbjct: 181 GTLSEVFPSGRVAPPLVISLPSGELILGKENIGVFVDQNGKLLQTERICWSEAPTSIVIQ 240
Query: 240 NPYALALLPRFVEIRSLRGPYPLIQTIVFRNVRHLRQSNNSVIIALENSIHCLFPVPLGA 299
NPYA+ALLPR VE+R LR PYPLIQTIV +N+R L +SNN+VI+ L+NS++ LFPV +GA
Sbjct: 241 NPYAIALLPRRVEVRLLRSPYPLIQTIVLQNIRRLVKSNNAVIVGLDNSVYVLFPVSIGA 300
Query: 300 Q-------------------------------------------IVQLTAAGNFEEALSL 316
Q IVQLTA+GNFEEAL+L
Sbjct: 301 QVFTLALRILMFLAAAHGFLWMNMKMCFILPNISVEYDTISVFFIVQLTASGNFEEALAL 360
Query: 317 CKLLPPEDSNLRAAKEDSIHIRYAHYLFDNGSYEESMEHFLASQVDITYVLSLYTSIILP 376
CK+LPP++S+LRAAKE SIH R+AHYLF+NGSYEE+MEHFLASQVDIT+VLS+Y SIILP
Sbjct: 361 CKVLPPDESSLRAAKESSIHTRFAHYLFENGSYEEAMEHFLASQVDITHVLSMYPSIILP 420
Query: 377 KTTIVHDSDKL-DIFGDPLHLSRGSS-MSDDMEPSSASNMSELDDNAELESKKMSHNMLM 434
KTTI+ DK+ DI GD LSRGSS +SDDME SS E +DNA+LESKKMSHN LM
Sbjct: 421 KTTIIPQPDKMVDISGDEASLSRGSSGISDDMESSSPRYFLESEDNADLESKKMSHNTLM 480
Query: 435 ALIKFLHKKRHSIIEKATAEGTEEVVFDAVGNNFESYNSNRFKKINKRHGSIPVSSEARE 494
ALIK+L K+R ++IEKAT+EGTEEV+ DAVG + + +S++ KK +K +I
Sbjct: 481 ALIKYLLKRRPAVIEKATSEGTEEVISDAVGKTYGANDSSKSKKSSKSGAAI-------- 532
Query: 495 MASILDTALLQAMLLTGQPSMAENLLRVLNYCDLKICEEILQEGSYHVSLVELYKCNSMH 554
LL+ +NY D+KICEEIL + + +L+EL+K NSMH
Sbjct: 533 -----------------------ELLKGVNYSDVKICEEILMKSKNYSALLELFKSNSMH 569
Query: 555 REALEIINKSVKESESSQSK--IAHRFKPEAIIEYLKPLCELDTTLVLEYSMLVLESCPT 612
EAL+++N+ ES+++QS+ + F PE IIEYLKPLC D LVLEYSMLVLESCPT
Sbjct: 570 HEALKLLNQLADESKTNQSQTDVTQIFSPELIIEYLKPLCRTDPMLVLEYSMLVLESCPT 629
Query: 613 QTIELFLSGNIPADMVNLYLKQHAPNLQATYLELVLSMNEGAVSGTLQNEMVHLYLSEVL 672
QTI+LFLSGNI AD+VN YLKQHAPN+Q YLEL+++MN+ AV +YLSEVL
Sbjct: 630 QTIDLFLSGNISADLVNSYLKQHAPNMQGRYLELMMAMNDTAVQ---------IYLSEVL 680
Query: 673 DWHADLSSEQKWDEKVYSPKRKKLLSALESISGYNPEALLKLLPSDALYEERAILLGKMN 732
D +A S++QKWDEK + P+RKKLLSALESISGY+P+ LLK LP DALYEERA++LGKMN
Sbjct: 681 DLYAAKSAQQKWDEKDHPPERKKLLSALESISGYSPQPLLKRLPRDALYEERAVILGKMN 740
Query: 733 QHELALSLYVHKLHVPELALSYCDHVYESA-HKSSVKSLSNIYLMLLQIYLNPRRTTKNY 791
QHELALS+YVHKLH P+LAL+YCD +YES + S K SNIYL +LQIYLNP+++ K++
Sbjct: 741 QHELALSIYVHKLHAPDLALAYCDRIYESVTYLPSGKPSSNIYLTVLQIYLNPKKSAKDF 800
Query: 792 EKKISNLLSPRNKSIRKVTSKSLSRTMSRG-SKKIAAIEIAEDAKA--SQSSDSGRSDAD 848
K+I L S + K+ LS G SKKI AIE AED + S S+DSGRSD D
Sbjct: 801 AKRIVALGSFESSDTTKMMDSVLSSKAKGGRSKKIVAIEGAEDMRVGLSSSTDSGRSDVD 860
Query: 849 TEEFTEEECTSIMLDEALDLLSRRWDRINGAQALKLLPKETKLQNLLPILGPLVRKSSEM 908
TEE EE +++M+ E LDLLS+RW+RINGAQALKLLP+ETKL NLLP L PL+R SSE
Sbjct: 861 TEEPLEEGDSTVMISEVLDLLSQRWERINGAQALKLLPRETKLHNLLPFLAPLLRNSSEA 920
Query: 909 YRNCSVVRSLRQSENLQVKDELYNKRKAVIKISDDNMCSLCHKKIGTSVFAVYPNGKTLV 968
+RN SV++SLRQSENLQVK+ELY RK V +++ ++MCSLC+KKIGTSVFAVYPNGKTLV
Sbjct: 921 HRNFSVIKSLRQSENLQVKEELYKHRKGVAQVTSESMCSLCNKKIGTSVFAVYPNGKTLV 980
Query: 969 HFVCFRDSQSMKAVAKVS 986
HFVCFRDSQ MKAV+K +
Sbjct: 981 HFVCFRDSQGMKAVSKTT 998
>At1g22860 unknown protein
Length = 947
Score = 48.9 bits (115), Expect = 1e-05
Identities = 52/231 (22%), Positives = 101/231 (43%), Gaps = 17/231 (7%)
Query: 142 FVEVKDFGVLDVVKSMSWCGENICLGIRKAYVILNATSGSISEVFTSGRLAPPLVVSLPS 201
FV +K+ + +K++ W + + G K Y +++ +G +FT ++ P ++ L
Sbjct: 200 FVVLKEILGIGGIKTLVWLDDYVIAGTVKGYSLISCVTGLSGVIFTLPDVSGPPLLKLLC 259
Query: 202 GE--LLLGKDNIGVIVDQNGKLRPEGRICWSEAPTEVVIQNPYALALLPRFVEIRSLRGP 259
E +LL DN+GV+VD NG+ G + + P V + Y + + +EI +
Sbjct: 260 KEWKVLLLVDNVGVVVDTNGQ-PIGGSLVFRRRPDSVGELSFYLVTVGDGKMEIHQKKSG 318
Query: 260 YPLIQTIVFRN--------VRHLRQSNNSVIIALENSIHCLFPVPLGAQIVQLTAAGNFE 311
+Q++ F N +++ + + VP QI L +
Sbjct: 319 -ACVQSVSFGPQGCGPSLLAADEAGDGNLLVVTTLSKLIFYRRVPYEEQIKDLLRKKRYR 377
Query: 312 EALSLCKLLPPEDSNLRAAKE--DSIHIRYAHYLFDNGSYEESMEHFLASQ 360
E +SL + L DS +K+ +H + + L + +EE++ FL S+
Sbjct: 378 ETISLVEEL---DSQGEISKDMLSFLHAQIGYLLLFDLRFEEAVNQFLKSE 425
Score = 44.7 bits (104), Expect = 3e-04
Identities = 29/135 (21%), Positives = 63/135 (46%), Gaps = 14/135 (10%)
Query: 861 MLDEALDLLSRRWDRINGAQALKLLPKETKLQNLLPILGP-------------LVRKSSE 907
M A+ LL + ++ Q L L + + L P ++R
Sbjct: 795 MFKAAVRLLHNHGESLDPLQVLDLFDDRADIFFYMQKLSPDMPLKLASDTILRMLRARVH 854
Query: 908 MYRNCSVVRSLRQSENLQVKDELYNKRKAVIKISDDNMCSLCHKKIGTSVFAVYPNGKTL 967
+R +V ++ ++ ++ + +R ++I+D+++C C+ ++GT +FA+YP+ T+
Sbjct: 855 HHRQGQIVHNISRALDVDSRLARLEERSRHMQINDESLCDSCYARLGTKLFAMYPD-DTI 913
Query: 968 VHFVCFRDSQSMKAV 982
V + C+R K+V
Sbjct: 914 VCYKCYRRLGESKSV 928
Score = 37.4 bits (85), Expect = 0.042
Identities = 61/293 (20%), Positives = 118/293 (39%), Gaps = 51/293 (17%)
Query: 490 SEAREMASILDTALLQAMLLTGQPSMAENLLRVLNYCDLKICEEILQEGSYHVSLVELYK 549
S+ A +LD+A+ + ENL N C ++ E +L E + +L LY
Sbjct: 494 SDPPSRADLLDSAIKNITRALNRVEDMENLASSGNNCVVEELETLLTESGHLRTLAFLYA 553
Query: 550 CNSMHREAL----------------------------EIINKSVKESESSQSK--IAHRF 579
M +AL E+I S KE+ ++++ +
Sbjct: 554 TKGMGAKALAIWRLFTKNYSSGLWQDSDDLVPYLHDNELIRLSGKEAAAAEAARILEEPC 613
Query: 580 KPEAIIEYLKPLCELDTTL---VLEYSMLVLESCPTQTIELFLSGNIPADMVNLYLK--- 633
PE +++L + +++ VL E P Q I+ + +++ Y +
Sbjct: 614 DPELALQHLSWIADVNPLFAIQVLTSDKRTEELSPEQVIQAIDPKKV--EIIQRYFQWLI 671
Query: 634 ----QHAPNLQATYLELVLSMNEGAVSGT-LQNEMVHLYLSEVLDWH-ADLSSEQKWDEK 687
P L +Y LS+ A+ +QN + + + H +++ S ++
Sbjct: 672 EERDYTDPQLHTSY---ALSLARSALECVEVQNGIQEADVPNGSEAHDSNVGSISLFEVD 728
Query: 688 VYSPKRKKLLSALESISGYNPEALLKLLPSDALYEERAILLGKMNQHELALSL 740
V R++L + L+S Y+PE +L+L+ L+ E+AIL ++ + L L +
Sbjct: 729 V----RERLQAFLQSSDLYDPEEILELVEGSELWLEKAILYRRIGKETLVLQI 777
>At3g16000 myosin heavy chain like protein
Length = 727
Score = 32.0 bits (71), Expect = 1.8
Identities = 35/170 (20%), Positives = 67/170 (38%), Gaps = 18/170 (10%)
Query: 789 KNYEKKISNLLSPRNKSIRKVTSKSLSRTMSRGSKKIAAIEIAEDAKASQSSDSGRSD-- 846
K++E K+ + R K + K + LS S K E+ + + + D
Sbjct: 178 KDFEAKLQHEQEERKKEVEKAKEEQLSLINQLNSAKDLVTELGRELSSEKKLCEKLKDQI 237
Query: 847 -----ADTEEFTEEECTSIMLDEALDLLSRRWDRINGAQALKLLPKETKLQNL------- 894
+ ++ ++E L E LDL+ DRIN +L+L E K Q
Sbjct: 238 ESLENSLSKAGEDKEALETKLREKLDLVEGLQDRIN-LLSLELKDSEEKAQRFNASLAKK 296
Query: 895 ---LPILGPLVRKSSEMYRNCSVVRSLRQSENLQVKDELYNKRKAVIKIS 941
L L + ++S + ++ E ++ + EL +K A+ +++
Sbjct: 297 EAELKELNSIYTQTSRDLAEAKLEIKQQKEELIRTQSELDSKNSAIEELN 346
>At1g78940 unknown protein
Length = 740
Score = 32.0 bits (71), Expect = 1.8
Identities = 59/263 (22%), Positives = 110/263 (41%), Gaps = 34/263 (12%)
Query: 384 SDKLDI-FGDPLHLSRGSSMSDDMEPSSASNMSELDD-NAELESKKMSHNMLMALIKFLH 441
S++L I F DP L+ S+ S++ +S+ + LDD AE++ ++ M +
Sbjct: 276 SNRLGIKFSDPDFLNESSTFSEESGRTSSYSSQSLDDVEAEMKRLRLELKQTMDMYSTAC 335
Query: 442 KKRHSIIEKAT----AEGTEEVVFDAVGNNFESYNSNRFKKINKRHGSIPVSSEAREMAS 497
K+ S ++AT EE + ++ E+ S K+ K ++ + A+ +A
Sbjct: 336 KEALSARQQATELQKLRTEEERRLEEAKSSEEAAMSIVEKERAKAKAALEAAEAAKRLAE 395
Query: 498 ILDTALLQAMLLT--GQPSMAENLLRVLNYCDLKICE--------EILQEGSYHVSLVEL 547
+ L A + T S + +R Y +I E + + EG Y
Sbjct: 396 VESKRRLTAEMKTMKESDSFSRGFVRYRKYTVDEIEEATSNFAESQKVGEGGY------- 448
Query: 548 YKCNSMHREALEIINKSVKESESSQSKIAHRFKPEAIIEYLKPLCELDTTLVL----EYS 603
+ R L+ + +VK ++ +F+ E +E L + + L+L E+
Sbjct: 449 ---GPVFRGFLDHTSVAVKVLRPDAAQGRSQFQKE--VEVLSCIRHPNMVLLLGACPEFG 503
Query: 604 MLVLESCPTQTIE--LFLSGNIP 624
+LV E ++E LF+ GN P
Sbjct: 504 ILVYEYMAKGSLEDRLFMRGNTP 526
>At1g03080 unknown protein
Length = 1744
Score = 32.0 bits (71), Expect = 1.8
Identities = 28/90 (31%), Positives = 45/90 (49%), Gaps = 6/90 (6%)
Query: 518 NLLRVLNYCDLKICEEILQEGSYHVSLVELYKCNSMHREALEIINKSVKESESSQSKIAH 577
N L+ YC + EE+ Q G H S+VE + +H E+ K ++E S +I
Sbjct: 594 NALQQEIYC---LKEELSQIGKKHQSMVEQVELVGLHPESFGSSVKELQEENSKLKEIRE 650
Query: 578 R--FKPEAIIEYLKPLCEL-DTTLVLEYSM 604
R + A+IE L+ + +L L+LE S+
Sbjct: 651 RESIEKTALIEKLEMMEKLVQKNLLLENSI 680
>At5g12400 putative protein
Length = 1595
Score = 31.6 bits (70), Expect = 2.3
Identities = 41/181 (22%), Positives = 74/181 (40%), Gaps = 21/181 (11%)
Query: 808 KVTSKSLSRTMSR--GSKKIAAIEIAEDAKASQSSDSGRSDADTEEFTEEECTSIMLDEA 865
KV ++L +R GS I I E++++++ S +A E T E + +
Sbjct: 1123 KVLPRALISKAARQGGSMNIPGIFYPENSESAKRSRRVAWEAAVESSTTSEQLGLQIRTL 1182
Query: 866 LDLLSRRWDRINGAQALKLLPKETKLQNLLPILGPLVRKSSEMYRNCSVVRSLRQSENLQ 925
+ +WD I + +LLP L RKS+ +++ V R + E ++
Sbjct: 1183 QSYI--KWDDIENS-------------HLLPTLDKESRKSARLFKKAIVRRKCTEEETVK 1227
Query: 926 VKDELYNKRKAVIKISDDNMCSLCHKKIGTSVFAVYPNGKTLVHFVCFRDSQSMKAVAKV 985
+ + KR+ + + N C + G F + + L H V + + KAV K
Sbjct: 1228 YLLD-FGKRRNIPDVVSKNGCMVEESSSGRKRFWLNESHVPL-HLV--KGFEEKKAVRKT 1283
Query: 986 S 986
S
Sbjct: 1284 S 1284
>At2g29040 hypothetical protein
Length = 720
Score = 31.6 bits (70), Expect = 2.3
Identities = 20/57 (35%), Positives = 31/57 (54%), Gaps = 10/57 (17%)
Query: 376 PKTTIVHDSDKLDIFGDPLHLSRGSSMSDDMEPSSASNMSELDDNAELESKKMSHNM 432
PK +V D+D D L RG + +D S+SN+ E D++ +LE+KK N+
Sbjct: 192 PKKALVDDND------DDLETKRGKELPND----SSSNVVEDDNDDDLETKKGKDNI 238
>At4g24270 unknown protein
Length = 816
Score = 31.2 bits (69), Expect = 3.0
Identities = 22/69 (31%), Positives = 31/69 (44%), Gaps = 2/69 (2%)
Query: 826 AAIEIAEDAKASQSSDSGRSDADTEEFTEEECTSIMLDEALDLLSRRWDRINGAQALKLL 885
A+ E A S DSG SD+D+E+ E + L+ L +D Q +KLL
Sbjct: 19 ASAENPARADPPSSDDSGDSDSDSEDEAESNQQIVTLESELSANPYNYDAY--VQYIKLL 76
Query: 886 PKETKLQNL 894
K L+ L
Sbjct: 77 RKTANLEKL 85
>At2g15860 unknown protein
Length = 512
Score = 31.2 bits (69), Expect = 3.0
Identities = 24/96 (25%), Positives = 47/96 (48%), Gaps = 10/96 (10%)
Query: 797 NLLSPRNKSIRKVTSKSLSRTMSRGSKKIAAI-EIAEDAKASQSSDSGRSDADTEEFTEE 855
++LS K+ + S++ + + +K IA + E+ ED+++S + +ADTE+ ++
Sbjct: 53 SVLSDLQKAAEDI-SRNAAAVAEKAAKSIAEMGEVDEDSESSAKEEEKTEEADTEQDSD- 110
Query: 856 ECTSIMLDEALDLLSRRWDRINGAQALKLLPKETKL 891
DE L +R+ GA LL + K+
Sbjct: 111 -------DENAKLKKSALERLEGASEESLLSQGLKV 139
>At4g32200 putative protein
Length = 1552
Score = 30.4 bits (67), Expect = 5.2
Identities = 35/157 (22%), Positives = 75/157 (47%), Gaps = 5/157 (3%)
Query: 786 RTTKNYEKKISNLLSPRNKSIRKVTSKSLSRTMSRGSKKIAAIEIAEDAKASQSSDSGRS 845
RTTK + ++ + + + V K + +RG+++I I + AK + S SG
Sbjct: 1271 RTTKKAQARLDRVKAYLKEQEDLVGPKVDAENQARGAEEIVGILMQRGAKIAASELSGLK 1330
Query: 846 DADTEEFTEEECTSIMLDEALDLLSRRWDRIN-GAQALKLLPKETKLQNLLPILGPLVRK 904
+ T+ T+E +++ D L+ D++ Q ++ P + + + +L ++R+
Sbjct: 1331 EL-TKRATDEVNALNVIELGDDDLNMSPDQLGFSRQISQVAPVADQHGSNVDLLAEVIRR 1389
Query: 905 -SSEMYRNCSVVRSLRQSENLQVKDEL--YNKRKAVI 938
SSE R S +S + + ++ L Y++R++ I
Sbjct: 1390 ASSEERREAEGFLSCVESRHKEYREMLRYYDERRSDI 1426
>At4g12740 adenine DNA glycosylase like protein
Length = 608
Score = 30.4 bits (67), Expect = 5.2
Identities = 21/66 (31%), Positives = 33/66 (49%), Gaps = 2/66 (3%)
Query: 793 KKISNLLSPRNKSIRKV-TSKSLSRTMSRGSKKIA-AIEIAEDAKASQSSDSGRSDADTE 850
K +S +PR K +RK K R R +++ A E AE+A+A + +++ E
Sbjct: 41 KTMSQSFAPREKLMRKCREKKEAEREAEREAEREAEEEEKAEEAEAEADKEEAEEESEEE 100
Query: 851 EFTEEE 856
E EEE
Sbjct: 101 EEEEEE 106
>At3g22790 unknown protein
Length = 1694
Score = 30.4 bits (67), Expect = 5.2
Identities = 51/217 (23%), Positives = 88/217 (40%), Gaps = 17/217 (7%)
Query: 424 ESKKMSHNMLMALIKFLHKKRHSIIEKATAEGTEEVVFDAVGN----NFESY--NSNRFK 477
+S + + L I L ++ +I+E+A A VV+ ++G+ E++ N N +
Sbjct: 1044 KSLHLKFSELKGEICILEEENGAILEEAIALNNVSVVYQSLGSEKAEQAEAFAKNLNSLQ 1103
Query: 478 KINKRHGSIPVSSEAREMASILDTALLQAMLLTGQPSMAE-NLLRVLNYCDLKICEEILQ 536
IN + E +D+ L + L Q S+ E N L L + + EE L+
Sbjct: 1104 NINSGLKQKVETLEEILKGKEVDSQELNSKLEKLQESLEEANELNDLLEHQILVKEETLR 1163
Query: 537 EGSYHV-----SLVELYKCNSMHREALEIINKSVKESESSQSKIAHRFKPEAII-----E 586
+ + + L + N+ EA+E + K KES + + R + E
Sbjct: 1164 QKAIELLEAEEMLKATHNANAELCEAVEELRKDCKESRKLKGNLEKRNSELCDLAGRQDE 1223
Query: 587 YLKPLCELDTTLVLEYSMLVLESCPTQTIELFLSGNI 623
+K L L L E +L E + E FLS +
Sbjct: 1224 EIKILSNLKENLESEVKLLHKEIQEHRVREEFLSSEL 1260
>At4g39920 unknown protein
Length = 345
Score = 30.0 bits (66), Expect = 6.8
Identities = 33/120 (27%), Positives = 53/120 (43%), Gaps = 9/120 (7%)
Query: 800 SPRNKSIRKVTSKSLSRTMSRGSKKIAAIE--IAEDAKASQSSDSGRSDADTEEFTEEEC 857
SP + S T +S S +++ S +IE IAE AS S+DS + +D E
Sbjct: 39 SPDSSSSSSSTLESTSSFLAKFSDSKRSIESRIAESRLASSSTDSSKLKSDLAEI----- 93
Query: 858 TSIMLDEALDLLSRRWDRINGAQALKLLPKETKLQNLLPIL-GPLVRKSSEMYRNCSVVR 916
S+ +D LL+ + + L + L+ L IL G LV K +++ S +
Sbjct: 94 -SVAIDNLEKLLAENSYFLPSYEVRSSLKIVSDLKQSLDILSGELVPKKKFSFKSKSTTK 152
>At1g65360
Length = 226
Score = 30.0 bits (66), Expect = 6.8
Identities = 21/56 (37%), Positives = 30/56 (53%), Gaps = 6/56 (10%)
Query: 910 RNCSVVRSLRQSENLQVKDELYNKRKAVIKISDDNMCSLCHKKIGTSVFAVYPNGK 965
R +V+ ++S NLQV ++KRKA + C+LC KI VF+ P GK
Sbjct: 9 RKVEIVKMTKES-NLQVT---FSKRKAGLFKKASEFCTLCDAKIAMIVFS--PAGK 58
>At5g56030 HEAT SHOCK PROTEIN 81-2 (HSP81-2) (sp|P55737)
Length = 699
Score = 29.6 bits (65), Expect = 8.8
Identities = 21/65 (32%), Positives = 33/65 (50%), Gaps = 5/65 (7%)
Query: 366 VLSLYTSIILPKTTIVHDSDKLDIFGDPLH--LSRGSSMSDDMEPSSASNMSELDDNAEL 423
VL L+ + +L T D+ + FG +H L G S+ DD + + M L+D+A+
Sbjct: 634 VLLLFETALL---TSGFSLDEPNTFGSRIHRMLKLGLSIDDDDAVEADAEMPPLEDDADA 690
Query: 424 ESKKM 428
E KM
Sbjct: 691 EGSKM 695
>At5g56010 heat shock protein 90
Length = 699
Score = 29.6 bits (65), Expect = 8.8
Identities = 21/65 (32%), Positives = 34/65 (52%), Gaps = 5/65 (7%)
Query: 366 VLSLYTSIILPKTTIVHDSDKLDIFGDPLH--LSRGSSMSDDMEPSSASNMSELDDNAEL 423
VL L+ + +L T D+ + FG +H L G S+ DD + ++M L+D+A+
Sbjct: 634 VLLLFETALL---TSGFSLDEPNTFGSRIHRMLKLGLSIDDDDVVEADADMPPLEDDADA 690
Query: 424 ESKKM 428
E KM
Sbjct: 691 EGSKM 695
>At5g06700 unknown protein
Length = 608
Score = 29.6 bits (65), Expect = 8.8
Identities = 22/86 (25%), Positives = 41/86 (47%), Gaps = 5/86 (5%)
Query: 830 IAEDAKASQSSDSGRSDADTEEFTEEECTSIMLDEA---LDLLSRRWDRINGAQALKLLP 886
+++ + + S++ R+ ADTE + TS +A +DL + N + A
Sbjct: 172 LSDKSSTTGSNNQSRTTADTETVNRNQTTSPAPSKAPVSVDLKTN--SSSNSSTASSTPK 229
Query: 887 KETKLQNLLPILGPLVRKSSEMYRNC 912
K+TK +L+ + + K SE +NC
Sbjct: 230 KQTKTVDLVSSVKQEIEKWSESLKNC 255
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.133 0.373
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,851,665
Number of Sequences: 26719
Number of extensions: 887461
Number of successful extensions: 2916
Number of sequences better than 10.0: 17
Number of HSP's better than 10.0 without gapping: 2
Number of HSP's successfully gapped in prelim test: 15
Number of HSP's that attempted gapping in prelim test: 2895
Number of HSP's gapped (non-prelim): 25
length of query: 992
length of database: 11,318,596
effective HSP length: 109
effective length of query: 883
effective length of database: 8,406,225
effective search space: 7422696675
effective search space used: 7422696675
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 65 (29.6 bits)
Medicago: description of AC147964.8


