

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147964.5 + phase: 0
(447 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
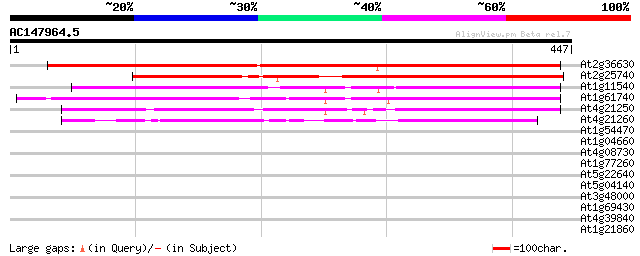
Score E
Sequences producing significant alignments: (bits) Value
At2g36630 unknown protein 419 e-117
At2g25740 unknown protein 405 e-113
At1g11540 hypothetical protein; similar to EST gb|AI998648.1 226 2e-59
At1g61740 unknown protein 220 1e-57
At4g21250 hypothetical protein 214 1e-55
At4g21260 hypothetical protein 145 4e-35
At1g54470 hypothetical protein 40 0.002
At1g04660 unknown protein 31 1.6
At4g08730 hypothetical protein 30 3.5
At1g77260 unknown protein 30 3.5
At5g22640 unknown protein 29 6.0
At5g04140 ferredoxin-dependent glutamate synthase 29 6.0
At3g48000 aldehyde dehydrogenase (NAD+)-like protein 29 6.0
At1g69430 hypothetical protein 29 6.0
At4g39840 unknown protein 28 7.9
At1g21860 pectinesterase, putative 28 7.9
>At2g36630 unknown protein
Length = 459
Score = 419 bits (1078), Expect = e-117
Identities = 202/417 (48%), Positives = 297/417 (70%), Gaps = 10/417 (2%)
Query: 31 SGESSYERVWPEMKFGWRIVVGSIVGFFGAALGSVGGVGGGGIFIPMLTLIIGFDPKSST 90
S S+ E++WP++KF W++V+ +++ F G+A G+VGGVGGGGIF+PMLTLI+GFD KS+
Sbjct: 42 SSLSATEKIWPDLKFSWKLVLATVIAFLGSACGTVGGVGGGGIFVPMLTLILGFDTKSAA 101
Query: 91 ALSKCMITGAAGSTVYYNLRLRHPTLDMPLIDYDLALLFQPMLMLGISIGVAFNVMFADW 150
A+SKCMI GA+ S+V+YN+R+RHPT ++P++DYDLALLFQPML+LGI++GV+ +V+F W
Sbjct: 102 AISKCMIMGASASSVWYNVRVRHPTKEVPILDYDLALLFQPMLLLGITVGVSLSVVFPYW 161
Query: 151 MVTILLIILFIGTSTKALVKGIDTWKKETIMKKEAFEEAAQMLESGSTPDYASEEDYKSL 210
++T+L+IILF+GTS+++ KGI+ WK+ET++K E ++ A M+ S + + +Y+ L
Sbjct: 162 LITVLIIILFVGTSSRSFFKGIEMWKEETLLKNEMAQQRANMVNSRG--ELLIDTEYEPL 219
Query: 211 -PADLQDEEVPLLDNIHWKELSVLMYVWVAFLIVQILKTYSKTCSIEYWLLNSLQVPIAI 269
P + + E + N+ WK L +L+ VW+ FL++QI+K K CS YW+L +Q P+A+
Sbjct: 220 YPREEKSELEIIRSNLKWKGLLILVTVWLTFLLIQIVKNEIKVCSTIYWILFIVQFPVAL 279
Query: 270 SVTLFEAICLCKGTRVIASRGK-------EITWKFHKICLYCFCGIIAGMVSGLLGLGGG 322
+V FEA L + + G I W + CG+I G+V GLLG GGG
Sbjct: 280 AVFGFEASKLYTANKKRLNSGNTECICEATIEWTPLSLIFCGLCGLIGGIVGGLLGSGGG 339
Query: 323 FILGPLFLELGIPPQVASATSTFAMVFSSSMSVVQYYHLDRFPIPYASYLVLVATIAALT 382
F+LGPL LE+G+ PQVASAT+TF M+FSSS+SVV++Y L RFPIPYA YL+ V+ +A
Sbjct: 340 FVLGPLLLEIGVIPQVASATATFVMMFSSSLSVVEFYLLKRFPIPYAMYLISVSILAGFW 399
Query: 383 GQHVVRKIIAIFGRASIIVFILAFTIFVSAISLGGVGIGNMVEKMENEEYMGFDNLC 439
GQ +RK++AI RASIIVF+L+ I SA+++G +GI ++ + N E+MGF C
Sbjct: 400 GQSFIRKLVAILRRASIIVFVLSGVICASALTMGVIGIEKSIKMIHNHEFMGFLGFC 456
>At2g25740 unknown protein
Length = 922
Score = 405 bits (1042), Expect = e-113
Identities = 211/351 (60%), Positives = 267/351 (75%), Gaps = 31/351 (8%)
Query: 99 GAAGSTVYYNLRLRHPTLDMPLIDYDLALLFQPMLMLGISIGVAFNVMFADWMVTILLII 158
GA+ STVYYNLRLRHPTLDMP+IDYDLALL QPMLMLGISIGVAFNV+F DW+VT+LLI+
Sbjct: 2 GASVSTVYYNLRLRHPTLDMPIIDYDLALLIQPMLMLGISIGVAFNVIFPDWLVTVLLIV 61
Query: 159 LFIGTSTKALVKGIDTWKKETIMKKEAFEEAAQMLESGSTPDYASEEDYKSLPA------ 212
LF+GTSTKA +KG +TW KETI KKEA A+ LES +E +Y LPA
Sbjct: 62 LFLGTSTKAFLKGSETWNKETIEKKEA----AKRLESNGVS--GTEVEYVPLPAAPSTNP 115
Query: 213 -DLQDEEVPLLDNIHWKELSVLMYVWVAFLIVQILKTYSKTCSIEYWLLNSLQVPIAISV 271
+ + EEV +++N++WKEL +L++VW+ FL +QI K +P+A+ V
Sbjct: 116 GNKKKEEVSIIENVYWKELGLLVFVWIVFLALQISK-----------------IPVAVGV 158
Query: 272 TLFEAICLCKGTRVIASRGK-EITWKFHKICLYCFCGIIAGMVSGLLGLGGGFILGPLFL 330
+ +EA+ L +G R+IAS+G+ + + ++ +YC GIIAG+V GLLGLGGGFI+GPLFL
Sbjct: 159 SGYEAVALYQGRRIIASKGQGDSNFTVGQLVMYCTFGIIAGIVGGLLGLGGGFIMGPLFL 218
Query: 331 ELGIPPQVASATSTFAMVFSSSMSVVQYYHLDRFPIPYASYLVLVATIAALTGQHVVRKI 390
ELG+PPQV+SAT+TFAM FSSSMSVV+YY L RFP+PYA YLV VATIAA GQHVVR++
Sbjct: 219 ELGVPPQVSSATATFAMTFSSSMSVVEYYLLKRFPVPYALYLVGVATIAAWVGQHVVRRL 278
Query: 391 IAIFGRASIIVFILAFTIFVSAISLGGVGIGNMVEKMENEEYMGFDNLCHH 441
IA GRAS+I+FILA IF+SAISLGGVGI NM+ K++ EYMGF+NLC +
Sbjct: 279 IAAIGRASLIIFILASMIFISAISLGGVGIVNMIGKIQRHEYMGFENLCKY 329
>At1g11540 hypothetical protein; similar to EST gb|AI998648.1
Length = 450
Score = 226 bits (577), Expect = 2e-59
Identities = 133/409 (32%), Positives = 218/409 (52%), Gaps = 33/409 (8%)
Query: 50 VVGSIVGFFGAALGSVGGVGGGGIFIPMLTLIIGFDPKSSTALSKCMITGAAGSTVYYNL 109
++ +++ FF A++ S GG+GGGG+F+ ++T+I G + K++++ S M+TG + + V NL
Sbjct: 56 IIAAVLSFFAASISSAGGIGGGGLFLSIMTIIAGLEMKTASSFSAFMVTGVSFANVGCNL 115
Query: 110 RLRHP-TLDMPLIDYDLALLFQPMLMLGISIGVAFNVMFADWMVTILLIILFIGTSTKAL 168
LR+P + D LID+DLAL QP L+LG+SIGV N MF +W+V L + ++ K
Sbjct: 116 FLRNPKSRDKTLIDFDLALTIQPCLLLGVSIGVICNRMFPNWLVLFLFAVFLAWSTMKTC 175
Query: 169 VKGIDTWKKETIMKKEAFEEAAQMLESGSTPDYASEEDYKSLPADLQDEEVPLLDNIHWK 228
KG+ W E+ K +E +P + E + D + W
Sbjct: 176 KKGVSYWNLESERAKIKSPRDVDGIEVARSPLLSEERE---------DVRQRGMIRFPWM 226
Query: 229 ELSVLMYVWVAFLIVQILKTYS--------KTCSIEYWLLNSLQVPIAISVTLFEAICLC 280
+L VL+ +W+ F + + + K C YW L+SLQ+P+ T+F +C+
Sbjct: 227 KLGVLVIIWLLFFSINLFRGNKYGQGIISIKPCGALYWFLSSLQIPL----TIFFTLCIY 282
Query: 281 KGTRVIASRGKE----------ITWKFHKICLYCFCGIIAGMVSGLLGLGGGFILGPLFL 330
V ++ + + +K+ L ++AG++ GL G+GGG ++ PL L
Sbjct: 283 FSDNVQSNHTSHSNQNSEQETGVGGRQNKLMLPVMA-LLAGVLGGLFGIGGGMLISPLLL 341
Query: 331 ELGIPPQVASATSTFAMVFSSSMSVVQYYHLDRFPIPYASYLVLVATIAALTGQHVVRKI 390
++GI P+V +AT +F ++FSSSMS +QY L A+ LV +A+L G VV+K+
Sbjct: 342 QIGIAPEVTAATCSFMVLFSSSMSAIQYLLLGMEHAGTAAIFALVCFVASLVGLMVVKKV 401
Query: 391 IAIFGRASIIVFILAFTIFVSAISLGGVGIGNMVEKMENEEYMGFDNLC 439
IA +GRASIIVF + + +S + + G N+ + YMGF C
Sbjct: 402 IAKYGRASIIVFAVGIVMALSTVLMTTHGAFNVWNDFVSGRYMGFKLPC 450
>At1g61740 unknown protein
Length = 458
Score = 220 bits (561), Expect = 1e-57
Identities = 134/457 (29%), Positives = 242/457 (52%), Gaps = 40/457 (8%)
Query: 6 SLAERVLEDEKPEIVVMKKTSFFWYSGESSYERVWPEMKFGWRIVVGSIVGFFGAALGSV 65
S+AE+ P ++ KTS + + S + P ++ ++ ++ F +++ S
Sbjct: 19 SIAEQEPSILSPVDQLLNKTSSYL---DFSTKFNQPRIELTTSTIIAGLLSFLASSISSA 75
Query: 66 GGVGGGGIFIPMLTLIIGFDPKSSTALSKCMITGAAGSTVYYNLRLRHPTLD-MPLIDYD 124
GG+GGGG+++P++T++ G D K++++ S M+TG + + V NL +R+P LID+D
Sbjct: 76 GGIGGGGLYVPIMTIVAGLDLKTASSFSAFMVTGGSIANVGCNLFVRNPKSGGKTLIDFD 135
Query: 125 LALLFQPMLMLGISIGVAFNVMFADWMVTILLIILFIGTSTKALVKGIDTWKKETIMKKE 184
LALL +P ++LG+SIGV N++F +W++T L + ++ K G+ W+ E+ M K
Sbjct: 136 LALLLEPCMLLGVSIGVICNLVFPNWLITSLFAVFLAWSTLKTFGNGLYYWRLESEMVK- 194
Query: 185 AFEEAAQMLESGSTPDYASEEDYKSLPADL-QDEEVPLLDNIHWKELSVLMYVWVAFLIV 243
+ ES + E+ +SL L +D + P W +L VL+ +W+++ V
Sbjct: 195 -------IRESNRIEEDDEEDKIESLKLPLLEDYQRP--KRFPWIKLGVLVIIWLSYFAV 245
Query: 244 QILKTYS--------KTCSIEYWLLNSLQVPIAISVTLFEAICLCKGTRVIASRGKEITW 295
+L+ + C YWL++S Q+P+ TLF + +C V + + +
Sbjct: 246 YLLRGNKYGEGIISIEPCGNAYWLISSSQIPL----TLFFTLWICFSDNVQSQQQSDYHV 301
Query: 296 KFHKI-------------CLYCFCGIIAGMVSGLLGLGGGFILGPLFLELGIPPQVASAT 342
+ C++ ++AG++ G+ G+GGG ++ PL L++GI P+V +AT
Sbjct: 302 SVKDVEDLRSNDGARSNKCMFPVMALLAGVLGGVFGIGGGMLISPLLLQVGIAPEVTAAT 361
Query: 343 STFAMVFSSSMSVVQYYHLDRFPIPYASYLVLVATIAALTGQHVVRKIIAIFGRASIIVF 402
+F ++FSS+MS +QY L AS ++ +A+L G VV+K+I +GRASIIVF
Sbjct: 362 CSFMVLFSSTMSAIQYLLLGMEHTGTASIFAVICFVASLVGLKVVQKVITEYGRASIIVF 421
Query: 403 ILAFTIFVSAISLGGVGIGNMVEKMENEEYMGFDNLC 439
+ + +S + + G ++ + YMGF C
Sbjct: 422 SVGIVMALSIVLMTSYGALDVWNDYVSGRYMGFKLPC 458
>At4g21250 hypothetical protein
Length = 449
Score = 214 bits (544), Expect = 1e-55
Identities = 133/423 (31%), Positives = 221/423 (51%), Gaps = 49/423 (11%)
Query: 42 EMKFGWRIVVGSIVGFFGAALGSVGGVGGGGIFIPMLTLIIGFDPKSSTALSKCMITGAA 101
E+K I++ ++ F A + S GG+GGGG+FIP++T++ G D K++++ S M+TG +
Sbjct: 51 ELKLSSAIIMAGVLCFLAALISSAGGIGGGGLFIPIMTIVAGVDLKTASSFSAFMVTGGS 110
Query: 102 GSTVYYNLRLRHPTLDMPLIDYDLALLFQPMLMLGISIGVAFNVMFADWMVTILLIILFI 161
+ V NL L+DYDLALL +P ++LG+SIGV N + +W++T+L +
Sbjct: 111 IANVISNL-----FGGKALLDYDLALLLEPCMLLGVSIGVICNRVLPEWLITVLFAVFLA 165
Query: 162 GTSTKALVKGIDTWKKETIMKKEAFEEAAQMLESGSTPDYASEEDYKSLPADLQDEEVPL 221
+ K G+ WK E+ + +E+ + + EE+ K+L A L + +
Sbjct: 166 WSILKTCRSGVKFWKLESEIARESGHGRPERGQG------QIEEETKNLKAPLLEAQATK 219
Query: 222 -LDNIHWKELSVLMYVWVAFLIVQILKTYS--------KTCSIEYWLLNSLQVPIAISVT 272
I W +L VL+ VW +F ++ +L+ K C +EYW+L SLQ+P+A+
Sbjct: 220 NKSKIPWTKLGVLVIVWASFFVIYLLRGNKDGKGIITIKPCGVEYWILLSLQIPLAL--- 276
Query: 273 LFEAICLCK----------------GTRVIASRGKEITWKFHKICLYCFCGIIAGMVSGL 316
+F + L + GTR+ K KF + +AG++ G+
Sbjct: 277 IFTKLALSRTESRQEQSPNDQKNQEGTRL----DKSTRLKFPAM------SFLAGLLGGI 326
Query: 317 LGLGGGFILGPLFLELGIPPQVASATSTFAMVFSSSMSVVQYYHLDRFPIPYASYLVLVA 376
G+GGG ++ PL L+ GIPPQ+ +AT++F + FS++MS VQY L A +
Sbjct: 327 FGIGGGMLISPLLLQSGIPPQITAATTSFMVFFSATMSAVQYLLLGMQNTDTAYVFSFIC 386
Query: 377 TIAALTGQHVVRKIIAIFGRASIIVFILAFTIFVSAISLGGVGIGNMVEKMENEEYMGFD 436
+A+L G +V+K +A FGRASIIVF + + +S + + G ++ + MGF
Sbjct: 387 FLASLLGLVLVQKAVAQFGRASIIVFSVGTVMSLSTVLMTSFGALDVWTDYVAGKDMGFK 446
Query: 437 NLC 439
C
Sbjct: 447 LPC 449
>At4g21260 hypothetical protein
Length = 393
Score = 145 bits (366), Expect = 4e-35
Identities = 104/379 (27%), Positives = 182/379 (47%), Gaps = 59/379 (15%)
Query: 42 EMKFGWRIVVGSIVGFFGAALGSVGGVGGGGIFIPMLTLIIGFDPKSSTALSKCMITGAA 101
E+K +VV ++ F A + S G+ D K++++ S M+TG +
Sbjct: 55 ELKLSPALVVAGVLCFTAALISSASGI----------------DLKAASSFSAFMVTGGS 98
Query: 102 GSTVYYNLRLRHPTLDMPLIDYDLALLFQPMLMLGISIGVAFNVMFADWMVTILLIILFI 161
+ + N H LIDYDLALL +P ++LG+S+GV N +F +W++T L ++ +
Sbjct: 99 IANLINN----HFGCKK-LIDYDLALLLEPCMLLGVSVGVICNKVFPEWLITGLFVVFLM 153
Query: 162 GTSTKALVKGIDTWKKETIMKKEAFEEAAQMLESGSTPDYASEEDYKSLPADLQDEEVPL 221
+S + G +WK I++++ +++ E +K L ++ E
Sbjct: 154 WSSMETCENGHTSWKLSLILREKEDMRDSRLAEVKRRRTIIF---FKHLYLKIKKTETK- 209
Query: 222 LDNIHWKELSVLMYVWVAFLIVQILKTYSKTCSIEYWLLNSLQVPIAISVTLFEAICLCK 281
+ + L ++ K CS+EYW+L SLQ+P+A+ T+ A+ +
Sbjct: 210 -QSFLGRNLGIISI---------------KPCSVEYWILLSLQIPLALVFTIL-ALSRTE 252
Query: 282 GTRVIASRGKEITWKFHKICLYCFCGIIAGMVSGLLGLGGGFILGPLFLELGIPPQVASA 341
+ + +E AG++ G+ G+GGG I+ PL L GIPPQV +A
Sbjct: 253 SLQEQSISNQE-----------------AGLLGGIFGIGGGMIISPLLLRAGIPPQVTAA 295
Query: 342 TSTFAMVFSSSMSVVQYYHLDRFPIPYASYLVLVATIAALTGQHVVRKIIAIFGRASIIV 401
T++F + FS++MS VQY L A ++ A+ G +K++ F RASIIV
Sbjct: 296 TTSFMVFFSATMSGVQYLLLGMQNTEAAYVFSVICFFASTLGLVFAQKVVPHFRRASIIV 355
Query: 402 FILAFTIFVSAISLGGVGI 420
F++ ++++ I + GI
Sbjct: 356 FLVGTMMYLTTIVMASFGI 374
>At1g54470 hypothetical protein
Length = 112
Score = 40.4 bits (93), Expect = 0.002
Identities = 17/50 (34%), Positives = 29/50 (58%)
Query: 42 EMKFGWRIVVGSIVGFFGAALGSVGGVGGGGIFIPMLTLIIGFDPKSSTA 91
E+K +V ++ F A + S G+G GG+FIP+ TL+ D K+ ++
Sbjct: 55 ELKLSLAAIVAGVLYFLAALISSACGIGSGGLFIPITTLVSRLDLKTGSS 104
>At1g04660 unknown protein
Length = 212
Score = 30.8 bits (68), Expect = 1.6
Identities = 13/21 (61%), Positives = 15/21 (70%)
Query: 51 VGSIVGFFGAALGSVGGVGGG 71
+G + G GA LG VGGVGGG
Sbjct: 139 LGGVGGLGGAGLGGVGGVGGG 159
>At4g08730 hypothetical protein
Length = 188
Score = 29.6 bits (65), Expect = 3.5
Identities = 13/21 (61%), Positives = 14/21 (65%)
Query: 52 GSIVGFFGAALGSVGGVGGGG 72
G I G FG GSVGG+ GGG
Sbjct: 156 GGIGGRFGGRFGSVGGIFGGG 176
>At1g77260 unknown protein
Length = 655
Score = 29.6 bits (65), Expect = 3.5
Identities = 19/48 (39%), Positives = 24/48 (49%), Gaps = 2/48 (4%)
Query: 21 VMKKTSFFWYSGESSYERVWPEMKFGWRIVVGSIVGF--FGAALGSVG 66
+MK S FW SY RV+ +F R V+ GF F AAL +G
Sbjct: 485 IMKAESRFWLEVVESYVRVFRWKEFKLRNVLDMRAGFGGFAAALNDLG 532
>At5g22640 unknown protein
Length = 871
Score = 28.9 bits (63), Expect = 6.0
Identities = 13/37 (35%), Positives = 24/37 (64%), Gaps = 3/37 (8%)
Query: 158 ILFIGTSTKALVKGIDTWKKETIMKKEAFEEAAQMLE 194
+L I S K ++ G++ W +E KK+A+EE +M++
Sbjct: 577 VLGIEKSVKPMLDGLEKWTEE---KKKAYEERKEMIQ 610
>At5g04140 ferredoxin-dependent glutamate synthase
Length = 1648
Score = 28.9 bits (63), Expect = 6.0
Identities = 18/63 (28%), Positives = 31/63 (48%), Gaps = 2/63 (3%)
Query: 45 FGWRIVVGSIVGFFGAALGSVG-GVGGGGIFIPMLTLIIGFDPKSSTALSKCMITGAAGS 103
FG ++ G + G + VG G+ GG I + + I GF P+ +T + + GA G
Sbjct: 1439 FGCFLIPGMNIRLIGESNDYVGKGMAGGEIVVTPVEKI-GFVPEEATIVGNTCLYGATGG 1497
Query: 104 TVY 106
++
Sbjct: 1498 QIF 1500
>At3g48000 aldehyde dehydrogenase (NAD+)-like protein
Length = 538
Score = 28.9 bits (63), Expect = 6.0
Identities = 20/81 (24%), Positives = 36/81 (43%), Gaps = 5/81 (6%)
Query: 323 FILGPLFLELGIPPQVASATSTFAMVFSSSMSVVQYYHLDRFPIPYASYLVLVATIAALT 382
F G LFLE G+PP V + S F ++++ H+D + + I L
Sbjct: 239 FYAGKLFLEAGLPPGVLNIVSGFGATAGAALA----SHMDVDKLAFTGSTDTGKVILGLA 294
Query: 383 GQHVVRKI-IAIFGRASIIVF 402
++ + + + G++ IVF
Sbjct: 295 ANSNLKPVTLELGGKSPFIVF 315
>At1g69430 hypothetical protein
Length = 350
Score = 28.9 bits (63), Expect = 6.0
Identities = 22/73 (30%), Positives = 33/73 (45%), Gaps = 13/73 (17%)
Query: 227 WKELSVLMYVWVAFLIVQILKTYS----KTCSIEYWL--------LNSLQVPIAISVTLF 274
WK L V+ Y+W+ +IV L ++ CS Y L ++ V + SV
Sbjct: 167 WKRL-VITYLWICTVIVVCLTSFCVFLVAVCSSFYVLGFSPDFNAYGAILVGLVFSVVFA 225
Query: 275 EAICLCKGTRVIA 287
AI +C T VI+
Sbjct: 226 NAIIICNTTIVIS 238
>At4g39840 unknown protein
Length = 451
Score = 28.5 bits (62), Expect = 7.9
Identities = 18/77 (23%), Positives = 35/77 (45%), Gaps = 3/77 (3%)
Query: 230 LSVLMYV-WVAFLIVQILKTYSKTCSIEYWLLNSLQVPIAISVTLFEAICLCKGTRVIAS 288
L L YV ++ L++ ++ +S C + +L Q + +V L + + RV+
Sbjct: 350 LQTLGYVFYLLLLLMYLVLVFSTDCGLGLKVLGLAQTFVGFAVGLHYYVAVFH--RVVLR 407
Query: 289 RGKEITWKFHKICLYCF 305
+ + WK H + CF
Sbjct: 408 QPPKTNWKIHGVYATCF 424
>At1g21860 pectinesterase, putative
Length = 538
Score = 28.5 bits (62), Expect = 7.9
Identities = 23/88 (26%), Positives = 42/88 (47%), Gaps = 6/88 (6%)
Query: 347 MVFSSSMSVVQYYHLDRFPIPYASYLVLVATIAALTGQHVVRKIIAIFGRASIIVFILAF 406
++F +S +VQ +HLD Y+ Y+V + V + R +I V+ ++
Sbjct: 422 VIFENSEDIVQSWHLD----GYSFYVVGMELGKWSPASRKVYNLNDAILRCTIQVYPRSW 477
Query: 407 TIFVSAISLGGVGIGNMVEKMENEEYMG 434
T I+L VG+ NM ++ +Y+G
Sbjct: 478 TAIY--IALDNVGMWNMRSEIWERQYLG 503
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.326 0.142 0.431
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,680,929
Number of Sequences: 26719
Number of extensions: 408442
Number of successful extensions: 1997
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 1948
Number of HSP's gapped (non-prelim): 29
length of query: 447
length of database: 11,318,596
effective HSP length: 102
effective length of query: 345
effective length of database: 8,593,258
effective search space: 2964674010
effective search space used: 2964674010
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 62 (28.5 bits)
Medicago: description of AC147964.5


