

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147964.4 + phase: 0 /pseudo
(276 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
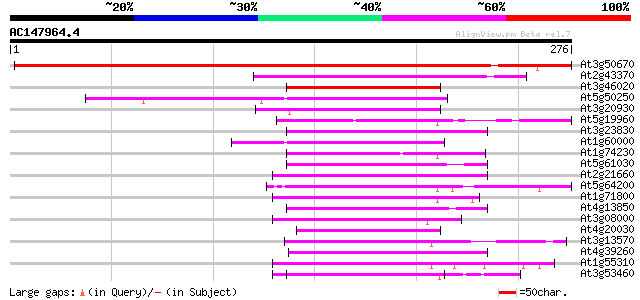
Score E
Sequences producing significant alignments: (bits) Value
At3g50670 U1 snRNP 70K protein 433 e-122
At2g43370 U1 small nuclear ribonucleoprotein 70 kDa like protein 112 2e-25
At3g46020 RNA binding protein -like 66 2e-11
At5g50250 RNA-binding protein-like 60 1e-09
At3g20930 unknown protein 60 1e-09
At5g19960 glycine-rich RNA-binding protein - like 58 5e-09
At3g23830 glycine-rich RNA binding protein, putative 58 5e-09
At1g60000 unknown protein 57 1e-08
At1g74230 putative RNA-binding protein 57 1e-08
At5g61030 RNA-binding protein - like 56 2e-08
At2g21660 glycine-rich RNA binding protein 56 2e-08
At5g64200 unknown protein 55 3e-08
At1g71800 cleavage stimulation factor like protein 55 3e-08
At4g13850 glycine-rich RNA-binding protein AtGRP2 - like 55 5e-08
At3g08000 RNA-binding protein like 55 5e-08
At4g20030 putative protein 54 7e-08
At3g13570 serine/arginine-rich protein 54 9e-08
At4g39260 glycine-rich protein (clone AtGRP8) 53 2e-07
At1g55310 Serine/arginine-rich protein 53 2e-07
At3g53460 RNA-binding protein cp29 protein 53 2e-07
>At3g50670 U1 snRNP 70K protein
Length = 427
Score = 433 bits (1113), Expect = e-122
Identities = 212/275 (77%), Positives = 238/275 (86%), Gaps = 4/275 (1%)
Query: 3 DPLLRNQNAAVQARTKAQNRANVLQLKLIGQSHPTGLTANLLKLFEPRPPLEYKPPPEKR 62
DP LRN NAAVQAR K QNRANVLQLKL+GQSHPTGLT NLLKLFEPRPPLEYKPPPEKR
Sbjct: 6 DPFLRNPNAAVQARAKVQNRANVLQLKLMGQSHPTGLTNNLLKLFEPRPPLEYKPPPEKR 65
Query: 63 KCPPLTGMAQFVSKFAEPGEPEYSPPVPVVETPAERRARVHKLRLEKGAAKAAEELEKYD 122
KCPP TGMAQFVS FAEPG+PEY+PP P VE P+++R R+HKLRLEKG KAAE+L+KYD
Sbjct: 66 KCPPYTGMAQFVSNFAEPGDPEYAPPKPEVELPSQKRERIHKLRLEKGVEKAAEDLKKYD 125
Query: 123 PHNDPNISGDPYKTLFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIE 182
P+NDPN +GDPYKTLFV+RL+YE++ES+IKREFESYG IKRV LVTD +NKP+GYAFIE
Sbjct: 126 PNNDPNATGDPYKTLFVSRLNYESSESKIKREFESYGPIKRVHLVTDQLTNKPKGYAFIE 185
Query: 183 YLHTRDMKAAYKQADGRKIEGRRVLVDVERGRTVPNWRPRRLGGGLGTTRVGGDEVNQRH 242
Y+HTRDMKAAYKQADG+KI+GRRVLVDVERGRTVPNWRPRRLGGGLGT+RVGG E
Sbjct: 186 YMHTRDMKAAYKQADGQKIDGRRVLVDVERGRTVPNWRPRRLGGGLGTSRVGGGE---EI 242
Query: 243 SGREQQQSGPSRSEEP-RVREERHADRDREKSRER 276
G +Q Q S+SEEP R REER R++ K RER
Sbjct: 243 VGEQQPQGRTSQSEEPSRPREEREKSREKGKERER 277
>At2g43370 U1 small nuclear ribonucleoprotein 70 kDa like protein
Length = 333
Score = 112 bits (281), Expect = 2e-25
Identities = 58/134 (43%), Positives = 75/134 (55%), Gaps = 3/134 (2%)
Query: 121 YDPHNDPNISGDPYKTLFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAF 180
YDP D GDPY TLFV RLS+ TTE ++ YG IK +RLV + RGY F
Sbjct: 50 YDPSGDSKAVGDPYCTLFVGRLSHHTTEDTLREVMSKYGRIKNLRLVRHIVTGASRGYGF 109
Query: 181 IEYLHTRDMKAAYKQADGRKIEGRRVLVDVERGRTVPNWRPRRLGGGLGTTRVGGDEVNQ 240
+EY ++M AY+ A I+GR ++VD R + +P W PRRLGGGLG + G
Sbjct: 110 VEYETEKEMLRAYEDAHHSLIDGREIIVDYNRQQLMPGWIPRRLGGGLGGRKESG---QL 166
Query: 241 RHSGREQQQSGPSR 254
R GR++ P R
Sbjct: 167 RFGGRDRPFRAPLR 180
>At3g46020 RNA binding protein -like
Length = 102
Score = 65.9 bits (159), Expect = 2e-11
Identities = 29/76 (38%), Positives = 49/76 (64%)
Query: 137 LFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHTRDMKAAYKQA 196
LFV+RLS TT+ +++ F +G IK RL+ D+E+ +P+G+ FI + D + A K
Sbjct: 9 LFVSRLSAYTTDQSLRQLFSPFGQIKEARLIRDSETQRPKGFGFITFDSEDDARKALKSL 68
Query: 197 DGRKIEGRRVLVDVER 212
DG+ ++GR + V+V +
Sbjct: 69 DGKIVDGRLIFVEVAK 84
>At5g50250 RNA-binding protein-like
Length = 289
Score = 60.1 bits (144), Expect = 1e-09
Identities = 43/200 (21%), Positives = 82/200 (40%), Gaps = 23/200 (11%)
Query: 38 GLTANLLKLFEPRPPLEYKPPPEKRKC-----PPLTGMAQFVSKFAEPGEPEYSPPVPVV 92
G + + + FE + + PPE+ K P F + G E S +
Sbjct: 89 GTSVTVDESFESEDGVGFPEPPEEAKLFVGNLPYDVDSQALAMLFEQAGTVEISEVIYNR 148
Query: 93 ETPAERRARVHKLRLEKGAAKAAEELEKYD-----------------PHNDPNISGDPYK 135
+T R + + A KA E+ ++ P P + ++
Sbjct: 149 DTDQSRGFGFVTMSTVEEAEKAVEKFNSFEVNGRRLTVNRAAPRGSRPERQPRVYDAAFR 208
Query: 136 TLFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHTRDMKAAYKQ 195
++V L ++ R++R F +G + R+V+D E+ + RG+ F++ + ++ A
Sbjct: 209 -IYVGNLPWDVDSGRLERLFSEHGKVVDARVVSDRETGRSRGFGFVQMSNENEVNVAIAA 267
Query: 196 ADGRKIEGRRVLVDVERGRT 215
DG+ +EGR + V+V RT
Sbjct: 268 LDGQNLEGRAIKVNVAEERT 287
>At3g20930 unknown protein
Length = 374
Score = 60.1 bits (144), Expect = 1e-09
Identities = 30/93 (32%), Positives = 51/93 (54%), Gaps = 2/93 (2%)
Query: 122 DPHNDPNISGDPYKT--LFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYA 179
D + + P KT LF+ LS+ T+E ++ FE +G + V+++ D S + +GYA
Sbjct: 267 DSRDQDDSESPPVKTKKLFITGLSFYTSEKTLRAAFEGFGELVEVKIIMDKISKRSKGYA 326
Query: 180 FIEYLHTRDMKAAYKQADGRKIEGRRVLVDVER 212
F+EY A K+ +G+ I G ++VDV +
Sbjct: 327 FLEYTTEEAAGTALKEMNGKIINGWMIVVDVAK 359
>At5g19960 glycine-rich RNA-binding protein - like
Length = 337
Score = 58.2 bits (139), Expect = 5e-09
Identities = 42/148 (28%), Positives = 68/148 (45%), Gaps = 24/148 (16%)
Query: 132 DPYKTLFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHTRDMKA 191
D +++V L Y+ TE ++R F YG++ V++V D S + + Y F+ + + R
Sbjct: 4 DDGNSVYVGGLPYDITEEAVRRVFSIYGSVLTVKIVND-RSVRGKCYGFVTFSNRRSADD 62
Query: 192 AYKQADGRKIEGRRVLVD---VERGRTVPNWRPRRLGGGLGTTRVGGDEVNQRHSGREQQ 248
A + DG+ I GR V V+ GR P P RL Q H G ++
Sbjct: 63 AIEDMDGKSIGGRAVRVNDVTTRGGRMNPG--PGRL---------------QPHGGWDR- 104
Query: 249 QSGPSRSEEPRVREERHADRDREKSRER 276
P R + +R++DR RE+ R +
Sbjct: 105 --SPDRRSDGNYERDRYSDRSRERDRSQ 130
>At3g23830 glycine-rich RNA binding protein, putative
Length = 136
Score = 58.2 bits (139), Expect = 5e-09
Identities = 30/99 (30%), Positives = 52/99 (52%)
Query: 137 LFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHTRDMKAAYKQA 196
LFV LS+ T +S +K+ F S+G + ++ D E+ + RG+ F+ + A K+
Sbjct: 37 LFVGGLSWGTDDSSLKQAFTSFGEVTEATVIADRETGRSRGFGFVSFSCEDSANNAIKEM 96
Query: 197 DGRKIEGRRVLVDVERGRTVPNWRPRRLGGGLGTTRVGG 235
DG+++ GR++ V++ R+ GGG G GG
Sbjct: 97 DGKELNGRQIRVNLATERSSAPRSSFGGGGGYGGGGGGG 135
>At1g60000 unknown protein
Length = 258
Score = 57.0 bits (136), Expect = 1e-08
Identities = 31/105 (29%), Positives = 53/105 (49%), Gaps = 1/105 (0%)
Query: 110 GAAKAAEELEKYDPHNDPNISGDPYKTLFVARLSYETTESRIKREFESYGAIKRVRLVTD 169
G A +K P+ +P +K LFV LS+ T + F G + R+V D
Sbjct: 153 GRALKVNFADKPKPNKEPLYPETEHK-LFVGNLSWTVTSESLAGAFRECGDVVGARVVFD 211
Query: 170 AESNKPRGYAFIEYLHTRDMKAAYKQADGRKIEGRRVLVDVERGR 214
++ + RGY F+ Y +M+ A + DG ++EGR + V++ +G+
Sbjct: 212 GDTGRSRGYGFVCYSSKAEMETALESLDGFELEGRAIRVNLAQGK 256
Score = 31.2 bits (69), Expect = 0.63
Identities = 32/142 (22%), Positives = 59/142 (41%), Gaps = 12/142 (8%)
Query: 85 YSPPVPVV--ETPAERRARVHKLRLEKGAAKAAEELEKYDPHN---DPNISGDPYKTLFV 139
+S P P++ RR RV L + EE EK D + DP + + L+
Sbjct: 36 FSTPKPLLISSRSCSRRFRV----LSETITVKLEEEEKDDGASAVLDPPAAVNT--KLYF 89
Query: 140 ARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHTRDMKAAYKQADGR 199
L Y + + + + + + V ++ + ++ + RG+AF+ + D DG
Sbjct: 90 GNLPYNVDSATLAQIIQDFANPELVEVLYNRDTGQSRGFAFVTMSNVEDCNIIIDNLDGT 149
Query: 200 KIEGRRVLVDVERGRTVPNWRP 221
+ GR + V+ + PN P
Sbjct: 150 EYLGRALKVNF-ADKPKPNKEP 170
>At1g74230 putative RNA-binding protein
Length = 289
Score = 56.6 bits (135), Expect = 1e-08
Identities = 32/100 (32%), Positives = 51/100 (51%), Gaps = 3/100 (3%)
Query: 137 LFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHTRDMKAAYKQA 196
+FV +SY T E ++ F YG + +++ D E+ + RG+AF+ + T + A Q
Sbjct: 36 IFVGGISYSTDEFGLREAFSKYGEVVDAKIIVDRETGRSRGFAFVTFTSTEEASNA-MQL 94
Query: 197 DGRKIEGRRVLVD--VERGRTVPNWRPRRLGGGLGTTRVG 234
DG+ + GRR+ V+ ERG GGG G + G
Sbjct: 95 DGQDLHGRRIRVNYATERGSGFGGRGFGGPGGGYGASDGG 134
>At5g61030 RNA-binding protein - like
Length = 309
Score = 56.2 bits (134), Expect = 2e-08
Identities = 29/99 (29%), Positives = 47/99 (47%), Gaps = 8/99 (8%)
Query: 137 LFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHTRDMKAAYKQA 196
LF+ ++Y E ++ F YG + R++ D E+ + RG+ F+ + + +A +
Sbjct: 42 LFIGGMAYSMDEDSLREAFTKYGEVVDTRVILDRETGRSRGFGFVTFTSSEAASSAIQAL 101
Query: 197 DGRKIEGRRVLVDVERGRTVPNWRPRRLGGGLGTTRVGG 235
DGR + GR V V+ RT GGG G GG
Sbjct: 102 DGRDLHGRVVKVNYANDRT--------SGGGFGGGGYGG 132
>At2g21660 glycine-rich RNA binding protein
Length = 176
Score = 55.8 bits (133), Expect = 2e-08
Identities = 30/106 (28%), Positives = 53/106 (49%)
Query: 130 SGDPYKTLFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHTRDM 189
SGD FV L++ T + ++ F YG + +++ D E+ + RG+ F+ + + M
Sbjct: 3 SGDVEYRCFVGGLAWATDDRALETAFAQYGDVIDSKIINDRETGRSRGFGFVTFKDEKAM 62
Query: 190 KAAYKQADGRKIEGRRVLVDVERGRTVPNWRPRRLGGGLGTTRVGG 235
K A + +G+ ++GR + V+ + R R GGG G GG
Sbjct: 63 KDAIEGMNGQDLDGRSITVNEAQSRGSGGGGGHRGGGGGGYRSGGG 108
>At5g64200 unknown protein
Length = 303
Score = 55.5 bits (132), Expect = 3e-08
Identities = 46/167 (27%), Positives = 75/167 (44%), Gaps = 24/167 (14%)
Query: 127 PNISGDPYKTLFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHT 186
P+IS D Y +L V +++ TT + F YG + V + D + RG+AF+ Y +
Sbjct: 10 PDIS-DTY-SLLVLNITFRTTADDLYPLFAKYGKVVDVFIPRDRRTGDSRGFAFVRYKYK 67
Query: 187 RDMKAAYKQADGRKIEGRRVLVD----------VERGRTV-PNWRPRRLGGGLGTTRVGG 235
+ A ++ DGR ++GR + V + +GR V P + RR +R
Sbjct: 68 DEAHKAVERLDGRVVDGREITVQFAKYGPNAEKISKGRVVEPPPKSRR-----SRSRSPR 122
Query: 236 DEVNQRHSGREQQQSGPSRSEEPR------VREERHADRDREKSRER 276
+ R S ++ P RS PR RE+ + R R +S +R
Sbjct: 123 RSRSPRRSRSPPRRRSPRRSRSPRRRSRDDYREKDYRKRSRSRSYDR 169
>At1g71800 cleavage stimulation factor like protein
Length = 461
Score = 55.5 bits (132), Expect = 3e-08
Identities = 33/104 (31%), Positives = 51/104 (48%), Gaps = 2/104 (1%)
Query: 130 SGDPYKTLFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHTRDM 189
S + +FV + Y+ TE +++ G + RLVTD E+ KP+GY F EY
Sbjct: 4 SSSQRRCVFVGNIPYDATEEQLREICGEVGPVVSFRLVTDRETGKPKGYGFCEYKDEETA 63
Query: 190 KAAYKQADGRKIEGRRVLVD-VERGRTVPNWRPRRLGG-GLGTT 231
+A + +I GR++ VD E + R + GG GL +T
Sbjct: 64 LSARRNLQSYEINGRQLRVDFAENDKGTDKTRDQSQGGPGLPST 107
>At4g13850 glycine-rich RNA-binding protein AtGRP2 - like
Length = 158
Score = 54.7 bits (130), Expect = 5e-08
Identities = 28/99 (28%), Positives = 51/99 (51%), Gaps = 3/99 (3%)
Query: 137 LFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHTRDMKAAYKQA 196
LF+ LS+ T ++ ++ F +G + +++ D E+ + RG+ F+ + AA +
Sbjct: 37 LFIGGLSWGTDDASLRDAFAHFGDVVDAKVIVDRETGRSRGFGFVNFNDEGAATAAISEM 96
Query: 197 DGRKIEGRRVLVDVERGRTVPNWRPRRLGGGLGTTRVGG 235
DG+++ GR + V+ R PR GGG G + GG
Sbjct: 97 DGKELNGRHIRVNPANDRPS---APRAYGGGGGYSGGGG 132
>At3g08000 RNA-binding protein like
Length = 143
Score = 54.7 bits (130), Expect = 5e-08
Identities = 31/95 (32%), Positives = 47/95 (48%), Gaps = 2/95 (2%)
Query: 130 SGDPYKTLFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHTRDM 189
S P LF+ LS+ E +K F S+G + VR+ D S + RG+ F+++ D
Sbjct: 36 SASPSSKLFIGGLSWSVDEQSLKDAFSSFGEVAEVRIAYDKGSGRSRGFGFVDFAEEGDA 95
Query: 190 KAAYKQADGRKIEGR--RVLVDVERGRTVPNWRPR 222
+A DG+ + GR R+ +ER R P PR
Sbjct: 96 LSAKDAMDGKGLLGRPLRISFALERVRGGPVVVPR 130
>At4g20030 putative protein
Length = 115
Score = 54.3 bits (129), Expect = 7e-08
Identities = 24/71 (33%), Positives = 40/71 (55%)
Query: 142 LSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHTRDMKAAYKQADGRKI 201
L + T+E +KREF ++G I V+L+ D + +GYAFI++ D A + D R
Sbjct: 10 LPFSTSEDFLKREFSAFGEIAEVKLIKDEAMKRSKGYAFIQFTSQDDAFLAIETMDRRMY 69
Query: 202 EGRRVLVDVER 212
GR + +D+ +
Sbjct: 70 NGRMIYIDIAK 80
>At3g13570 serine/arginine-rich protein
Length = 262
Score = 53.9 bits (128), Expect = 9e-08
Identities = 39/140 (27%), Positives = 64/140 (44%), Gaps = 16/140 (11%)
Query: 136 TLFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHTRDMKAAYKQ 195
+L V L ++ + ++R FE +G +K + L D + PRG+ FI+++ D A Q
Sbjct: 38 SLLVRNLRHDCRQEDLRRPFEQFGPVKDIYLPRDYYTGDPRGFGFIQFMDPADAAEAKHQ 97
Query: 196 ADGRKIEGRRV-LVDVERGRTVPNWRPRRLGGGLGTTRVGGDEVNQRHSGREQQQSGPSR 254
DG + GR + +V E R P R GG + + +++++S P
Sbjct: 98 MDGYLLLGRELTVVFAEENRKKPTEMRTRDRGG------------RSNRFQDRRRSPPRY 145
Query: 255 SEEPRVREERHADRDREKSR 274
S P R R R R +SR
Sbjct: 146 SRSPPPRRGR---RSRSRSR 162
>At4g39260 glycine-rich protein (clone AtGRP8)
Length = 169
Score = 53.1 bits (126), Expect = 2e-07
Identities = 26/98 (26%), Positives = 52/98 (52%)
Query: 138 FVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHTRDMKAAYKQAD 197
FV L++ T + ++R F +G + +++ D ES + RG+ F+ + + M+ A ++ +
Sbjct: 9 FVGGLAWATNDEDLQRTFSQFGDVIDSKIINDRESGRSRGFGFVTFKDEKAMRDAIEEMN 68
Query: 198 GRKIEGRRVLVDVERGRTVPNWRPRRLGGGLGTTRVGG 235
G++++GR + V+ + R R G G G GG
Sbjct: 69 GKELDGRVITVNEAQSRGSGGGGGGRGGSGGGYRSGGG 106
>At1g55310 Serine/arginine-rich protein
Length = 220
Score = 53.1 bits (126), Expect = 2e-07
Identities = 44/156 (28%), Positives = 66/156 (42%), Gaps = 17/156 (10%)
Query: 130 SGDPYKTLFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHTRDM 189
S D +L V L ++ + +++ FE +G +K + L D + PRG+ F++++ D
Sbjct: 31 SRDLPTSLLVRNLRHDCRQEDLRKSFEQFGPVKDIYLPRDYYTGDPRGFGFVQFMDPADA 90
Query: 190 KAAYKQADGRKIEGRRV-LVDVERGRTVP-NWRPRRLGGGLGTTR-------VGGDEVNQ 240
A DG + GR + +V E R P R R GGG R
Sbjct: 91 ADAKHHMDGYLLLGRELTVVFAEENRKKPTEMRARERGGGRFRDRRRTPPRYYSRSRSPP 150
Query: 241 RHSGREQQQSG-----PSRSEEPR---VREERHADR 268
GR + +SG P R PR REER+ R
Sbjct: 151 PRRGRSRSRSGDYYSPPPRRHHPRSISPREERYDGR 186
>At3g53460 RNA-binding protein cp29 protein
Length = 342
Score = 52.8 bits (125), Expect = 2e-07
Identities = 37/133 (27%), Positives = 64/133 (47%), Gaps = 13/133 (9%)
Query: 130 SGDPYKTLFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHTRDM 189
S P LFV LS+ +++ + FES G ++ V ++ D + + RG+ F+ ++
Sbjct: 94 SFSPDLKLFVGNLSFNVDSAQLAQLFESAGNVEMVEVIYDKVTGRSRGFGFVTMSTAAEV 153
Query: 190 KAAYKQADGRKIEGRRVLVDV-----------ERGRTVPNWRPRRLGGGLGTTRVGGDEV 238
+AA +Q +G + EGR + V+ RG + R GGG G+ R GG
Sbjct: 154 EAAAQQFNGYEFEGRPLRVNAGPPPPKREESFSRGPRSGGYGSER-GGGYGSER-GGGYG 211
Query: 239 NQRHSGREQQQSG 251
++R G ++ G
Sbjct: 212 SERGGGYGSERGG 224
Score = 42.4 bits (98), Expect = 3e-04
Identities = 17/78 (21%), Positives = 40/78 (50%)
Query: 137 LFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHTRDMKAAYKQA 196
L+V LS+ + ++ F G + R++ D +S + +G+ F+ +++++ A
Sbjct: 259 LYVGNLSWGVDDMALENLFNEQGKVVEARVIYDRDSGRSKGFGFVTLSSSQEVQKAINSL 318
Query: 197 DGRKIEGRRVLVDVERGR 214
+G ++GR++ V R
Sbjct: 319 NGADLDGRQIRVSEAEAR 336
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.313 0.133 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,447,634
Number of Sequences: 26719
Number of extensions: 298841
Number of successful extensions: 1967
Number of sequences better than 10.0: 250
Number of HSP's better than 10.0 without gapping: 161
Number of HSP's successfully gapped in prelim test: 91
Number of HSP's that attempted gapping in prelim test: 1133
Number of HSP's gapped (non-prelim): 674
length of query: 276
length of database: 11,318,596
effective HSP length: 98
effective length of query: 178
effective length of database: 8,700,134
effective search space: 1548623852
effective search space used: 1548623852
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC147964.4


