

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147964.1 + phase: 0
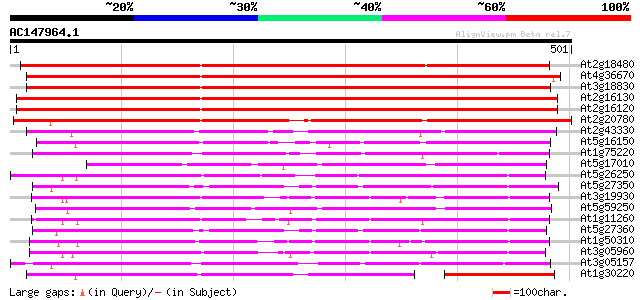
(501 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g18480 putative sugar transporter 657 0.0
At4g36670 sugar transporter like protein 647 0.0
At3g18830 sugar transporter like protein 571 e-163
At2g16130 putative sugar transporter 554 e-158
At2g16120 putative sugar transporter 554 e-158
At2g20780 putative sugar transporter 422 e-118
At2g43330 membrane transporter like protein 245 4e-65
At5g16150 sugar transporter like protein 233 2e-61
At1g75220 integral membrane protein, putative 224 7e-59
At5g17010 sugar transporter - like protein 213 3e-55
At5g26250 hexose transporter - like protein 209 2e-54
At5g27350 sugar transporter like protein 206 3e-53
At3g19930 monosaccharide transport protein, STP4 204 7e-53
At5g59250 D-xylose-H+ symporter - like protein 204 9e-53
At1g11260 glucose transporter 203 2e-52
At5g27360 sugar-porter family protein 2 (SFP2) 199 2e-51
At1g50310 hexose transporter, putative 199 3e-51
At3g05960 putative hexose transporter 199 4e-51
At3g05157 sugar transporter like protein 197 9e-51
At1g30220 unknown protein 193 2e-49
>At2g18480 putative sugar transporter
Length = 508
Score = 657 bits (1696), Expect = 0.0
Identities = 333/473 (70%), Positives = 398/473 (83%), Gaps = 2/473 (0%)
Query: 10 DQTNTFNKYAFACAIVASMVSIVSGYDTGVMSGAMLFIKEDLGISDTQQEVLAGILNLCA 69
D NK+AF CAIVAS++SI+ GYDTGVMSGA +FI++DL I+DTQ EVLAGILNLCA
Sbjct: 13 DPNPHMNKFAFGCAIVASIISIIFGYDTGVMSGAQIFIRDDLKINDTQIEVLAGILNLCA 72
Query: 70 LVGSLTAGRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVCGVGVGFALMI 129
LVGSLTAG+TSD IGRRYTI L++++F++G+ LMGYGPNY +LMVGRC+ GVGVGFALMI
Sbjct: 73 LVGSLTAGKTSDVIGRRYTIALSAVIFLVGSVLMGYGPNYPVLMVGRCIAGVGVGFALMI 132
Query: 130 APVYSAEISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLGWRLMLGIAALPS 189
APVYSAEISSAS RGFLTSLPELCI +GILLGY+SNY GK L+LKLGWRLMLGIAA PS
Sbjct: 133 APVYSAEISSASHRGFLTSLPELCISLGILLGYVSNYCFGK-LTLKLGWRLMLGIAAFPS 191
Query: 190 FVVAFCILTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIKIAAGLDENCNDE 249
++AF I MPESPRWLVMQG+L +AKK+++ VSNT +EAE R +DI AA +D E
Sbjct: 192 LILAFGITRMPESPRWLVMQGRLEEAKKIMVLVSNTEEEAEERFRDILTAAEVDVTEIKE 251
Query: 250 TVKLPQKSHQGEGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSPRIFRKAG 309
+K + G+ VW+EL+++P P+VR +LIAAVGIHFFEHATGIEAV+LYSPRIF+KAG
Sbjct: 252 VGGGVKKKNHGKSVWRELVIKPRPAVRLILIAAVGIHFFEHATGIEAVVLYSPRIFKKAG 311
Query: 310 ITSKEKLLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLGLSLTVVD 369
+ SK+KLLLAT+GVGLTK F++IA FLLDK+GRR+LL STGGM+ LT L +SLT+V
Sbjct: 312 VVSKDKLLLATVGVGLTKAFFIIIATFLLDKVGRRKLLLTSTGGMVFALTSLAVSLTMVQ 371
Query: 370 KSNGNVLWALILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVAVNRTMN 429
+ G + WAL LSIV+TYA+VAFF+IGLGPITWVYSSEIFPL+LRAQGASIGVAVNR MN
Sbjct: 372 RF-GRLAWALSLSIVSTYAFVAFFSIGLGPITWVYSSEIFPLRLRAQGASIGVAVNRIMN 430
Query: 430 AVVSMTFISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGKALEEMEMVF 482
A VSM+F+S+ KAIT GG FF+FAGI+V AW FF+F LPETKG LEEME +F
Sbjct: 431 ATVSMSFLSMTKAITTGGVFFVFAGIAVAAWWFFFFMLPETKGLPLEEMEKLF 483
>At4g36670 sugar transporter like protein
Length = 493
Score = 647 bits (1670), Expect = 0.0
Identities = 323/480 (67%), Positives = 392/480 (81%), Gaps = 4/480 (0%)
Query: 16 NKYAFACAIVASMVSIVSGYDTGVMSGAMLFIKEDLGISDTQQEVLAGILNLCALVGSLT 75
N++A CAIVAS+VSI+ GYDTGVMSGAM+FI+EDL +D Q EVL GILNLCALVGSL
Sbjct: 14 NRFALQCAIVASIVSIIFGYDTGVMSGAMVFIEEDLKTNDVQIEVLTGILNLCALVGSLL 73
Query: 76 AGRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVCGVGVGFALMIAPVYSA 135
AGRTSD IGRRYTI LASILF+LG+ LMG+GPNY +L+ GRC G+GVGFALM+APVYSA
Sbjct: 74 AGRTSDIIGRRYTIVLASILFMLGSILMGWGPNYPVLLSGRCTAGLGVGFALMVAPVYSA 133
Query: 136 EISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLGWRLMLGIAALPSFVVAFC 195
EI++AS RG L SLP LCI IGILLGYI NY K L + +GWRLMLGIAA+PS V+AF
Sbjct: 134 EIATASHRGLLASLPHLCISIGILLGYIVNYFFSK-LPMHIGWRLMLGIAAVPSLVLAFG 192
Query: 196 ILTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIKIAAGLDENCNDETVKLPQ 255
IL MPESPRWL+MQG+L + K++L VSN+ +EAELR +DIK AAG+D C D+ VK+
Sbjct: 193 ILKMPESPRWLIMQGRLKEGKEILELVSNSPEEAELRFQDIKAAAGIDPKCVDDVVKMEG 252
Query: 256 KSHQGEGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSPRIFRKAGITSKEK 315
K GEGVWKELILRPTP+VR +L+ A+GIHFF+HA+GIEAV+LY PRIF+KAGIT+K+K
Sbjct: 253 KKTHGEGVWKELILRPTPAVRRVLLTALGIHFFQHASGIEAVLLYGPRIFKKAGITTKDK 312
Query: 316 LLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLGLSLTVVDKSNGNV 375
L L TIGVG+ K F+ A LLDK+GRR+LL S GGM+I LT+LG LT+ + G +
Sbjct: 313 LFLVTIGVGIMKTTFIFTATLLLDKVGRRKLLLTSVGGMVIALTMLGFGLTMAQNAGGKL 372
Query: 376 LWALILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVAVNRTMNAVVSMT 435
WAL+LSIVA Y++VAFF+IGLGPITWVYSSE+FPLKLRAQGAS+GVAVNR MNA VSM+
Sbjct: 373 AWALVLSIVAAYSFVAFFSIGLGPITWVYSSEVFPLKLRAQGASLGVAVNRVMNATVSMS 432
Query: 436 FISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGKALEEMEMVFTK---KSSGKNVA 492
F+S+ AIT GG+FFMFAG++ +AW FF+F LPETKGK+LEE+E +F + K G+N A
Sbjct: 433 FLSLTSAITTGGAFFMFAGVAAVAWNFFFFLLPETKGKSLEEIEALFQRDGDKVRGENGA 492
>At3g18830 sugar transporter like protein
Length = 539
Score = 571 bits (1471), Expect = e-163
Identities = 274/468 (58%), Positives = 370/468 (78%), Gaps = 1/468 (0%)
Query: 16 NKYAFACAIVASMVSIVSGYDTGVMSGAMLFIKEDLGISDTQQEVLAGILNLCALVGSLT 75
N YAFACAI+ASM SI+ GYD GVMSGAM++IK DL I+D Q +LAG LN+ +L+GS
Sbjct: 33 NNYAFACAILASMTSILLGYDIGVMSGAMIYIKRDLKINDLQIGILAGSLNIYSLIGSCA 92
Query: 76 AGRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVCGVGVGFALMIAPVYSA 135
AGRTSD+IGRRYTI LA +F GA LMG PNYA LM GR + G+GVG+ALMIAPVY+A
Sbjct: 93 AGRTSDWIGRRYTIVLAGAIFFAGAILMGLSPNYAFLMFGRFIAGIGVGYALMIAPVYTA 152
Query: 136 EISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLGWRLMLGIAALPSFVVAFC 195
E+S ASSRGFL S PE+ I GI+LGY+SN L LK+GWRLMLGI A+PS ++A
Sbjct: 153 EVSPASSRGFLNSFPEVFINAGIMLGYVSNLAFSN-LPLKVGWRLMLGIGAVPSVILAIG 211
Query: 196 ILTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIKIAAGLDENCNDETVKLPQ 255
+L MPESPRWLVMQG+LG AK+VL + S++ EA LRL+DIK AAG+ +C+D+ V++ +
Sbjct: 212 VLAMPESPRWLVMQGRLGDAKRVLDKTSDSPTEATLRLEDIKHAAGIPADCHDDVVQVSR 271
Query: 256 KSHQGEGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSPRIFRKAGITSKEK 315
++ GEGVW+EL++RPTP+VR ++IAA+GIHFF+ A+GI+AV+L+SPRIF+ AG+ + +
Sbjct: 272 RNSHGEGVWRELLIRPTPAVRRVMIAAIGIHFFQQASGIDAVVLFSPRIFKTAGLKTDHQ 331
Query: 316 LLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLGLSLTVVDKSNGNV 375
LLAT+ VG+ K F+++A FLLD++GRR LL S GGM++ L LG SLT++D+S V
Sbjct: 332 QLLATVAVGVVKTSFILVATFLLDRIGRRPLLLTSVGGMVLSLAALGTSLTIIDQSEKKV 391
Query: 376 LWALILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVAVNRTMNAVVSMT 435
+WA++++I YVA F+IG GPITWVYSSEIFPL+LR+QG+S+GV VNR + V+S++
Sbjct: 392 MWAVVVAIATVMTYVATFSIGAGPITWVYSSEIFPLRLRSQGSSMGVVVNRVTSGVISIS 451
Query: 436 FISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGKALEEMEMVFT 483
F+ + KA+T GG+F++F GI+ +AW+FFY FLPET+G+ LE+M+ +F+
Sbjct: 452 FLPMSKAMTTGGAFYLFGGIATVAWVFFYTFLPETQGRMLEDMDELFS 499
>At2g16130 putative sugar transporter
Length = 511
Score = 554 bits (1427), Expect = e-158
Identities = 273/484 (56%), Positives = 361/484 (74%), Gaps = 2/484 (0%)
Query: 7 DKEDQTNTFNKYAFACAIVASMVSIVSGYDTGVMSGAMLFIKEDLGISDTQQEVLAGILN 66
+ E +++AFACAI+ASM SI+ GYD GVMSGA +FIK+DL +SD Q E+L GILN
Sbjct: 14 ESEPPRGNRSRFAFACAILASMTSIILGYDIGVMSGAAIFIKDDLKLSDVQLEILMGILN 73
Query: 67 LCALVGSLTAGRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVCGVGVGFA 126
+ +L+GS AGRTSD+IGRRYTI LA F GA LMG+ NY +MVGR V G+GVG+A
Sbjct: 74 IYSLIGSGAAGRTSDWIGRRYTIVLAGFFFFCGALLMGFATNYPFIMVGRFVAGIGVGYA 133
Query: 127 LMIAPVYSAEISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLGWRLMLGIAA 186
+MIAPVY+ E++ ASSRGFL+S PE+ I IGILLGY+SNY K L +GWR MLGI A
Sbjct: 134 MMIAPVYTTEVAPASSRGFLSSFPEIFINIGILLGYVSNYFFAK-LPEHIGWRFMLGIGA 192
Query: 187 LPSFVVAFCILTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIKIAAGLDENC 246
+PS +A +L MPESPRWLVMQG+LG A KVL + SNT +EA RL DIK A G+ ++
Sbjct: 193 VPSVFLAIGVLAMPESPRWLVMQGRLGDAFKVLDKTSNTKEEAISRLNDIKRAVGIPDDM 252
Query: 247 NDETVKLPQKSHQGEGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSPRIFR 306
D+ + +P K G+GVWK+L++RPTPSVR +LIA +GIHF + A+GI+AV+LYSP IF
Sbjct: 253 TDDVIVVPNKKSAGKGVWKDLLVRPTPSVRHILIACLGIHFSQQASGIDAVVLYSPTIFS 312
Query: 307 KAGITSKEKLLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLGLSLT 366
+AG+ SK LLAT+ VG+ K +F+V+ L+D+ GRR LL S GGM LT LG SLT
Sbjct: 313 RAGLKSKNDQLLATVAVGVVKTLFIVVGTCLVDRFGRRALLLTSMGGMFFSLTALGTSLT 372
Query: 367 VVDKSNGNVL-WALILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVAVN 425
V+D++ G L WA+ L++ +VA F++G GP+TWVY+SEIFP++LRAQGAS+GV +N
Sbjct: 373 VIDRNPGQTLKWAIGLAVTTVMTFVATFSLGAGPVTWVYASEIFPVRLRAQGASLGVMLN 432
Query: 426 RTMNAVVSMTFISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGKALEEMEMVFTKK 485
R M+ ++ MTF+S+ K +TIGG+F +FAG++V AW+FF+ FLPET+G LEE+E +F
Sbjct: 433 RLMSGIIGMTFLSLSKGLTIGGAFLLFAGVAVAAWVFFFTFLPETRGVPLEEIESLFGSY 492
Query: 486 SSGK 489
S+ K
Sbjct: 493 SANK 496
>At2g16120 putative sugar transporter
Length = 511
Score = 554 bits (1427), Expect = e-158
Identities = 274/484 (56%), Positives = 361/484 (73%), Gaps = 2/484 (0%)
Query: 7 DKEDQTNTFNKYAFACAIVASMVSIVSGYDTGVMSGAMLFIKEDLGISDTQQEVLAGILN 66
+ E ++YAFACAI+ASM SI+ GYD GVMSGA +FIK+DL +SD Q E+L GILN
Sbjct: 14 ESEPPRGNRSRYAFACAILASMTSIILGYDIGVMSGASIFIKDDLKLSDVQLEILMGILN 73
Query: 67 LCALVGSLTAGRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVCGVGVGFA 126
+ +LVGS AGRTSD++GRRYTI LA F GA LMG+ NY +MVGR V G+GVG+A
Sbjct: 74 IYSLVGSGAAGRTSDWLGRRYTIVLAGAFFFCGALLMGFATNYPFIMVGRFVAGIGVGYA 133
Query: 127 LMIAPVYSAEISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLGWRLMLGIAA 186
+MIAPVY+AE++ ASSRGFLTS PE+ I IGILLGY+SNY K L LGWR MLG+ A
Sbjct: 134 MMIAPVYTAEVAPASSRGFLTSFPEIFINIGILLGYVSNYFFSK-LPEHLGWRFMLGVGA 192
Query: 187 LPSFVVAFCILTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIKIAAGLDENC 246
+PS +A +L MPESPRWLV+QG+LG A KVL + SNT +EA RL DIK A G+ ++
Sbjct: 193 VPSVFLAIGVLAMPESPRWLVLQGRLGDAFKVLDKTSNTKEEAISRLDDIKRAVGIPDDM 252
Query: 247 NDETVKLPQKSHQGEGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSPRIFR 306
D+ + +P K G+GVWK+L++RPTPSVR +LIA +GIHF + A+GI+AV+LYSP IF
Sbjct: 253 TDDVIVVPNKKSAGKGVWKDLLVRPTPSVRHILIACLGIHFAQQASGIDAVVLYSPTIFS 312
Query: 307 KAGITSKEKLLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLGLSLT 366
KAG+ SK LLAT+ VG+ K +F+V+ ++D+ GRR LL S GGM + LT LG SLT
Sbjct: 313 KAGLKSKNDQLLATVAVGVVKTLFIVVGTCVVDRFGRRALLLTSMGGMFLSLTALGTSLT 372
Query: 367 VVDKSNGNVL-WALILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVAVN 425
V++++ G L WA+ L++ +VA F+IG GP+TWVY SEIFP++LRAQGAS+GV +N
Sbjct: 373 VINRNPGQTLKWAIGLAVTTVMTFVATFSIGAGPVTWVYCSEIFPVRLRAQGASLGVMLN 432
Query: 426 RTMNAVVSMTFISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGKALEEMEMVFTKK 485
R M+ ++ MTF+S+ K +TIGG+F +FAG++ AW+FF+ FLPET+G LEEME +F
Sbjct: 433 RLMSGIIGMTFLSLSKGLTIGGAFLLFAGVAAAAWVFFFTFLPETRGIPLEEMETLFGSY 492
Query: 486 SSGK 489
++ K
Sbjct: 493 TANK 496
>At2g20780 putative sugar transporter
Length = 547
Score = 422 bits (1086), Expect = e-118
Identities = 223/519 (42%), Positives = 331/519 (62%), Gaps = 39/519 (7%)
Query: 4 NQGDKEDQTNTFNKYAFACAIVASMVSIVSGY---------------------DTGVMSG 42
N + E + + KY ACA AS+ +++ GY D GVMSG
Sbjct: 40 NHREAEARNSRTRKYVMACAFFASLNNVLLGYGRFYLYNRILLLLLYFVDLQKDVGVMSG 99
Query: 43 AMLFIKEDLGISDTQQEVLAGILNLCALVGSLTAGRTSDYIGRRYTIFLASILFILGAGL 102
A+LFI++DL I++ Q EVL G L++ +L GSL GRTSD IGR++T+ LA+++F GA +
Sbjct: 100 AVLFIQQDLKITEVQTEVLIGSLSIISLFGSLAGGRTSDSIGRKWTMALAALVFQTGAAV 159
Query: 103 MGYGPNYAILMVGRCVCGVGVGFALMIAPVYSAEISSASSRGFLTSLPELCIGIGILLGY 162
M P++ +LM+GR + G+G+G +MIAPVY AEIS +RGF TS PE+ I +GILLGY
Sbjct: 160 MAVAPSFEVLMIGRTLAGIGIGLGVMIAPVYIAEISPTVARGFFTSFPEIFINLGILLGY 219
Query: 163 ISNYVLGKYLSLKLGWRLMLGIAALPSFVVAFCILTMPESPRWLVMQGQLGKAKKVLMQV 222
+SNY LS+ + WR+ML + LPS + F + +PESPRWLVM+G++ A++VLM+
Sbjct: 220 VSNYAFSG-LSVHISWRIMLAVGILPSVFIGFALCVIPESPRWLVMKGRVDSAREVLMKT 278
Query: 223 SNTTQEAELRLKDIKIAAGLDENCNDETVKLPQKSHQGEGVWKELILRPTPSVRWMLIAA 282
+ EAE RL +I++AA E D VW+EL L P+P VR MLI
Sbjct: 279 NERDDEAEERLAEIQLAAAHTEGSEDRP------------VWREL-LSPSPVVRKMLIVG 325
Query: 283 VGIHFFEHATGIEAVMLYSPRIFRKAGITSKEKLLLATIGVGLTKIVFLVIALFLLDKLG 342
GI F+ TGI+A + YSP I ++AGI + KLL AT+ VG+TK VF++ A FL+D +G
Sbjct: 326 FGIQCFQQITGIDATVYYSPEILKEAGIQDETKLLAATVAVGVTKTVFILFATFLIDSVG 385
Query: 343 RRRLLQISTGGMIIGLTLLGLSLTVVDKSNGNVLWALILSIVATYAYVAFFNIGLGPITW 402
R+ LL +ST GM + L L +LT + G + L+++ VAFF+IG+GP+ W
Sbjct: 386 RKPLLYVSTIGMTLCLFCLSFTLTFL----GQGTLGITLALLFVCGNVAFFSIGMGPVCW 441
Query: 403 VYSSEIFPLKLRAQGASIGVAVNRTMNAVVSMTFISIYKAITIGGSFFMFAGISVLAWLF 462
V +SEIFPL+LRAQ +++G NR + +V+M+F+S+ +AIT+GG+FF+F+ +S L+ +F
Sbjct: 442 VLTSEIFPLRLRAQASALGAVGNRVCSGLVAMSFLSVSRAITVGGTFFVFSLVSALSVIF 501
Query: 463 FYFFLPETKGKALEEMEMVFTKKSSGKNVAIEMDPIQKV 501
Y +PET GK+LE++E++F K+ +E+ +++
Sbjct: 502 VYVLVPETSGKSLEQIELMFQGGLERKDGEVELGDAERL 540
>At2g43330 membrane transporter like protein
Length = 509
Score = 245 bits (626), Expect = 4e-65
Identities = 155/480 (32%), Positives = 247/480 (51%), Gaps = 29/480 (6%)
Query: 16 NKYAFACAIVASMVSIVSGYDTGVMSGAMLFIKEDLGI---SDTQQEVLAGILNLCALVG 72
N Y + A + ++ GYDTGV+SGA+L+IK+D + S QE + + + A++G
Sbjct: 28 NSYILGLTVTAGIGGLLFGYDTGVISGALLYIKDDFEVVKQSSFLQETIVSMALVGAMIG 87
Query: 73 SLTAGRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVCGVGVGFALMIAPV 132
+ G +DY GR+ A ++F GA +M P+ +L+ GR + G+GVG A + APV
Sbjct: 88 AAAGGWINDYYGRKKATLFADVVFAAGAIVMAAAPDPYVLISGRLLVGLGVGVASVTAPV 147
Query: 133 YSAEISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLGWRLMLGIAALPSFVV 192
Y AE S + RG L S L I G L Y+ N + + WR MLG++ +P+ +
Sbjct: 148 YIAEASPSEVRGGLVSTNVLMITGGQFLSYLVN---SAFTQVPGTWRWMLGVSGVPAVIQ 204
Query: 193 AFCILTMPESPRWLVMQGQLGKAKKVLMQVSNTTQ-EAELRLKDIKIAAGLDENCNDETV 251
+L MPESPRWL M+ + +A +VL + + ++ E E+ D AA +E TV
Sbjct: 205 FILMLFMPESPRWLFMKNRKAEAIQVLARTYDISRLEDEI---DHLSAAEEEEKQRKRTV 261
Query: 252 KLPQKSHQGEGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSPRIFRKAGIT 311
L + + +R +A G+ F+ TGI VM YSP I + AG
Sbjct: 262 GY-------------LDVFRSKELRLAFLAGAGLQAFQQFTGINTVMYYSPTIVQMAGFH 308
Query: 312 SKEKLLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLGLSL---TVV 368
S + L ++ V V+ ++ +D GR++L S G+II L +L +S +
Sbjct: 309 SNQLALFLSLIVAAMNAAGTVVGIYFIDHCGRKKLALSSLFGVIISLLILSVSFFKQSET 368
Query: 369 DKSNGNVLWALILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVAVNRTM 428
G W +L + Y+ FF G+GP+ W +SEI+P + R + VN
Sbjct: 369 SSDGGLYGWLAVLGLAL---YIVFFAPGMGPVPWTVNSEIYPQQYRGICGGMSATVNWIS 425
Query: 429 NAVVSMTFISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGKALEEMEMVFTKKSSG 488
N +V+ TF++I +A G +F + AGI+VLA +F F+PET+G E+E ++ +++ G
Sbjct: 426 NLIVAQTFLTIAEAAGTGMTFLILAGIAVLAVIFVIVFVPETQGLTFSEVEQIWKERAYG 485
>At5g16150 sugar transporter like protein
Length = 546
Score = 233 bits (594), Expect = 2e-61
Identities = 155/463 (33%), Positives = 246/463 (52%), Gaps = 34/463 (7%)
Query: 25 VASMVSIVSGYDTGVMSGAMLFIKEDLGISDTQ--QEVLAGILNLCALVGSLTAGRTSDY 82
VA + +I+ GY GV++GA+ ++ +DLGI++ Q + L A VGS T G +D
Sbjct: 111 VACLGAILFGYHLGVVNGALEYLAKDLGIAENTVLQGWIVSSLLAGATVGSFTGGALADK 170
Query: 83 IGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVCGVGVGFALMIAPVYSAEISSASS 142
GR T L +I +GA L + ++VGR + G+G+G + I P+Y +EIS
Sbjct: 171 FGRTRTFQLDAIPLAIGAFLCATAQSVQTMIVGRLLAGIGIGISSAIVPLYISEISPTEI 230
Query: 143 RGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLGWRLMLGIAALPSFVVAFCILTMPES 202
RG L S+ +L I IGIL I+ L + L WR M G+A +PS ++A + PES
Sbjct: 231 RGALGSVNQLFICIGILAALIAGLPLA---ANPLWWRTMFGVAVIPSVLLAIGMAFSPES 287
Query: 203 PRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIKIAAGLDENCNDETVKLPQKSHQGEG 262
PRWLV QG++ +A+K + + + EL ++D+ + Q S + E
Sbjct: 288 PRWLVQQGKVSEAEKAIKTLYGKERVVEL-VRDLSASG--------------QGSSEPEA 332
Query: 263 VWKELILRPTPSVRWMLIAAVG--IHFFEHATGIEAVMLYSPRIFRKAGITSKEKLLLAT 320
W +L S R+ + +VG + F+ GI AV+ YS +FR AGI S + A+
Sbjct: 333 GWFDLF-----SSRYWKVVSVGAALFLFQQLAGINAVVYYSTSVFRSAGIQSD---VAAS 384
Query: 321 IGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLGLSLTVVDKSNGNVLWALI 380
VG + + +A L+DK+GR+ LL S GGM + + LL LS T ++
Sbjct: 385 ALVGASNVFGTAVASSLMDKMGRKSLLLTSFGGMALSMLLLSLSFTW----KALAAYSGT 440
Query: 381 LSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVAVNRTMNAVVSMTFISIY 440
L++V T YV F++G GP+ + EIF ++RA+ ++ + ++ N V+ + F+S+
Sbjct: 441 LAVVGTVLYVLSFSLGAGPVPALLLPEIFASRIRAKAVALSLGMHWISNFVIGLYFLSVV 500
Query: 441 KAITIGGSFFMFAGISVLAWLFFYFFLPETKGKALEEMEMVFT 483
I + FAG+ VLA L+ + ETKG++LEE+E+ T
Sbjct: 501 TKFGISSVYLGFAGVCVLAVLYIAGNVVETKGRSLEEIELALT 543
>At1g75220 integral membrane protein, putative
Length = 487
Score = 224 bits (572), Expect = 7e-59
Identities = 140/465 (30%), Positives = 239/465 (51%), Gaps = 31/465 (6%)
Query: 21 ACAIVASMVSIVSGYDTGVMSGAMLFIKEDLGISDTQQEVLAGILNLCALVGSLTAGRTS 80
AC ++ ++ I G+ G S I +DLG++ ++ V + N+ A+VG++ +G+ +
Sbjct: 50 ACVLIVALGPIQFGFTCGYSSPTQAAITKDLGLTVSEYSVFGSLSNVGAMVGAIASGQIA 109
Query: 81 DYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVCGVGVGFALMIAPVYSAEISSA 140
+YIGR+ ++ +A+I I+G + + + + L +GR + G GVG PVY AEI+
Sbjct: 110 EYIGRKGSLMIAAIPNIIGWLCISFAKDTSFLYMGRLLEGFGVGIISYTVPVYIAEIAPQ 169
Query: 141 SSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLGWRLMLGIAALPSFVVAFCILTMP 200
+ RG L S+ +L + IGI+L Y+ L L + WR++ + LP ++ + +P
Sbjct: 170 NMRGGLGSVNQLSVTIGIMLAYL--------LGLFVPWRILAVLGILPCTLLIPGLFFIP 221
Query: 201 ESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIKIAAGLDENCNDETVKLPQKSHQG 260
ESPRWL G + + L + + + + +IK + N TV+ +
Sbjct: 222 ESPRWLAKMGMTDEFETSLQVLRGFETDITVEVNEIKRSVASSTKRN--TVRFVDLKRR- 278
Query: 261 EGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSPRIFRKAGITSKEKLLLAT 320
+ L+ +G+ + GI V+ YS IF AG+TS AT
Sbjct: 279 -------------RYYFPLMVGIGLLVLQQLGGINGVLFYSSTIFESAGVTSSN---AAT 322
Query: 321 IGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLGLSLTV---VDKSNGNVLW 377
GVG ++V I+ +L+DK GRR LL IS+ GM I L ++ + + V + W
Sbjct: 323 FGVGAIQVVATAISTWLVDKAGRRLLLTISSVGMTISLVIVAAAFYLKEFVSPDSDMYSW 382
Query: 378 ALILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVAVNRTMNAVVSMTFI 437
ILS+V A V FF++G+GPI W+ SEI P+ ++ SI N + +++MT
Sbjct: 383 LSILSVVGVVAMVVFFSLGMGPIPWLIMSEILPVNIKGLAGSIATLANWFFSWLITMT-A 441
Query: 438 SIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGKALEEMEMVF 482
++ A + GG+F ++ + +F ++PETKGK LEE++ +F
Sbjct: 442 NLLLAWSSGGTFTLYGLVCAFTVVFVTLWVPETKGKTLEELQSLF 486
>At5g17010 sugar transporter - like protein
Length = 503
Score = 213 bits (541), Expect = 3e-55
Identities = 140/415 (33%), Positives = 218/415 (51%), Gaps = 23/415 (5%)
Query: 69 ALVGSLTAGRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVCGVGVGFALM 128
AL GS+ A +D IGRR + LA++L+++GA + P Y++L++GR + GV VG A+
Sbjct: 104 ALFGSIVAFTIADVIGRRKELILAALLYLVGALVTALAPTYSVLIIGRVIYGVSVGLAMH 163
Query: 129 IAPVYSAEISSASSRGFLTSLPELCIGIGILLGYISNYVLGKY-LSLKLGWRLMLGIAAL 187
AP+Y AE + + RG L SL E I +LG + Y +G +++ GWR M +
Sbjct: 164 AAPMYIAETAPSPIRGQLVSLKEFFI----VLGMVGGYGIGSLTVNVHSGWRYMYATSVP 219
Query: 188 PSFVVAFCILTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIKIAAGLD---E 244
+ ++ + +P SPRWL+++ GK V N + A L ++ A +D E
Sbjct: 220 LAVIMGIGMWWLPASPRWLLLRVIQGKGN-----VENQREAAIKSLCCLRGPAFVDSAAE 274
Query: 245 NCNDETVKLPQKSHQGEGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSPRI 304
N+ +L E + EL LI G+ F+ TG +V+ Y+P I
Sbjct: 275 QVNEILAELTFVGEDKEVTFGELFQGKCLKA---LIIGGGLVLFQQITGQPSVLYYAPSI 331
Query: 305 FRKAGITSKEKLLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLGLS 364
+ AG ++ +I +GL K++ +A+ ++D+LGRR LL GGM++ L LLG
Sbjct: 332 LQTAGFSAAGDATRVSILLGLLKLIMTGVAVVVIDRLGRRPLLLGGVGGMVVSLFLLGSY 391
Query: 365 LTVVDKSNGNVLWALILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVAV 424
S ++++VA YV + + GPI W+ SEIFPLKLR +G S+ V V
Sbjct: 392 YLFFSASP-------VVAVVALLLYVGCYQLSFGPIGWLMISEIFPLKLRGRGLSLAVLV 444
Query: 425 NRTMNAVVSMTFISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGKALEEME 479
N NA+V+ F + + + G F F I VL+ +F +F +PETKG LEE+E
Sbjct: 445 NFGANALVTFAFSPLKELLGAGILFCGFGVICVLSLVFIFFIVPETKGLTLEEIE 499
>At5g26250 hexose transporter - like protein
Length = 507
Score = 209 bits (533), Expect = 2e-54
Identities = 144/498 (28%), Positives = 244/498 (48%), Gaps = 42/498 (8%)
Query: 1 MIINQGDKEDQTNTFNKYAFACAIVASMVSIVSGYDTGVMSGAML---FIKEDL-GISDT 56
+I + G+ + Y F C I+A++ ++ GYD G+ G F+KE + +
Sbjct: 4 VISSNGNSKSFDAKMTVYVFICVIIAAVGGLIFGYDIGISGGVTAMDDFLKEFFPSVYER 63
Query: 57 QQ---------------EVLAGILNLCALVGSLTAGRTSDYIGRRYTIFLASILFILGAG 101
++ ++ L L ALV S A T +GRR T+ LASI F++G G
Sbjct: 64 KKHAHENNYCKYDNQFLQLFTSSLYLAALVASFFASATCSKLGRRPTMQLASIFFLIGVG 123
Query: 102 LMGYGPNYAILMVGRCVCGVGVGFALMIAPVYSAEISSASSRGFLTSLPELCIGIGILLG 161
L N +L++GR + G GVGF P++ +EI+ A RG L + +L + IGIL+
Sbjct: 124 LAAGAVNIYMLIIGRILLGFGVGFGNQAVPLFLSEIAPARLRGGLNIVFQLMVTIGILIA 183
Query: 162 YISNYVLGKYLSLKLGWRLMLGIAALPSFVVAFCILTMPESPRWLVMQGQLGKAKKVLMQ 221
I NY GWR+ LG A +P+ ++ F L + E+P L+ + + + K+ L +
Sbjct: 184 NIVNYFTSSI--HPYGWRIALGGAGIPALILLFGSLLICETPTSLIERNKTKEGKETLKK 241
Query: 222 VSNTTQEAELRLKDIKIAAGLDENCNDETVKLPQKSHQGEGVWKELILRPTPSVRWMLIA 281
+ ++ + + I A + D KL + P+ R +
Sbjct: 242 IRG-VEDVDEEYESIVHACDIARQVKDPYTKLMK-----------------PASRPPFVI 283
Query: 282 AVGIHFFEHATGIEAVMLYSPRIFRKAGITSKEKLLLATIGVGLTKIVFLVIALFLLDKL 341
+ + FF+ TGI A+M Y+P +F+ G + LL+ + G ++ + +FL+DK
Sbjct: 284 GMLLQFFQQFTGINAIMFYAPVLFQTVGF-GNDAALLSAVVTGTINVLSTFVGIFLVDKT 342
Query: 342 GRRRLLQISTGGMIIGLTLLGLSLTV-VDKSNGNVLWALILSIVATYAYVAFFNIGLGPI 400
GRR LL S+ M+I ++G+ L +D + ++ ++ YV F GP+
Sbjct: 343 GRRFLLLQSSVHMLICQLVIGIILAKDLDVTGTLARPQALVVVIFVCVYVMGFAWSWGPL 402
Query: 401 TWVYSSEIFPLKLRAQGASIGVAVNRTMNAVVSMTFISIYKAITIGGSFFMFAGISVLAW 460
W+ SE FPL+ R +G ++ V+ N V++ F+S+ A+ G FF F+G V+
Sbjct: 403 GWLIPSETFPLETRTEGFALAVSCNMFFTFVIAQAFLSMLCAMK-SGIFFFFSGWIVVMG 461
Query: 461 LFFYFFLPETKGKALEEM 478
LF FF+PETKG ++++M
Sbjct: 462 LFALFFVPETKGVSIDDM 479
>At5g27350 sugar transporter like protein
Length = 474
Score = 206 bits (523), Expect = 3e-53
Identities = 140/474 (29%), Positives = 232/474 (48%), Gaps = 34/474 (7%)
Query: 21 ACAIVASMVSIVSGYD----TGVMSGAMLFIKEDLGISDTQQEVLAGILNLCALVGSLTA 76
AC I+++ V++ + TG SGA + +DL +S Q L A +G+L
Sbjct: 29 ACVILSTFVAVCGSFSFGVATGYTSGAETGVMKDLDLSIAQFSAFGSFATLGAAIGALFC 88
Query: 77 GRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVCGVGVGFALMIAPVYSAE 136
G + IGRR T++++ L I G + + +L GR + G+G G + PVY AE
Sbjct: 89 GNLAMVIGRRGTMWVSDFLCITGWLSIAFAKEVVLLNFGRIISGIGFGLTSYVVPVYIAE 148
Query: 137 ISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLGWRLMLGIAALPSFVVAFCI 196
I+ RG T +L G+ + Y G +++ WR + + ALP F+ +
Sbjct: 149 ITPKHVRGTFTFSNQLLQNAGLAM----IYFCGNFIT----WRTLALLGALPCFIQVIGL 200
Query: 197 LTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIKIAAGLDENCNDETVKLPQK 256
+PESPRWL G + + L ++ + +I++ + EN
Sbjct: 201 FFVPESPRWLAKVGSDKELENSLFRLRGRDADISREASEIQVMTKMVEN----------- 249
Query: 257 SHQGEGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSPRIFRKAGITSKEKL 316
+ + +L R R+ L+ +G+ + +G AV+ Y+ IFRKAG + +
Sbjct: 250 --DSKSSFSDLFQR---KYRYTLVVGIGLMLIQQFSGSAAVISYASTIFRKAGFS----V 300
Query: 317 LLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLGLSLTVVDKSNGNVL 376
+ T +G+ I +I L L+DK GRR LL S GM + LLG++ T+ + L
Sbjct: 301 AIGTTMLGIFVIPKAMIGLILVDKWGRRPLLMTSAFGMSMTCMLLGVAFTLQKMQLLSEL 360
Query: 377 WALILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVAVNRTMNAVVSMTF 436
ILS + Y+A + IGLG + WV SEIFP+ ++ SI V+ + +++V+ F
Sbjct: 361 -TPILSFICVMMYIATYAIGLGGLPWVIMSEIFPINIKVTAGSIVTLVSFSSSSIVTYAF 419
Query: 437 ISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGKALEEMEMVFTKKSSGKN 490
+++ T G+FF+FAGI A LF + +PETKG +LEE+++ + +N
Sbjct: 420 NFLFEWST-QGTFFIFAGIGGAALLFIWLLVPETKGLSLEEIQVSLIHQPDERN 472
>At3g19930 monosaccharide transport protein, STP4
Length = 514
Score = 204 bits (520), Expect = 7e-53
Identities = 146/492 (29%), Positives = 241/492 (48%), Gaps = 57/492 (11%)
Query: 20 FACAIVASMVSIVSGYDTGVMSGAML---FIKE---------------DLGISDTQQEVL 61
F + + ++ GYD G+ G F++E + D+Q L
Sbjct: 24 FVTCFIGAFGGLIFGYDLGISGGVTSMEPFLEEFFPYVYKKMKSAHENEYCRFDSQLLTL 83
Query: 62 -AGILNLCALVGSLTAGRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVCG 120
L + ALV SL A + GR++++FL F +G+ G+ N A+L++GR + G
Sbjct: 84 FTSSLYVAALVSSLFASTITRVFGRKWSMFLGGFTFFIGSAFNGFAQNIAMLLIGRILLG 143
Query: 121 VGVGFALMIAPVYSAEISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLGWRL 180
GVGFA PVY +E++ + RG + ++ I GI++ I NY + + +GWR+
Sbjct: 144 FGVGFANQSVPVYLSEMAPPNLRGAFNNGFQVAIIFGIVVATIINYFTAQ-MKGNIGWRI 202
Query: 181 MLGIAALPSFVVAFCILTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIKIAA 240
LG+A +P+ ++ L +P++P L+ +G +AK++L + T +
Sbjct: 203 SLGLACVPAVMIMIGALILPDTPNSLIERGYTEEAKEMLQSIRGTNE------------- 249
Query: 241 GLDENCNDETVKLPQKSHQGEGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLY 300
+DE D + ++S Q + WK ++L P R LI I FF+ TGI + Y
Sbjct: 250 -VDEEFQD-LIDASEESKQVKHPWKNIML---PRYRPQLIMTCFIPFFQQLTGINVITFY 304
Query: 301 SPRIFRKAGITSKEKLLLATIGVGLTKIVFLVIALFLLDKLGRRRLL----------QIS 350
+P +F+ G SK LL A + G+ +++ +++F +D+ GRR L QI+
Sbjct: 305 APVLFQTLGFGSKASLLSAMV-TGIIELLCTFVSVFTVDRFGRRILFLQGGIQMLVSQIA 363
Query: 351 TGGMIIGLTLLGLSLTVVDKSNGNVLWALILSIVATYAYVAFFNIGLGPITWVYSSEIFP 410
G M IG+ + KS+ N++ ALI YVA F GP+ W+ SEI P
Sbjct: 364 IGAM-IGVKFGVAGTGNIGKSDANLIVALIC------IYVAGFAWSWGPLGWLVPSEISP 416
Query: 411 LKLRAQGASIGVAVNRTMNAVVSMTFISIYKAITIGGSFFMFAGISVLAWLFFYFFLPET 470
L++R+ +I V+VN +V+ F+++ + G FF FA V+ +F Y LPET
Sbjct: 417 LEIRSAAQAINVSVNMFFTFLVAQLFLTMLCHMKF-GLFFFFAFFVVIMTIFIYLMLPET 475
Query: 471 KGKALEEMEMVF 482
K +EEM V+
Sbjct: 476 KNVPIEEMNRVW 487
>At5g59250 D-xylose-H+ symporter - like protein
Length = 558
Score = 204 bits (519), Expect = 9e-53
Identities = 141/472 (29%), Positives = 237/472 (49%), Gaps = 27/472 (5%)
Query: 24 IVASMVSIVSGYDTGVMSGAMLFIKED-------LGISDTQQEVLAGILNLCALVGSLTA 76
I ++ ++ GYD G SGA L ++ S Q ++ AL+GS++
Sbjct: 103 IFPALGGLLFGYDIGATSGATLSLQSPALSGTTWFNFSPVQLGLVVSGSLYGALLGSISV 162
Query: 77 GRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVCGVGVGFALMIAPVYSAE 136
+D++GRR + +A++L++LG+ + G P+ IL+VGR + G G+G A+ AP+Y AE
Sbjct: 163 YGVADFLGRRRELIIAAVLYLLGSLITGCAPDLNILLVGRLLYGFGIGLAMHGAPLYIAE 222
Query: 137 ISSASSRGFLTSLPELCIGIGILLGYISNYVLGKY-LSLKLGWRLMLGIAALPSFVVAFC 195
+ RG L SL EL I +GILLG + +G + + + GWR M G + ++
Sbjct: 223 TCPSQIRGTLISLKELFIVLGILLG----FSVGSFQIDVVGGWRYMYGFGTPVALLMGLG 278
Query: 196 ILTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIKIAAGLDENCNDE---TVK 252
+ ++P SPRWL+++ GK + L + A +L+ + E D+ +VK
Sbjct: 279 MWSLPASPRWLLLRAVQGKGQ--LQEYKEKAMLALSKLRGRPPGDKISEKLVDDAYLSVK 336
Query: 253 LPQKSHQGEGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSPRIFRKAGITS 312
+ + G + E+ P L G+ F+ TG +V+ Y+ I + AG ++
Sbjct: 337 TAYEDEKSGGNFLEVFQGPNLKA---LTIGGGLVLFQQITGQPSVLYYAGSILQTAGFSA 393
Query: 313 KEKLLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLGLSLTVVDKSN 372
++ +G+ K++ +A+ +D LGRR LL G+ + L LL +
Sbjct: 394 AADATRVSVIIGVFKLLMTWVAVAKVDDLGRRPLLIGGVSGIALSLFLLSAYYKFLGGFP 453
Query: 373 GNVLWALILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVAVNRTMNAVV 432
+ AL+L YV + I GPI+W+ SEIFPL+ R +G S+ V N NA+V
Sbjct: 454 LVAVGALLL-------YVGCYQISFGPISWLMVSEIFPLRTRGRGISLAVLTNFGSNAIV 506
Query: 433 SMTFISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGKALEEMEMVFTK 484
+ F + + + F +F GI++++ LF +PETKG +LEE+E K
Sbjct: 507 TFAFSPLKEFLGAENLFLLFGGIALVSLLFVILVVPETKGLSLEEIESKILK 558
>At1g11260 glucose transporter
Length = 522
Score = 203 bits (516), Expect = 2e-52
Identities = 150/489 (30%), Positives = 240/489 (48%), Gaps = 50/489 (10%)
Query: 20 FACAIVASMVSIVSGYDTGVMSGAML---FIKEDL-GISDTQQE---------------- 59
F C +VA+M ++ GYD G+ G F+K + QQE
Sbjct: 25 FTC-VVAAMGGLIFGYDIGISGGVTSMPSFLKRFFPSVYRKQQEDASTNQYCQYDSPTLT 83
Query: 60 VLAGILNLCALVGSLTAGRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVC 119
+ L L AL+ SL A + GRR ++ ILF GA + G+ + +L+VGR +
Sbjct: 84 MFTSSLYLAALISSLVASTVTRKFGRRLSMLFGGILFCAGALINGFAKHVWMLIVGRILL 143
Query: 120 GVGVGFALMIAPVYSAEISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLGWR 179
G G+GFA P+Y +E++ RG L +L I IGIL+ + NY K + GWR
Sbjct: 144 GFGIGFANQAVPLYLSEMAPYKYRGALNIGFQLSITIGILVAEVLNYFFAK-IKGGWGWR 202
Query: 180 LMLGIAALPSFVVAFCILTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIKIA 239
L LG A +P+ ++ L +P++P ++ +GQ +EA+ +L+ I+
Sbjct: 203 LSLGGAVVPALIITIGSLVLPDTPNSMIERGQ--------------HEEAKTKLRRIR-- 246
Query: 240 AGLDENCN--DETVKLPQKSHQGEGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAV 297
G+D+ D+ V ++S E W+ L+ R R L AV I FF+ TGI +
Sbjct: 247 -GVDDVSQEFDDLVAASKESQSIEHPWRNLLRR---KYRPHLTMAVMIPFFQQLTGINVI 302
Query: 298 MLYSPRIFRKAGITSKEKLLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIG 357
M Y+P +F G T+ L+ A + G + +++++ +D+ GRR L M+I
Sbjct: 303 MFYAPVLFNTIGFTTDASLMSAVV-TGSVNVAATLVSIYGVDRWGRRFLFLEGGTQMLIC 361
Query: 358 LTLLGLSLTV---VDKSNGNV-LWALILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKL 413
++ + VD + G + W I+ + YVA F GP+ W+ SEIFPL++
Sbjct: 362 QAVVAACIGAKFGVDGTPGELPKWYAIVVVTFICIYVAGFAWSWGPLGWLVPSEIFPLEI 421
Query: 414 RAQGASIGVAVNRTMNAVVSMTFISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGK 473
R+ SI V+VN +++ F+++ + G F +FA V+ +F Y FLPETKG
Sbjct: 422 RSAAQSITVSVNMIFTFIIAQIFLTMLCHLKF-GLFLVFAFFVVVMSIFVYIFLPETKGI 480
Query: 474 ALEEMEMVF 482
+EEM V+
Sbjct: 481 PIEEMGQVW 489
>At5g27360 sugar-porter family protein 2 (SFP2)
Length = 478
Score = 199 bits (507), Expect = 2e-51
Identities = 134/463 (28%), Positives = 232/463 (49%), Gaps = 34/463 (7%)
Query: 21 ACAIVASMVSIVSGYDTGVM----SGAMLFIKEDLGISDTQQEVLAGILNLCALVGSLTA 76
AC I+++ +++ + GV SGA + I +DL +S Q A + L A +G+L +
Sbjct: 33 ACVILSTFIAVCGSFSFGVSLGYTSGAEIGIMKDLDLSIAQFSAFASLSTLGAAIGALFS 92
Query: 77 GRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVCGVGVGFALMIAPVYSAE 136
G+ + +GRR T++++ +L I+G + + + L GR G+G+G + PVY AE
Sbjct: 93 GKMAIILGRRKTMWVSDLLCIIGWFSIAFAKDVMWLNFGRISSGIGLGLISYVVPVYIAE 152
Query: 137 ISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLGWRLMLGIAALPSFVVAFCI 196
IS RG T +L G+ + Y S G +L+ WR++ + ALP F+ +
Sbjct: 153 ISPKHVRGTFTFTNQLLQNSGLAMVYFS----GNFLN----WRILALLGALPCFIQVIGL 204
Query: 197 LTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIKIAAGLDENCNDETVKLPQK 256
+PESPRWL G + + L+++ + DI++ + EN
Sbjct: 205 FFVPESPRWLAKVGSDKELENSLLRLRGGNADISREASDIEVMTKMVEN----------- 253
Query: 257 SHQGEGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSPRIFRKAGITSKEKL 316
+ + +L R R+ L+ +G+ + +G AV+ Y+ I RKAG + +
Sbjct: 254 --DSKSSFCDLFQR---KYRYTLVVGIGLMLIQQFSGSSAVLSYASTILRKAGFS----V 304
Query: 317 LLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLGLSLTVVDKSNGNVL 376
+ + +GL I +I + L+DK GRR LL S GM I L+G++ T + K
Sbjct: 305 TIGSTLLGLFMIPKAMIGVILVDKWGRRPLLLTSVSGMCITSMLIGVAFT-LQKMQLLPE 363
Query: 377 WALILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVAVNRTMNAVVSMTF 436
+ + + Y+ + IGLG + WV SEIFP+ ++ SI V+ + +++V+ F
Sbjct: 364 LTPVFTFICVTLYIGTYAIGLGGLPWVIMSEIFPMNIKVTAGSIVTLVSWSSSSIVTYAF 423
Query: 437 ISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGKALEEME 479
+ + T G+F++F + LA LF + +PETKG +LEE++
Sbjct: 424 NFLLEWST-QGTFYVFGAVGGLALLFIWLLVPETKGLSLEEIQ 465
>At1g50310 hexose transporter, putative
Length = 517
Score = 199 bits (506), Expect = 3e-51
Identities = 141/491 (28%), Positives = 242/491 (48%), Gaps = 50/491 (10%)
Query: 18 YAFACAIVASMVSIVSGYDTGVMSG--------AMLFIKEDLGISDTQQE---------- 59
+ IVA+M ++ GYD G+ G + F + D + + ++E
Sbjct: 24 FVIMTCIVAAMGGLLFGYDLGISGGVTSMEEFLSKFFPEVDKQMHEARRETAYCKFDNQL 83
Query: 60 --VLAGILNLCALVGSLTAGRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRC 117
+ L L AL S A + GR+ ++F+ + F++G+ + N A+L+VGR
Sbjct: 84 LQLFTSSLYLAALASSFVASAVTRKYGRKISMFVGGVAFLIGSLFNAFATNVAMLIVGRL 143
Query: 118 VCGVGVGFALMIAPVYSAEISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLG 177
+ GVGVGFA PVY +E++ A RG L ++ I IGIL+ + NY G K G
Sbjct: 144 LLGVGVGFANQSTPVYLSEMAPAKIRGALNIGFQMAITIGILIANLINY--GTSQMAKNG 201
Query: 178 WRLMLGIAALPSFVVAFCILTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIK 237
WR+ LG+AA+P+ ++ +P++P ++ +G+ +A+++L + I+
Sbjct: 202 WRVSLGLAAVPAVIMVIGSFVLPDTPNSMLERGKYEQAREMLQK--------------IR 247
Query: 238 IAAGLDENCNDETVKLPQKSHQGEGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAV 297
A +DE D + + + + WK + + R L+ I FF+ TGI +
Sbjct: 248 GADNVDEEFQD-LCDACEAAKKVDNPWKNIFQQ--AKYRPALVFCSAIPFFQQITGINVI 304
Query: 298 MLYSPRIFRKAGITSKEKLLLATIGVGLTKIVFLVIALFLLDKLGRRRL-----LQISTG 352
M Y+P +F+ G L+ A I G +V +++++ +D+ GRR L +Q+
Sbjct: 305 MFYAPVLFKTLGFADDASLISAVI-TGAVNVVSTLVSIYAVDRYGRRILFLEGGIQMIVS 363
Query: 353 GMIIGLTLLGLSLTVVDKSNGNVLWALILSIVATYA-YVAFFNIGLGPITWVYSSEIFPL 411
+++G TL+G+ +G + A I+A YVA F GP+ W+ SEI PL
Sbjct: 364 QIVVG-TLIGMKFGTT--GSGTLTPATADWILAFICLYVAGFAWSWGPLGWLVPSEICPL 420
Query: 412 KLRAQGASIGVAVNRTMNAVVSMTFISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETK 471
++R G +I V+VN ++ F+++ + G F+ F G+ + +F YF LPETK
Sbjct: 421 EIRPAGQAINVSVNMFFTFLIGQFFLTMLCHMKF-GLFYFFGGMVAVMTVFIYFLLPETK 479
Query: 472 GKALEEMEMVF 482
G +EEM V+
Sbjct: 480 GVPIEEMGRVW 490
>At3g05960 putative hexose transporter
Length = 507
Score = 199 bits (505), Expect = 4e-51
Identities = 147/485 (30%), Positives = 239/485 (48%), Gaps = 50/485 (10%)
Query: 18 YAFACAIVASMVSIVSGYDTGVMSGAML---FIKEDLGIS---------------DTQ-Q 58
Y F C ++A++ ++ GYD G+ G F+KE D Q
Sbjct: 20 YVFICVMIAAVGGLIFGYDIGISGGVSAMDDFLKEFFPAVWERKKHVHENNYCKYDNQFL 79
Query: 59 EVLAGILNLCALVGSLTAGRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCV 118
++ L L ALV S A T +GRR T+ ASI F++G GL N +L++GR
Sbjct: 80 QLFTSSLYLAALVASFVASATCSKLGRRPTMQFASIFFLIGVGLTAGAVNLVMLIIGRLF 139
Query: 119 CGVGVGFALMIAPVYSAEISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLGW 178
G GVGF P++ +EI+ A RG L + +L + IGIL+ I NY GW
Sbjct: 140 LGFGVGFGNQAVPLFLSEIAPAQLRGGLNIVFQLMVTIGILIANIVNYFTATV--HPYGW 197
Query: 179 RLMLGIAALPSFVVAFCILTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIKI 238
R+ LG A +P+ ++ F L + E+P L+ + + + K+ L ++
Sbjct: 198 RIALGGAGIPAVILLFGSLLIIETPTSLIERNKNEEGKEALRKI---------------- 241
Query: 239 AAGLDENCNDE---TVKLPQKSHQGEGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIE 295
G+D+ NDE V + Q + +++L+ P+ R I + + F+ TGI
Sbjct: 242 -RGVDD-INDEYESIVHACDIASQVKDPYRKLL---KPASRPPFIIGMLLQLFQQFTGIN 296
Query: 296 AVMLYSPRIFRKAGITSKEKLLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMI 355
A+M Y+P +F+ G S LL A I G ++ + ++L+D+ GRR LL S+ M+
Sbjct: 297 AIMFYAPVLFQTVGFGSDAALLSAVI-TGSINVLATFVGIYLVDRTGRRFLLLQSSVHML 355
Query: 356 IGLTLLGLSLTVVDKSNGNV--LWALILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKL 413
I ++G+ L G + AL++ ++ YV F GP+ W+ SE FPL+
Sbjct: 356 ICQLIIGIILAKDLGVTGTLGRPQALVV-VIFVCVYVMGFAWSWGPLGWLIPSETFPLET 414
Query: 414 RAQGASIGVAVNRTMNAVVSMTFISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGK 473
R+ G ++ V+ N V++ F+S+ + G FF F+G ++ LF +FF+PETKG
Sbjct: 415 RSAGFAVAVSCNMFFTFVIAQAFLSMLCGMR-SGIFFFFSGWIIVMGLFAFFFIPETKGI 473
Query: 474 ALEEM 478
A+++M
Sbjct: 474 AIDDM 478
>At3g05157 sugar transporter like protein
Length = 458
Score = 197 bits (502), Expect = 9e-51
Identities = 139/488 (28%), Positives = 234/488 (47%), Gaps = 42/488 (8%)
Query: 1 MIINQGDKEDQTNTFNKYAFACAIVASMVSIVSGYD----TGVMSGAMLFIKEDLGISDT 56
++ ++ D++D+ T AC I+++ V++ S + G SGA I ++L +S
Sbjct: 5 LLRHENDRDDRRIT------ACVILSTFVAVCSSFSYGCANGYTSGAETAIMKELDLSMA 58
Query: 57 QQEVLAGILNLCALVGSLTAGRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGR 116
Q LNL VG+L +G+ + +GRR T++ + I G + + N L +GR
Sbjct: 59 QFSAFGSFLNLGGAVGALFSGQLAVILGRRRTLWACDLFCIFGWLSIAFAKNVLWLDLGR 118
Query: 117 CVCGVGVGFALMIAPVYSAEISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKL 176
G+GVG + PVY AEI+ RG ++ L GI L Y + +
Sbjct: 119 ISLGIGVGLTSYVVPVYIAEITPKHVRGAFSASTLLLQNSGISLIY--------FFGTVI 170
Query: 177 GWRLMLGIAALPSFVVAFCILTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDI 236
WR++ I ALP F+ I +PESPRWL G + + + L ++ + +I
Sbjct: 171 NWRVLAVIGALPCFIPVIGIYFIPESPRWLAKIGSVKEVENSLHRLRGKDADVSDEAAEI 230
Query: 237 KIAAG-LDENCNDETVKLPQKSHQGEGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIE 295
++ L+E+ + QK + R L+ +G+ + +G
Sbjct: 231 QVMTKMLEEDSKSSFCDMFQKKY-----------------RRTLVVGIGLMLIQQLSGAS 273
Query: 296 AVMLYSPRIFRKAGITSKEKLLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMI 355
+ YS IFRKAG + + L ++ G+ I ++ L L+D+ GRR LL S GM
Sbjct: 274 GITYYSNAIFRKAGFSER----LGSMIFGVFVIPKALVGLILVDRWGRRPLLLASAVGMS 329
Query: 356 IGLTLLGLSLTVVDKSNGNVLWALILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRA 415
IG L+G+S T + + N + + + Y FF IG+G + W+ SEIFP+ ++
Sbjct: 330 IGSLLIGVSFT-LQEMNLFPEFIPVFVFINILVYFGFFAIGIGGLPWIIMSEIFPINIKV 388
Query: 416 QGASIGVAVNRTMNAVVSMTFISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGKAL 475
SI + T VS F +++ + G+F++FA + L+ LF + +PETKG++L
Sbjct: 389 SAGSIVALTSWTTGWFVSYGFNFMFE-WSAQGTFYIFAMVGGLSLLFIWMLVPETKGQSL 447
Query: 476 EEMEMVFT 483
EE++ T
Sbjct: 448 EELQASLT 455
>At1g30220 unknown protein
Length = 580
Score = 193 bits (491), Expect = 2e-49
Identities = 123/350 (35%), Positives = 185/350 (52%), Gaps = 20/350 (5%)
Query: 16 NKYAFACAIVASMVSIVSGYDTGVMSGAMLFIKEDLGISDTQ---QEVLAGILNLCALVG 72
N Y A A + ++ GYDTGV+SGA+L+I++D D QE++ + A+VG
Sbjct: 25 NPYVLRLAFSAGIGGLLFGYDTGVISGALLYIRDDFKSVDRNTWLQEMIVSMAVAGAIVG 84
Query: 73 SLTAGRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVCGVGVGFALMIAPV 132
+ G +D +GRR I +A LF+LGA +M PN ++L+VGR G+GVG A M AP+
Sbjct: 85 AAIGGWANDKLGRRSAILMADFLFLLGAIIMAAAPNPSLLVVGRVFVGLGVGMASMTAPL 144
Query: 133 YSAEISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLGWRLMLGIAALPSFVV 192
Y +E S A RG L S I G L Y+ N + + WR MLGIA +P+ +
Sbjct: 145 YISEASPAKIRGALVSTNGFLITGGQFLSYLINLA---FTDVTGTWRWMLGIAGIPALLQ 201
Query: 193 AFCILTMPESPRWLVMQGQLGKAKKVLMQV-SNTTQEAELRLKDIKIAAGLDENCNDETV 251
+ T+PESPRWL +G+ +AK +L ++ S E E+R + + E + E +
Sbjct: 202 FVLMFTLPESPRWLYRKGREEEAKAILRRIYSAEDVEQEIRALKDSVETEILEEGSSEKI 261
Query: 252 KLPQKSHQGEGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSPRIFRKAGIT 311
+ + L +VR LIA VG+ F+ GI VM YSP I + AG
Sbjct: 262 NM-------------IKLCKAKTVRRGLIAGVGLQVFQQFVGINTVMYYSPTIVQLAGFA 308
Query: 312 SKEKLLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLL 361
S LL ++ +I+++ +D++GR++LL IS G+II L +L
Sbjct: 309 SNRTALLLSLVTAGLNAFGSIISIYFIDRIGRKKLLIISLFGVIISLGIL 358
Score = 79.7 bits (195), Expect = 3e-15
Identities = 37/98 (37%), Positives = 60/98 (60%)
Query: 389 YVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVAVNRTMNAVVSMTFISIYKAITIGGS 448
Y+ FF+ G+G + W+ +SEI+PL+ R I N N +V+ +F+S+ +AI +
Sbjct: 463 YIIFFSPGMGTVPWIVNSEIYPLRFRGICGGIAATANWISNLIVAQSFLSLTEAIGTSWT 522
Query: 449 FFMFAGISVLAWLFFYFFLPETKGKALEEMEMVFTKKS 486
F +F ISV+A LF +PETKG +EE+E + ++S
Sbjct: 523 FLIFGVISVIALLFVMVCVPETKGMPMEEIEKMLERRS 560
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.325 0.141 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,345,856
Number of Sequences: 26719
Number of extensions: 425499
Number of successful extensions: 1820
Number of sequences better than 10.0: 105
Number of HSP's better than 10.0 without gapping: 65
Number of HSP's successfully gapped in prelim test: 40
Number of HSP's that attempted gapping in prelim test: 1432
Number of HSP's gapped (non-prelim): 151
length of query: 501
length of database: 11,318,596
effective HSP length: 103
effective length of query: 398
effective length of database: 8,566,539
effective search space: 3409482522
effective search space used: 3409482522
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 62 (28.5 bits)
Medicago: description of AC147964.1


