

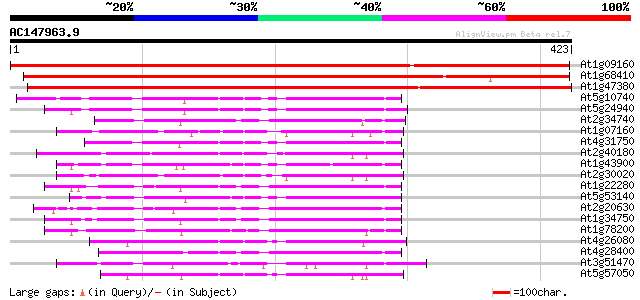
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147963.9 - phase: 0
(423 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g09160 putative protein phosphatase 2C 635 0.0
At1g68410 putative protein phosphatase 569 e-163
At1g47380 unknown protein 478 e-135
At5g10740 protein phosphatase 2C -like protein 112 3e-25
At5g24940 Protein phosphatase 2C-like protein 109 2e-24
At2g34740 putative protein phosphatase 2C 109 2e-24
At1g07160 unknown protein 109 3e-24
At4g31750 unknown protein 108 4e-24
At2g40180 protein phosphatase 2C (AthPP2C5) 106 3e-23
At1g43900 protein phosphatase type 2C like protein 104 8e-23
At2g30020 putative protein phosphatase 2C 103 2e-22
At1g22280 protein phosphatase type 2C like protein 100 2e-21
At5g53140 protein phosphatase 2C-like 100 3e-21
At2g20630 putative protein phosphatase 2C 99 3e-21
At1g34750 protein phosphatase type 2C, putative 99 6e-21
At1g78200 putative protein phosphatase 2C 98 8e-21
At4g26080 abscisic acid insensitive protein (ABI1) 96 4e-20
At4g28400 protein phosphatase 2C like protein 96 5e-20
At3g51470 protein phosphatase 2C-like protein 93 2e-19
At5g57050 protein phosphatase 2C ABI2 (PP2C) (sp|O04719) 91 9e-19
>At1g09160 putative protein phosphatase 2C
Length = 428
Score = 635 bits (1639), Expect = 0.0
Identities = 317/422 (75%), Positives = 359/422 (84%), Gaps = 2/422 (0%)
Query: 1 MSKAELSRMKQPLVPLATLIGRELRNGKTEKPFVKYGQAGLAKKGEDYFLIKTDCHRVPG 60
MS ++ SR + LVPLATLIGRELR+ K EKPFVKYGQA LAKKGEDYFLIKTDC RVPG
Sbjct: 1 MSVSKASRTQHSLVPLATLIGRELRSEKVEKPFVKYGQAALAKKGEDYFLIKTDCERVPG 60
Query: 61 DSSTSFSVFAILDGHNGISAAIFAKENIINNVMSAIPQGVSREEWLQALPRALVVAFVKT 120
D S++FSVF I DGHNG SAAI+ KE+++ NV+SAIPQG SR+EWLQALPRALV FVKT
Sbjct: 61 DPSSAFSVFGIFDGHNGNSAAIYTKEHLLENVVSAIPQGASRDEWLQALPRALVAGFVKT 120
Query: 121 DMEFQKKGETSGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEER 180
D+EFQ+KGETSGTT TFVIIDGWT+TVASVGDSRCILDTQGGVVSLLTVDHRLEENVEER
Sbjct: 121 DIEFQQKGETSGTTVTFVIIDGWTITVASVGDSRCILDTQGGVVSLLTVDHRLEENVEER 180
Query: 181 ERVTASGGEVGRLNVFGGNEVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQVKLSNA 240
ER+TASGGEVGRLNVFGGNEVGPLRCWPGGLCLSRSIGDTDVGE+IVPIPHVKQVKL +A
Sbjct: 181 ERITASGGEVGRLNVFGGNEVGPLRCWPGGLCLSRSIGDTDVGEFIVPIPHVKQVKLPDA 240
Query: 241 GGRLIIASDGIWDTLSSDMAAKSCRGVPAELAAKLVVKEALRSRGLKDDTTCLVVDIIPS 300
GGRLIIASDGIWD LSSD+AAK+CRG+ A+LAAKLVVKEALR++GLKDDTTC+VVDI+PS
Sbjct: 241 GGRLIIASDGIWDILSSDVAAKACRGLSADLAAKLVVKEALRTKGLKDDTTCVVVDIVPS 300
Query: 301 DYPVLPMPATPRKKHNVLSSLLFGKKSQNLANKGTNKLSAVGVVEELFEEGSAMLTERLG 360
+ L + P KK N +S L K + NK NKLSAVGVVEELFEEGSA+L +RLG
Sbjct: 301 GH--LSLAPAPMKKQNPFTSFLSRKNHMDTNNKNGNKLSAVGVVEELFEEGSAVLADRLG 358
Query: 361 NNVPSDTNSGIHRCAVCLADQPSGDGLSVNNDHFITPVSKPWEGPFLCTNCQKKKDAMEG 420
++ S+T +G+ +CAVC D+ + LS N I+ SK WEGPFLCT C+KKKDAMEG
Sbjct: 359 KDLLSNTETGLLKCAVCQIDESPSEDLSSNGGSIISSASKRWEGPFLCTICKKKKDAMEG 418
Query: 421 KR 422
KR
Sbjct: 419 KR 420
>At1g68410 putative protein phosphatase
Length = 436
Score = 569 bits (1467), Expect = e-163
Identities = 284/415 (68%), Positives = 336/415 (80%), Gaps = 4/415 (0%)
Query: 11 QPLVPLATLIGRELRNGKTEKPFVKYGQAGLAKKGEDYFLIKTDCHRVPGDSSTSFSVFA 70
+ LVPLA LI RE + K EKP V++GQA ++KGEDY LIKTD RVP +SST+FSVFA
Sbjct: 16 EKLVPLAALISRETKAAKMEKPIVRFGQAAQSRKGEDYVLIKTDSLRVPSNSSTAFSVFA 75
Query: 71 ILDGHNGISAAIFAKENIINNVMSAIPQGVSREEWLQALPRALVVAFVKTDMEFQKKGET 130
+ DGHNG +AA++ +EN++N+V+SA+P G+SR+EWL ALPRALV FVKTD EFQ +GET
Sbjct: 76 VFDGHNGKAAAVYTRENLLNHVISALPSGLSRDEWLHALPRALVSGFVKTDKEFQSRGET 135
Query: 131 SGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEERERVTASGGEV 190
SGTTATFVI+DGWTVTVA VGDSRCILDT+GG VS LTVDHRLE+N EERERVTASGGEV
Sbjct: 136 SGTTATFVIVDGWTVTVACVGDSRCILDTKGGSVSNLTVDHRLEDNTEERERVTASGGEV 195
Query: 191 GRLNVFGGNEVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQVKLSNAGGRLIIASDG 250
GRL++ GG E+GPLRCWPGGLCLSRSIGD DVGE+IVP+P VKQVKLSN GGRLIIASDG
Sbjct: 196 GRLSIVGGVEIGPLRCWPGGLCLSRSIGDMDVGEFIVPVPFVKQVKLSNLGGRLIIASDG 255
Query: 251 IWDTLSSDMAAKSCRGVPAELAAKLVVKEALRSRGLKDDTTCLVVDIIPSDYPVLPMPAT 310
IWD LSS++AAK+CRG+ AELAA+ VVKEALR RGLKDDTTC+VVDIIP + P P+
Sbjct: 256 IWDALSSEVAAKTCRGLSAELAARQVVKEALRRRGLKDDTTCIVVDIIPPENFQEPPPSP 315
Query: 311 PRKKHNVLSSLLFGKKSQNLANKGTNKLSAVGVVEELFEEGSAMLTERLGN---NVPSDT 367
P+K +N SLLF KKS N +NK + KLS VG+VEELFEEGSAML ERLG+ + S T
Sbjct: 316 PKKHNNFFKSLLFRKKS-NSSNKLSKKLSTVGIVEELFEEGSAMLAERLGSGDCSKESTT 374
Query: 368 NSGIHRCAVCLADQPSGDGLSVNNDHFITPVSKPWEGPFLCTNCQKKKDAMEGKR 422
GI CA+C D +G+SV+ + KPW+GPFLCT+C+ KKDAMEGKR
Sbjct: 375 GGGIFTCAICQLDLAPSEGISVHAGSIFSTSLKPWQGPFLCTDCRDKKDAMEGKR 429
>At1g47380 unknown protein
Length = 428
Score = 478 bits (1231), Expect = e-135
Identities = 232/410 (56%), Positives = 304/410 (73%), Gaps = 1/410 (0%)
Query: 14 VPLATLIGRELRNGKTEKPFVKYGQAGLAKKGEDYFLIKTDCHRVPGDSSTSFSVFAILD 73
VPL+ L+ RE N K + P + +GQ +KKGED+ L+KT+C RV GD T+FSVF + D
Sbjct: 10 VPLSVLLKRESANEKIDNPELIHGQHNQSKKGEDFTLVKTECQRVMGDGVTTFSVFGLFD 69
Query: 74 GHNGISAAIFAKENIINNVMSAIPQGVSREEWLQALPRALVVAFVKTDMEFQKKGETSGT 133
GHNG +AAI+ KEN++NNV++AIP ++R+EW+ ALPRALV FVKTD +FQ++ TSGT
Sbjct: 70 GHNGSAAAIYTKENLLNNVLAAIPSDLNRDEWVAALPRALVAGFVKTDKDFQERARTSGT 129
Query: 134 TATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEERERVTASGGEVGRL 193
T TFVI++GW V+VASVGDSRCIL+ G V L+ DHRLE N EER+RVTASGGEVGRL
Sbjct: 130 TVTFVIVEGWVVSVASVGDSRCILEPAEGGVYYLSADHRLEINEEERDRVTASGGEVGRL 189
Query: 194 NVFGGNEVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQVKLSNAGGRLIIASDGIWD 253
N GG E+GPLRCWPGGLCLSRSIGD DVGEYIVP+P+VKQVKLS+AGGRLII+SDG+WD
Sbjct: 190 NTGGGTEIGPLRCWPGGLCLSRSIGDLDVGEYIVPVPYVKQVKLSSAGGRLIISSDGVWD 249
Query: 254 TLSSDMAAKSCRGVPAELAAKLVVKEALRSRGLKDDTTCLVVDIIPSDYPVLPMPATPRK 313
+S++ A CRG+P E +A+ +VKEA+ +G++DDTTC+VVDI+P + P +P P+K
Sbjct: 250 AISAEEALDCCRGLPPESSAEHIVKEAVGKKGIRDDTTCIVVDILPLEKPAASVP-PPKK 308
Query: 314 KHNVLSSLLFGKKSQNLANKGTNKLSAVGVVEELFEEGSAMLTERLGNNVPSDTNSGIHR 373
+ + +F +K+ + ++ + + VVEELFEEGSAML+ERL P +
Sbjct: 309 QGKGMLKSMFKRKTSDSSSNIEKEYAEPDVVEELFEEGSAMLSERLDTKYPLCNMFKLFM 368
Query: 374 CAVCLADQPSGDGLSVNNDHFITPVSKPWEGPFLCTNCQKKKDAMEGKRS 423
CAVC + G+G+S++ +PW+GPFLC +CQ KKDAMEGKRS
Sbjct: 369 CAVCQVEVKPGEGVSIHAGSDNCRKLRPWDGPFLCASCQDKKDAMEGKRS 418
>At5g10740 protein phosphatase 2C -like protein
Length = 354
Score = 112 bits (281), Expect = 3e-25
Identities = 92/297 (30%), Positives = 154/297 (50%), Gaps = 34/297 (11%)
Query: 6 LSRMKQPLVPLATLIGREL-RNGKTEKPFVKYGQAGLAKKGEDYFLIKTDCHRVPGDSST 64
LS QP A G L +NGK + AG ED+F + D G +
Sbjct: 8 LSYSNQPQTVEAPASGGGLSQNGKFSYGYAS--SAGKRSSMEDFFETRID-----GINGE 60
Query: 65 SFSVFAILDGHNGISAAIFAKENIINNVMSAIPQGVSREEWLQALPRALVVAFVKTDMEF 124
+F + DGH G AA + K ++ +N+ ++ +++ A+ A+ TD E
Sbjct: 61 IVGLFGVFDGHGGARAAEYVKRHLFSNL-------ITHPKFISDTKSAITDAYNHTDSEL 113
Query: 125 QKKGET----SGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEER 180
K + +G+TA+ I+ G + VA+VGDSR ++ ++GG ++ DH+ +++ +ER
Sbjct: 114 LKSENSHNRDAGSTASTAILVGDRLVVANVGDSRAVI-SRGGKAIAVSRDHKPDQS-DER 171
Query: 181 ERVTASGGEVGRLNVFGGN-EVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQVKLSN 239
ER+ +GG V ++ G VG G L +SR+ GD + +Y+V P +++ K+ +
Sbjct: 172 ERIENAGGFV----MWAGTWRVG------GVLAVSRAFGDRLLKQYVVADPEIQEEKIDD 221
Query: 240 AGGRLIIASDGIWDTLSSDMAAKSCRGV-PAELAAKLVVKEALRSRGLKDDTTCLVV 295
LI+ASDG+WD S++ A + V E +AK +V EA++ RG D+ TC+VV
Sbjct: 222 TLEFLILASDGLWDVFSNEAAVAMVKEVEDPEDSAKKLVGEAIK-RGSADNITCVVV 277
>At5g24940 Protein phosphatase 2C-like protein
Length = 447
Score = 109 bits (273), Expect = 2e-24
Identities = 86/283 (30%), Positives = 148/283 (51%), Gaps = 34/283 (12%)
Query: 27 GKTEKPFVKYGQAGLAKKG---EDYFLIKTDCHRVPGDSSTSFSVFAILDGHNGISAAIF 83
G ++ YG A A K ED+F + D G +F + DGH G AA +
Sbjct: 25 GLSQNGKFSYGYASSAGKRSSMEDFFETRID-----GIDGEIVGLFGVFDGHGGSRAAEY 79
Query: 84 AKENIINNVMSAIPQGVSREEWLQALPRALVVAFVKTDMEFQKKGET----SGTTATFVI 139
K ++ +N+ ++ +++ A+ A+ TD E K + +G+TA+ I
Sbjct: 80 VKRHLFSNL-------ITHPKFISDTKSAIADAYTHTDSELLKSENSHTRDAGSTASTAI 132
Query: 140 IDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEERERVTASGGEVGRLNVFGGN 199
+ G + VA+VGDSR ++ +GG ++ DH+ +++ +ERER+ +GG V ++ G
Sbjct: 133 LVGDRLLVANVGDSRAVI-CRGGNAFAVSRDHKPDQS-DERERIENAGGFV----MWAGT 186
Query: 200 -EVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQVKLSNAGGRLIIASDGIWDTLSSD 258
VG G L +SR+ GD + +Y+V P +++ K+ ++ LI+ASDG+WD S++
Sbjct: 187 WRVG------GVLAVSRAFGDRLLKQYVVADPEIQEEKIDDSLEFLILASDGLWDVFSNE 240
Query: 259 MAAKSCRGV-PAELAAKLVVKEALRSRGLKDDTTCLVVDIIPS 300
A + V E + K +V EA++ RG D+ TC+VV + S
Sbjct: 241 EAVAVVKEVEDPEESTKKLVGEAIK-RGSADNITCVVVRFLES 282
>At2g34740 putative protein phosphatase 2C
Length = 239
Score = 109 bits (273), Expect = 2e-24
Identities = 79/239 (33%), Positives = 133/239 (55%), Gaps = 24/239 (10%)
Query: 65 SFSVFAILDGHNGISAAIFAKENIINNVMSAIPQGVSREEWLQALPRALVVAFVKTDMEF 124
+ ++AI DGH+G A + + ++ +N++S + ++ + +A+ A+ TD
Sbjct: 16 NLGLYAIFDGHSGSDVADYLQNHLFDNILS-------QPDFWRNPKKAIKRAYKSTDDYI 68
Query: 125 QKK--GETSGTTA-TFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEERE 181
+ G G+TA T ++IDG + VA+VGDSR IL + VV +TVDH E +ER+
Sbjct: 69 LQNVVGPRGGSTAVTAIVIDGKKIVVANVGDSRAILCRESDVVKQITVDH---EPDKERD 125
Query: 182 RVTASGGEVGRLNVFGGNEVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQVKLSNAG 241
V + GG F + G + G L ++R+ GD + E+I IP+++ ++ +
Sbjct: 126 LVKSKGG-------FVSQKPGNVPRVDGQLAMTRAFGDGGLKEHISVIPNIEIAEIHDDT 178
Query: 242 GRLIIASDGIWDTLSSDMAAKSC--RGVPAELAAKLVVKEALRSRGLKDDTTCLVVDII 298
LI+ASDG+W +S+D RG AE AAK+++ +AL +RG KDD +C+VV +
Sbjct: 179 KFLILASDGLWKVMSNDEVWDQIKKRG-NAEEAAKMLIDKAL-ARGSKDDISCVVVSFL 235
>At1g07160 unknown protein
Length = 380
Score = 109 bits (272), Expect = 3e-24
Identities = 85/274 (31%), Positives = 138/274 (50%), Gaps = 35/274 (12%)
Query: 36 YGQAGLAKKGEDYFLIKTDCHRVPGDSSTSFSVFAILDGHNGISAAIFAKENIINNVMSA 95
Y + G + ED F T+ P + +F + DGH G +AA FA +N+ +N++
Sbjct: 126 YCKRGKREAMEDRFSAITNLQGDPKQA-----IFGVYDGHGGPTAAEFAAKNLCSNILGE 180
Query: 96 IPQGVSREEWLQALPRALVVAFVKTDMEFQKKGETSGTTA--TFVIIDGWTVTVASVGDS 153
I G + + +A+ R ++ TD EF K+ G + T +I DG + VA+ GD
Sbjct: 181 IVGGRNESKIEEAVKRG----YLATDSEFLKEKNVKGGSCCVTALISDG-NLVVANAGDC 235
Query: 154 RCILDTQGGVVSLLTVDHRLEENVEERERVTASGGEVGRLNVFGGNEVGPLRCW--PGGL 211
R +L GG LT DHR + +ER R+ +SGG V N W G L
Sbjct: 236 RAVLSV-GGFAEALTSDHRPSRD-DERNRIESSGGYVDTFN----------SVWRIQGSL 283
Query: 212 CLSRSIGDTDVGEYIVPIPHVKQVKLSNAGGRLIIASDGIWDTLSS----DMAAKSCRGV 267
+SR IGD + ++I+ P + ++++ LI+ASDG+WD +S+ D+A C+G
Sbjct: 284 AVSRGIGDAHLKQWIISEPEINILRINPQHEFLILASDGLWDKVSNQEAVDIARPFCKGT 343
Query: 268 PAE----LAAKLVVKEALRSRGLKDDTTCLVVDI 297
+ LA K +V ++ SRG DD + +++ +
Sbjct: 344 DQKRKPLLACKKLVDLSV-SRGSLDDISVMLIQL 376
>At4g31750 unknown protein
Length = 311
Score = 108 bits (271), Expect = 4e-24
Identities = 76/245 (31%), Positives = 136/245 (55%), Gaps = 26/245 (10%)
Query: 57 RVPGDSSTSFSVFAILDGHNGISAAIFAKENIINNVMSAIPQGVSREEWLQALPRALVVA 116
R+ G +F + DGH G AA + K+N+ +N+ + +++ A+ A
Sbjct: 53 RIDGVEGEIVGLFGVFDGHGGARAAEYVKQNLFSNL-------IRHPKFISDTTAAIADA 105
Query: 117 FVKTDMEFQK----KGETSGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHR 172
+ +TD EF K + +G+TA+ I+ G + VA+VGDSR ++ +GG ++ DH+
Sbjct: 106 YNQTDSEFLKSENSQNRDAGSTASTAILVGDRLLVANVGDSRAVI-CRGGNAIAVSRDHK 164
Query: 173 LEENVEERERVTASGGEVGRLNVFGGN-EVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPH 231
+++ +ER+R+ +GG V ++ G VG G L +SR+ GD + +Y+V P
Sbjct: 165 PDQS-DERQRIEDAGGFV----MWAGTWRVG------GVLAVSRAFGDRLLKQYVVADPE 213
Query: 232 VKQVKLSNAGGRLIIASDGIWDTLSSDMAAKSCRGV-PAELAAKLVVKEALRSRGLKDDT 290
+++ K+ ++ LI+ASDG+WD +S++ A + + E AK ++ EA + RG D+
Sbjct: 214 IQEEKVDSSLEFLILASDGLWDVVSNEEAVGMIKAIEDPEEGAKRLMMEAYQ-RGSADNI 272
Query: 291 TCLVV 295
TC+VV
Sbjct: 273 TCVVV 277
>At2g40180 protein phosphatase 2C (AthPP2C5)
Length = 390
Score = 106 bits (264), Expect = 3e-23
Identities = 82/283 (28%), Positives = 135/283 (46%), Gaps = 20/283 (7%)
Query: 21 GRELRNGKTEKPFVKYGQAGLAKKGEDYFLIKTDCHRVPGDSSTSFSVFAILDGHNGISA 80
G E+ + + + Y + G ED + D + G + F VF DGH G A
Sbjct: 117 GAEVVEAEEDGYYSVYCKRGRRGPMEDRYFAAVDRNDDGGYKNAFFGVF---DGHGGSKA 173
Query: 81 AIFAKENIINNVMSAIPQGVSREEWLQALPRALVVAFVKTDMEFQKKGETSGTTATFVII 140
A FA N+ NN+ +A+ S E+ ++ A+ ++KTD +F K+G G +I
Sbjct: 174 AEFAAMNLGNNIEAAMASARSGEDGC-SMESAIREGYIKTDEDFLKEGSRGGACCVTALI 232
Query: 141 DGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEERERVTASGGEVGRLNVFGGNE 200
+ V++ GD R ++ ++GG LT DH + E +R+ A GG V N
Sbjct: 233 SKGELAVSNAGDCRAVM-SRGGTAEALTSDHNPSQ-ANELKRIEALGGYVDCCN------ 284
Query: 201 VGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQVKLSNAGGRLIIASDGIWDTLSS--- 257
G R G L +SR IGD + E+++ P + +++ LI+ASDG+WD +++
Sbjct: 285 -GVWRI-QGTLAVSRGIGDRYLKEWVIAEPETRTLRIKPEFEFLILASDGLWDKVTNQEA 342
Query: 258 -DMAAKSCRGV--PAELAAKLVVKEALRSRGLKDDTTCLVVDI 297
D+ C GV P L+A + E RG DD + +++ +
Sbjct: 343 VDVVRPYCVGVENPMTLSACKKLAELSVKRGSLDDISLIIIQL 385
>At1g43900 protein phosphatase type 2C like protein
Length = 371
Score = 104 bits (260), Expect = 8e-23
Identities = 86/269 (31%), Positives = 141/269 (51%), Gaps = 35/269 (13%)
Query: 36 YGQAGLAKKG---EDYFLIKTDCHRVPGDSSTSFSVFAILDGHNGISAAIFAKENIINNV 92
YG + L K EDYF +T V G F VF DGH G A + K N+ N+
Sbjct: 124 YGYSSLKGKRATMEDYF--ETRISDVNGQMVAFFGVF---DGHGGARTAEYLKNNLFKNL 178
Query: 93 MSAIPQGVSREEWLQALPRALVVAFVKTDMEF--QKKGE--TSGTTATFVIIDGWTVTVA 148
VS ++++ +A+V F +TD E+ ++ G+ +G+TA + G + VA
Sbjct: 179 -------VSHDDFISDTKKAIVEVFKQTDEEYLIEEAGQPKNAGSTAATAFLIGDKLIVA 231
Query: 149 SVGDSRCILDTQGGVVSLLTVDHRLEENVEERERVTASGGEVGRLNVFGGN-EVGPLRCW 207
+VGDSR + G V L + DH+ + + +ER+R+ +GG + ++ G VG
Sbjct: 232 NVGDSRVVASRNGSAVPL-SDDHKPDRS-DERQRIEDAGGFI----IWAGTWRVG----- 280
Query: 208 PGGLCLSRSIGDTDVGEYIVPIPHVKQVKLSNAGGRLIIASDGIWDTLSSDMAAKSCRGV 267
G L +SR+ GD + Y++ P +++ +S +++ASDG+W+ LS+ A R +
Sbjct: 281 -GILAVSRAFGDKQLKPYVIAEPEIQEEDISTLEF-IVVASDGLWNVLSNKDAVAIVRDI 338
Query: 268 -PAELAAKLVVKEALRSRGLKDDTTCLVV 295
AE AA+ +V+E +RG D+ TC+VV
Sbjct: 339 SDAETAARKLVQEGY-ARGSCDNITCIVV 366
>At2g30020 putative protein phosphatase 2C
Length = 396
Score = 103 bits (256), Expect = 2e-22
Identities = 78/271 (28%), Positives = 133/271 (48%), Gaps = 30/271 (11%)
Query: 36 YGQAGLAKKGEDYFLIKTDCHRVPGDSSTSFSVFAILDGHNGISAAIFAKENIINNVMSA 95
Y + G + ED F T+ H GD + +F + DGH G+ AA FA +N+ N++
Sbjct: 143 YCKRGRREAMEDRFSAITNLH---GDRKQA--IFGVYDGHGGVKAAEFAAKNLDKNIVEE 197
Query: 96 IPQGVSREEWLQALPRALVVAFVKTDMEFQKKGETSG-TTATFVIIDGWTVTVASVGDSR 154
+ E +A+ ++ TD F K+ + G + +++ + V++ GD R
Sbjct: 198 VVGKRDESEIAEAVKHG----YLATDASFLKEEDVKGGSCCVTALVNEGNLVVSNAGDCR 253
Query: 155 CILDTQGGVVSLLTVDHRLEENVEERERVTASGGEVGRLNVFGGNEVGPLRCW--PGGLC 212
++ GGV L+ DHR + +ER+R+ +GG V + F G W G L
Sbjct: 254 AVMSV-GGVAKALSSDHRPSRD-DERKRIETTGGYV---DTFHG-------VWRIQGSLA 301
Query: 213 LSRSIGDTDVGEYIVPIPHVKQVKLSNAGGRLIIASDGIWDTLSS----DMAAKSCRGV- 267
+SR IGD + ++++ P K ++ + LI+ASDG+WD +S+ D+A C G
Sbjct: 302 VSRGIGDAQLKKWVIAEPETKISRIEHDHEFLILASDGLWDKVSNQEAVDIARPLCLGTE 361
Query: 268 -PAELAAKLVVKEALRSRGLKDDTTCLVVDI 297
P LAA + + SRG DD + +++ +
Sbjct: 362 KPLLLAACKKLVDLSASRGSSDDISVMLIPL 392
>At1g22280 protein phosphatase type 2C like protein
Length = 281
Score = 100 bits (249), Expect = 2e-21
Identities = 82/281 (29%), Positives = 137/281 (48%), Gaps = 39/281 (13%)
Query: 27 GKTEKPFVKYGQAGLAKKG----EDYFL---IKTDCHRVPGDSSTSFSVFAILDGHNGIS 79
G++++ +KYG + + K EDY + I H + +FAI DGH G S
Sbjct: 25 GRSDEGMIKYGFSLVKGKANHPMEDYHVANFINIQDHEL--------GLFAIYDGHMGDS 76
Query: 80 AAIFAKENIINNVMSAIPQGVSREEWLQALPRALVVAFVKTDMEFQKK----GETSGTTA 135
+ ++ + +N++ E W+ R++ A+ KTD G T
Sbjct: 77 VPAYLQKRLFSNILK------EGEFWVDPR-RSIAKAYEKTDQAILSNSSDLGRGGSTAV 129
Query: 136 TFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEERERVTASGGEVGRLNV 195
T ++I+G + +A+VGDSR +L + GG ++ ++ DH E ER + GG
Sbjct: 130 TAILINGRKLWIANVGDSRAVL-SHGGAITQMSTDH---EPRTERSSIEDRGG------- 178
Query: 196 FGGNEVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQVKLSNAGGRLIIASDGIWDTL 255
F N G + G L +SR+ GD + ++ P +K+ + + L++ASDGIW +
Sbjct: 179 FVSNLPGDVPRVNGQLAVSRAFGDKGLKTHLSSEPDIKEATVDSQTDVLLLASDGIWKVM 238
Query: 256 SSDMAAKSCRGV-PAELAAKLVVKEALRSRGLKDDTTCLVV 295
+++ A + R V + AAK + EALR R KDD +C+VV
Sbjct: 239 TNEEAMEIARRVKDPQKAAKELTAEALR-RESKDDISCVVV 278
>At5g53140 protein phosphatase 2C-like
Length = 420
Score = 99.8 bits (247), Expect = 3e-21
Identities = 73/255 (28%), Positives = 134/255 (51%), Gaps = 29/255 (11%)
Query: 46 EDYFLIKTDCHRVPGDSSTSFSVFAILDGHNGISAAIFAKENIINNVMSAIPQGVSREEW 105
ED++ IK + G + F +F DGH G AA + KE++ NN+M ++
Sbjct: 115 EDFYDIKAST--IEGQAVCMFGIF---DGHGGSRAAEYLKEHLFNNLMK-------HPQF 162
Query: 106 LQALPRALVVAFVKTDMEFQKKGETS----GTTATFVIIDGWTVTVASVGDSRCILDTQG 161
L AL + +TD+ F + + + G+TA+ ++ G + VA+VGDSR I+ G
Sbjct: 163 LTDTKLALNETYKQTDVAFLESEKDTYRDDGSTASAAVLVGNHLYVANVGDSRTIVSKAG 222
Query: 162 GVVSLLTVDHRLEENVEERERVTASGGEVGRLNVFGGN-EVGPLRCWPGGLCLSRSIGDT 220
++L + DH+ + +ER+R+ ++GG + ++ G VG G L +SR+ G+
Sbjct: 223 KAIAL-SDDHKPNRS-DERKRIESAGGVI----MWAGTWRVG------GVLAMSRAFGNR 270
Query: 221 DVGEYIVPIPHVKQVKLSNAGGRLIIASDGIWDTLSSDMAAKSCRGVPAELAAKLVVKEA 280
+ +++V P ++ +++ + L++ASDG+WD + ++ A + AA + +
Sbjct: 271 MLKQFVVAEPEIQDLEIDHEAELLVLASDGLWDVVPNEDAVALAQSEEEPEAAARKLTDT 330
Query: 281 LRSRGLKDDTTCLVV 295
SRG D+ TC+VV
Sbjct: 331 AFSRGSADNITCIVV 345
>At2g20630 putative protein phosphatase 2C
Length = 279
Score = 99.4 bits (246), Expect = 3e-21
Identities = 83/290 (28%), Positives = 145/290 (49%), Gaps = 40/290 (13%)
Query: 19 LIGRELRNGKTEK--------PFVKYGQAGLAKKGEDYFLIKTDCHRVPGDSSTSFSVFA 70
L G + GKT+ FVK G+AG EDY + ++ +V G +FA
Sbjct: 14 LCGSDTGRGKTKVWKNIAHGYDFVK-GKAGHPM--EDY--VVSEFKKVDGHD---LGLFA 65
Query: 71 ILDGHNGISAAIFAKENIINNVMSAIPQGVSREEWLQALPRALVVAFVKTDM----EFQK 126
I DGH G A + + N+ +N++ ++ W A+ A++ TD + K
Sbjct: 66 IFDGHLGHDVAKYLQTNLFDNILK------EKDFWTDT-KNAIRNAYISTDAVILEQSLK 118
Query: 127 KGETSGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEERERVTAS 186
G+ T T ++IDG T+ +A+VGDSR ++ ++ GV S L+VDH E +E++ + +
Sbjct: 119 LGKGGSTAVTGILIDGKTLVIANVGDSRAVM-SKNGVASQLSVDH---EPSKEQKEIESR 174
Query: 187 GGEVGRLNVFGGNEVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQVKLSNAGGRLII 246
GG F N G + G L ++R+ GD + ++ P ++ + + ++
Sbjct: 175 GG-------FVSNIPGDVPRVDGQLAVARAFGDKSLKIHLSSDPDIRDENIDHETEFILF 227
Query: 247 ASDGIWDTLSSDMAAKSCRGV-PAELAAKLVVKEALRSRGLKDDTTCLVV 295
ASDG+W +S+ A + + + AAK +++EA+ S+ DD +C+VV
Sbjct: 228 ASDGVWKVMSNQEAVDLIKSIKDPQAAAKELIEEAV-SKQSTDDISCIVV 276
>At1g34750 protein phosphatase type 2C, putative
Length = 282
Score = 98.6 bits (244), Expect = 6e-21
Identities = 81/278 (29%), Positives = 134/278 (48%), Gaps = 33/278 (11%)
Query: 27 GKTEKPFVKYGQAGLAKKG----EDYFLIKTDCHRVPGDSSTSFSVFAILDGHNGISAAI 82
G+ +K+G + + K EDY + K ++ G+ +FAI DGH G
Sbjct: 26 GRNNDGEIKFGYSLVKGKANHPMEDYHVSKFV--KIDGNE---LGLFAIYDGHLGERVPA 80
Query: 83 FAKENIINNVMSAIPQGVSREEWLQALPRALVVAFVKTDMEFQKK----GETSGTTATFV 138
+ ++++ +N++ E++ R+++ A+ KTD G T T +
Sbjct: 81 YLQKHLFSNILK-------EEQFRYDPQRSIIAAYEKTDQAILSHSSDLGRGGSTAVTAI 133
Query: 139 IIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEERERVTASGGEVGRLNVFGG 198
+++G + VA+VGDSR +L +QGG +T+DH E ER + GG F
Sbjct: 134 LMNGRRLWVANVGDSRAVL-SQGGQAIQMTIDH---EPHTERLSIEGKGG-------FVS 182
Query: 199 NEVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQVKLSNAGGRLIIASDGIWDTLSSD 258
N G + G L +SR+ GD + ++ P VK + + L++ASDG+W +++
Sbjct: 183 NMPGDVPRVNGQLAVSRAFGDKSLKTHLRSDPDVKDSSIDDHTDVLVLASDGLWKVMANQ 242
Query: 259 MAAKSCRGVPAEL-AAKLVVKEALRSRGLKDDTTCLVV 295
A R + L AAK + EALR R KDD +C+VV
Sbjct: 243 EAIDIARRIKDPLKAAKELTTEALR-RDSKDDISCIVV 279
>At1g78200 putative protein phosphatase 2C
Length = 283
Score = 98.2 bits (243), Expect = 8e-21
Identities = 85/280 (30%), Positives = 140/280 (49%), Gaps = 34/280 (12%)
Query: 27 GKTEKPFVKYGQAGLAKKG----EDYFLIKTDCHRVPGDSSTSFSVFAILDGHNGISAAI 82
G+ + +KYG + + K EDY + K + +FAI DGH G A
Sbjct: 24 GRNGEGGIKYGFSLIKGKSNHSMEDYHVAK-----FTNFNGNELGLFAIFDGHKGDHVAA 78
Query: 83 FAKENIINNVMSAIPQGVSREEWLQALPRALVVAFVKTDMEFQKKG----ETSGTTA-TF 137
+ ++++ +N++ E+L RA+ A+ TD + E+ G+TA T
Sbjct: 79 YLQKHLFSNILKD-------GEFLVDPRRAIAKAYENTDQKILADNRTDLESGGSTAVTA 131
Query: 138 VIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEERERVTASGGEVGRLNVFG 197
++I+G + +A+VGDSR I+ ++G + +VDH +++ E R + + GG F
Sbjct: 132 ILINGKALWIANVGDSRAIVSSRGKAKQM-SVDHDPDDDTE-RSMIESKGG-------FV 182
Query: 198 GNEVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQVKLSNAGGRLIIASDGIWDTLSS 257
N G + G L +SR GD ++ Y+ P +K V + + LI+ASDGI +S+
Sbjct: 183 TNRPGDVPRVNGLLAVSRVFGDKNLKAYLNSEPEIKDVTIDSHTDFLILASDGISKVMSN 242
Query: 258 DMAAKSCRGV--PAELAAKLVVKEALRSRGLKDDTTCLVV 295
A + + P E AA+ VV EAL+ R KDD +C+VV
Sbjct: 243 QEAVDVAKKLKDPKE-AARQVVAEALK-RNSKDDISCIVV 280
>At4g26080 abscisic acid insensitive protein (ABI1)
Length = 434
Score = 95.9 bits (237), Expect = 4e-20
Identities = 76/271 (28%), Positives = 123/271 (45%), Gaps = 43/271 (15%)
Query: 61 DSSTSFSVFAILDGHNGISAAIFAKEN----IINNVMSAIPQGVSREEWLQALPRALVVA 116
D ++ F + DGH G A + +E + + P + WL+ +AL +
Sbjct: 165 DPQSAAHFFGVYDGHGGSQVANYCRERMHLALAEEIAKEKPMLCDGDTWLEKWKKALFNS 224
Query: 117 FVKTDMEFQKKG-ETSGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEE 175
F++ D E + ET G+T+ ++ + VA+ GDSR +L +G L+VDH+ +
Sbjct: 225 FLRVDSEIESVAPETVGSTSVVAVVFPSHIFVANCGDSRAVL-CRGKTALPLSVDHKPDR 283
Query: 176 NVEERERVTASGGEVGRLNVFGGNEVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQV 235
+E R+ A+GG+V + N G V G L +SRSIGD + I+P P V V
Sbjct: 284 E-DEAARIEAAGGKVIQWN---GARVF------GVLAMSRSIGDRYLKPSIIPDPEVTAV 333
Query: 236 KLSNAGGRLIIASDGIWDTLSSDMAAKSCR---------------------------GVP 268
K LI+ASDG+WD ++ + A + R P
Sbjct: 334 KRVKEDDCLILASDGVWDVMTDEEACEMARKRILLWHKKNAVAGDASLLADERRKEGKDP 393
Query: 269 AELAAKLVVKEALRSRGLKDDTTCLVVDIIP 299
A ++A + + RG KD+ + +VVD+ P
Sbjct: 394 AAMSAAEYLSKLAIQRGSKDNISVVVVDLKP 424
>At4g28400 protein phosphatase 2C like protein
Length = 283
Score = 95.5 bits (236), Expect = 5e-20
Identities = 67/229 (29%), Positives = 118/229 (51%), Gaps = 16/229 (6%)
Query: 68 VFAILDGHNGISAAIFAKENIINNVMSAIPQGVSREEWLQALPRALVVAFVKTDMEFQKK 127
+FAI DGH G A + + N+ +N++ E ++ R+ ++ ++ K
Sbjct: 67 LFAIFDGHLGHDVAKYLQTNLFDNILKEKDFWTDTENAIRNAYRSTDAVILQQSLKLGKG 126
Query: 128 GETSGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEERERVTASG 187
G T+ T ++IDG + VA+VGDSR ++ ++ GV L+VDH E +E++ + + G
Sbjct: 127 GSTA---VTGILIDGKKLVVANVGDSRAVM-SKNGVAHQLSVDH---EPSKEKKEIESRG 179
Query: 188 GEVGRLNVFGGNEVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQVKLSNAGGRLIIA 247
G F N G + G L ++R+ GD + ++ P + + + ++ A
Sbjct: 180 G-------FVSNIPGDVPRVDGQLAVARAFGDKSLKLHLSSEPDITHQTIDDHTEFILFA 232
Query: 248 SDGIWDTLSSDMAAKSCRGV-PAELAAKLVVKEALRSRGLKDDTTCLVV 295
SDGIW LS+ A + + + AAK +++EA+ SR KDD +C+VV
Sbjct: 233 SDGIWKVLSNQEAVDAIKSIKDPHAAAKHLIEEAI-SRKSKDDISCIVV 280
>At3g51470 protein phosphatase 2C-like protein
Length = 361
Score = 93.2 bits (230), Expect = 2e-19
Identities = 84/295 (28%), Positives = 138/295 (46%), Gaps = 42/295 (14%)
Query: 36 YGQAGLAKKGEDYFLIKTDCHRVPGDSSTSFSVFAILDGHNGISAAIFAKENIINNVMSA 95
+ G + ED F+ D G S+ +F + + DGH G+ AA F K+NI+ VM
Sbjct: 76 WSDKGPKQSMEDEFICVDDLTEYIGSSTGAF--YGVFDGHGGVDAASFTKKNIMKLVM-- 131
Query: 96 IPQGVSREEWLQALPRALVVAFVKTD---MEFQKKGETSGTTATFVIIDGWTVTVASVGD 152
+ + + +A AFVKTD + +SGTTA +I T+ +A+ GD
Sbjct: 132 -----EDKHFPTSTKKATRSAFVKTDHALADASSLDRSSGTTALTALILDKTMLIANAGD 186
Query: 153 SRCILDTQGGVVSLLTVDHRLEENVEERERVTASGGEV--GRLNVFGGNEVGPLRCWPGG 210
SR +L +G + L+ DH+ ER R+ GG + G LN G
Sbjct: 187 SRAVLGKRGRAIE-LSKDHK-PNCTSERLRIEKLGGVIYDGYLN--------------GQ 230
Query: 211 LCLSRSIGDTDV---GEYIVPI---PHVKQVKLSNAGGRLIIASDGIWDTLSSDMAAKSC 264
L ++R++GD + + P+ P ++++ L+ LI+ DG+WD +SS A
Sbjct: 231 LSVARALGDWHIKGTKGSLCPLSCEPELEEIVLTEEDEYLIMGCDGLWDVMSSQCAVTMV 290
Query: 265 RGV-----PAELAAKLVVKEALRSRGLKDDTTCLVVDIIPSDYPVLPMPATPRKK 314
R E ++ +VKEAL+ R D+ T +VV P P + +P + +++
Sbjct: 291 RRELMQHNDPERCSQALVKEALQ-RNSCDNLTVVVVCFSPEAPPRIEIPKSHKRR 344
>At5g57050 protein phosphatase 2C ABI2 (PP2C) (sp|O04719)
Length = 423
Score = 91.3 bits (225), Expect = 9e-19
Identities = 74/262 (28%), Positives = 124/262 (47%), Gaps = 44/262 (16%)
Query: 69 FAILDGHNGISAAIFAKEN----IINNVMSAIPQGVSREEWLQALPRALVVAFVKTDMEF 124
F + DGH G A + +E + ++ P+ + W + +AL +F++ D E
Sbjct: 161 FGVYDGHGGSQVANYCRERMHLALTEEIVKEKPEFCDGDTWQEKWKKALFNSFMRVDSEI 220
Query: 125 QKKG---ETSGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEERE 181
+ ET G+T+ ++ + VA+ GDSR +L +G L+VDH+ + + +E
Sbjct: 221 ETVAHAPETVGSTSVVAVVFPTHIFVANCGDSRAVL-CRGKTPLALSVDHKPDRD-DEAA 278
Query: 182 RVTASGGEVGRLNVFGGNEVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQVKLSNAG 241
R+ A+GG+V R N G V G L +SRSIGD + ++P P V V+
Sbjct: 279 RIEAAGGKVIRWN---GARVF------GVLAMSRSIGDRYLKPSVIPDPEVTSVRRVKED 329
Query: 242 GRLIIASDGIWDTLSS----DMAAKSC-------------------RGV---PAELAAKL 275
LI+ASDG+WD +++ D+A K RG PA ++A
Sbjct: 330 DCLILASDGLWDVMTNEEVCDLARKRILLWHKKNAMAGEALLPAEKRGEGKDPAAMSAAE 389
Query: 276 VVKEALRSRGLKDDTTCLVVDI 297
+ + +G KD+ + +VVD+
Sbjct: 390 YLSKMALQKGSKDNISVVVVDL 411
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.135 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,569,492
Number of Sequences: 26719
Number of extensions: 424814
Number of successful extensions: 1114
Number of sequences better than 10.0: 75
Number of HSP's better than 10.0 without gapping: 67
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 937
Number of HSP's gapped (non-prelim): 89
length of query: 423
length of database: 11,318,596
effective HSP length: 102
effective length of query: 321
effective length of database: 8,593,258
effective search space: 2758435818
effective search space used: 2758435818
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC147963.9


