

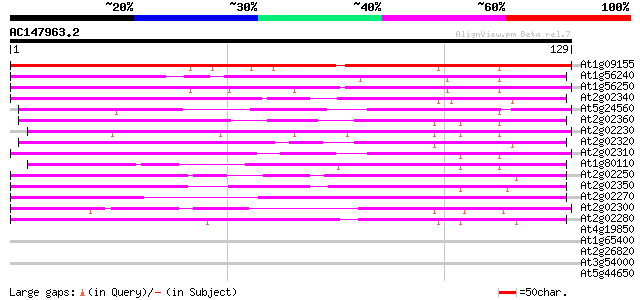
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147963.2 - phase: 0
(129 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g09155 unknown protein 104 1e-23
At1g56240 hypothetical protein 100 3e-22
At1g56250 hypothetical protein 97 3e-21
At2g02340 putative phloem-specific lectin 69 5e-13
At5g24560 phloem-specific lectin-like protein 69 9e-13
At2g02360 putative phloem-specific lectin 65 9e-12
At2g02230 putative phloem-specific lectin 65 9e-12
At2g02320 unknown protein 64 2e-11
At2g02310 putative phloem-specific lectin 64 2e-11
At1g80110 unknown protein 62 8e-11
At2g02250 lectin-like protein 62 1e-10
At2g02350 SKP1 interacting partner 3 (SKIP3) 58 1e-09
At2g02270 putative phloem-specific lectin 54 2e-08
At2g02300 putative phloem-specific lectin 53 4e-08
At2g02280 putative phloem-specific lectin 47 3e-06
At4g19850 lectin like protein 39 0.001
At1g65400 hypothetical protein 33 0.040
At2g26820 similar to avrRpt2-induced protein 1 30 0.26
At3g54000 unknown protein 29 0.75
At5g44650 unknown protein 28 0.97
>At1g09155 unknown protein
Length = 289
Score = 104 bits (259), Expect = 1e-23
Identities = 64/141 (45%), Positives = 93/141 (65%), Gaps = 14/141 (9%)
Query: 1 MRTHILTPNTTYVVYLITKVSHRVYGLDSAPAEVSVAMAN---KVQNG-MAYLYNKYE-- 54
++T L+PNT Y YLI KV+ R YGLD PAE S+ + N K+++ ++ L NK +
Sbjct: 142 IQTGALSPNTNYGAYLIMKVTSRAYGLDLVPAETSIKVGNGEKKIKSTYLSCLDNKKQQM 201
Query: 55 DTMFY-ENHRKMERNKLMEDNKEIRVPSKRDDGWMEIELGEFFC----GEVDMEVKMSVM 109
+ +FY + ++M ++++ ++ R P RDDGWMEIELGEF G+ D EV MS+
Sbjct: 202 ERVFYGQREQRMATHEVVRSHR--REPEVRDDGWMEIELGEFETGSGEGDDDKEVVMSLT 259
Query: 110 EV-GYRLKGGLIVEGIEVRPK 129
EV GY+LKGG+ ++GIEVRPK
Sbjct: 260 EVKGYQLKGGIAIDGIEVRPK 280
>At1g56240 hypothetical protein
Length = 284
Score = 99.8 bits (247), Expect = 3e-22
Identities = 63/145 (43%), Positives = 84/145 (57%), Gaps = 24/145 (16%)
Query: 1 MRTHILTPNTTYVVYLITKVSHRVYGLDSAPAEVSVAMANKVQNGMAYLYNKYEDTMFYE 60
++T +L+PNT Y YLI KV++ YGLD PAE SV K +NG NK +
Sbjct: 143 IQTTVLSPNTKYGAYLIMKVTNGAYGLDLVPAETSV----KSKNGQ---NNKNTTYLCCL 195
Query: 61 NHRKMERNKLMEDNKEIRV---------------PSKRDDGWMEIELGEFFCGE-VDMEV 104
+ +K + +L N+E R+ P RDDGW+EIELGEF E D EV
Sbjct: 196 DEKKQQMKRLFYGNREERMAMTVEAVGGDGKRREPKARDDGWLEIELGEFVTREGEDDEV 255
Query: 105 KMSVMEV-GYRLKGGLIVEGIEVRP 128
MS+ EV GY+LKGG++++GIEVRP
Sbjct: 256 NMSLTEVKGYQLKGGIVIDGIEVRP 280
>At1g56250 hypothetical protein
Length = 282
Score = 96.7 bits (239), Expect = 3e-21
Identities = 62/140 (44%), Positives = 81/140 (57%), Gaps = 12/140 (8%)
Query: 1 MRTHILTPNTTYVVYLITKVSHRVYGLDSAPAEVSVAMAN-KVQNGMAYL-----YNKYE 54
++T +L+ NT Y YLI KV+ YGLD PAE S+ N ++ YL +
Sbjct: 143 IQTRVLSANTRYGAYLIVKVTKGAYGLDLVPAETSIKSKNGQISKSATYLCCLDEKKQQM 202
Query: 55 DTMFYENHRK---MERNKLMEDNKEIRVPSKRDDGWMEIELGEFFCGE-VDMEVKMSVME 110
+FY N + M + D K R P RDDGWMEIELGEF E D EV M++ E
Sbjct: 203 KRLFYGNREERMAMTVEAVGGDGKR-REPKCRDDGWMEIELGEFETREGEDDEVNMTLTE 261
Query: 111 V-GYRLKGGLIVEGIEVRPK 129
V GY+LKGG++++GIEVRPK
Sbjct: 262 VKGYQLKGGILIDGIEVRPK 281
>At2g02340 putative phloem-specific lectin
Length = 305
Score = 69.3 bits (168), Expect = 5e-13
Identities = 44/132 (33%), Positives = 72/132 (54%), Gaps = 11/132 (8%)
Query: 1 MRTHILTPNTTYVVYLITKVSHRVYGLDSAPAEVSVAMANKVQNGMAYLYNKYEDTMFYE 60
+ + +++P T Y Y++ K + YG ++ EV V + + Y +++TM E
Sbjct: 177 INSRVISPKTRYSAYIVYKKLNICYGFENVAVEVVVGVVGQDLEESCRRYICFDETMD-E 235
Query: 61 NHRKMERNKLMEDNKEIRVPSKRDDGWMEIELGEFFC-GEV--DMEVKMSVMEVGYR-LK 116
R+ +R K + P +R DGWMEI++GEFF G + D E++M +E R K
Sbjct: 236 QFRRRDRGK------NLVKPERRKDGWMEIKIGEFFNEGGLLNDDEIEMVALEAKQRHWK 289
Query: 117 GGLIVEGIEVRP 128
GLI++GIE+RP
Sbjct: 290 RGLIIQGIEIRP 301
>At5g24560 phloem-specific lectin-like protein
Length = 251
Score = 68.6 bits (166), Expect = 9e-13
Identities = 47/131 (35%), Positives = 65/131 (48%), Gaps = 30/131 (22%)
Query: 3 THILTPNTTYVVYLITKVSHR-VYGLDSAPAEVSVAMANKVQNGMAYLYNKYEDTMFYEN 61
T +L+ T Y YL+ K +G +S P EVS T Y N
Sbjct: 144 TSLLSKATNYSAYLVFKEQEMGSFGFESLPLEVSFRSTR---------------TEVYNN 188
Query: 62 HRKMERNKLMEDNKEIRVPSKRDDGWMEIELGEFFCGEVDMEVKMSVMEV---GYRLKGG 118
R ++ E R+DGW+EIELGE++ G D E++MSV+E G+ KGG
Sbjct: 189 RRVFLKSGTQES---------REDGWLEIELGEYYVGFDDEEIEMSVLETREGGW--KGG 237
Query: 119 LIVEGIEVRPK 129
+IV+GIE+RPK
Sbjct: 238 IIVQGIEIRPK 248
>At2g02360 putative phloem-specific lectin
Length = 272
Score = 65.1 bits (157), Expect = 9e-12
Identities = 44/130 (33%), Positives = 67/130 (50%), Gaps = 20/130 (15%)
Query: 3 THILTPNTTYVVYLITKVSHRVYGLDSAPAEVSVAMANKVQNGMAYLYNKYEDTMFYENH 62
T +L+P T Y Y++ K + YG + E +V + + + ++ E
Sbjct: 155 TRVLSPRTRYSAYIVFKGVDKCYGFQNVAIEAAVGVVGQEPSRRLICFS--------EAI 206
Query: 63 RKMERNKLMEDNKEIRVPSKRDDGWMEIELGEFF--CGEVDM-EVKMSVMEV-GYRLKGG 118
R+ RN + P +R+DGWMEIELGEFF G +D E++MS +E K G
Sbjct: 207 RRGRRNVVK--------PKQREDGWMEIELGEFFNDGGIMDNDEIEMSALETKQLNRKCG 258
Query: 119 LIVEGIEVRP 128
LI++GIE+RP
Sbjct: 259 LIIQGIEIRP 268
>At2g02230 putative phloem-specific lectin
Length = 317
Score = 65.1 bits (157), Expect = 9e-12
Identities = 46/140 (32%), Positives = 73/140 (51%), Gaps = 15/140 (10%)
Query: 5 ILTPNTTYVVYLITKVSH-RVYGLDSAPAEVSVAMANKVQNGMA-YLYNKYEDTMFYENH 62
+L+ T Y VY++ K ++ R YG D P E V KV + Y + D+ +H
Sbjct: 175 MLSKGTHYSVYVVFKTANGRSYGFDLVPVEAGVGFVGKVATKKSVYFESGNADSRSATSH 234
Query: 63 RK--MERNKLMEDNKE----IRVPSKRDDGWMEIELGEFF-----CGEVDM-EVKMSVME 110
E + +E +E + P +R DGW E+ELG+F+ CG+ E+++S+ME
Sbjct: 235 YSGISEEEEEVEGERERGMNVVGPKERVDGWSEVELGKFYINNGGCGDDGSDEIEISIME 294
Query: 111 V-GYRLKGGLIVEGIEVRPK 129
K GLI++GIE+RP+
Sbjct: 295 TQNGNWKSGLIIQGIEIRPE 314
>At2g02320 unknown protein
Length = 307
Score = 64.3 bits (155), Expect = 2e-11
Identities = 44/130 (33%), Positives = 64/130 (48%), Gaps = 14/130 (10%)
Query: 3 THILTPNTTYVVYLITKVSHRVYGLDSAPAEVSVAMANKVQNGMAYLYNKYEDTMFYENH 62
T L+P T Y VY++ + YG E V M + Y + + M ++
Sbjct: 179 TRFLSPRTRYSVYIVFLKADICYGFAYVAMEAVVRMVGHELSESCRRYVCFHEAMEWQF- 237
Query: 63 RKMERNKLMEDNKEIRVPSKRDDGWMEIELGEFF---CGEVDMEVKMSVMEVGYR-LKGG 118
+ R L+ P +R+DGWMEIE+GEFF + E++MSV E R K G
Sbjct: 238 --LTRKNLVN-------PERREDGWMEIEIGEFFNEGAFRNNDEIEMSVSETTQRNTKRG 288
Query: 119 LIVEGIEVRP 128
LI++GIE+RP
Sbjct: 289 LIIQGIEIRP 298
>At2g02310 putative phloem-specific lectin
Length = 307
Score = 64.3 bits (155), Expect = 2e-11
Identities = 44/133 (33%), Positives = 68/133 (51%), Gaps = 16/133 (12%)
Query: 1 MRTHILTPNTTYVVYLITKVSHRVYGLDSAPAEVSVAMANKVQNGMAYLYNKYEDTMFYE 60
M T IL+ T Y VY++ K+ +GL P +V V + +++ D
Sbjct: 184 MSTQILSLGTHYSVYIVYKIKDERHGLRDLPIQVGVGFKGQEMPKQFICFDESTDKT--- 240
Query: 61 NHRKMERNKLMEDNKEIRVPSKRDDGWMEIELGEFFCGEVDM---EVKMSVMEV-GYRLK 116
++ + KLM+ K R DGWME E+G+FF M EV++S+++V LK
Sbjct: 241 --KEWPKKKLMKSKK-------RGDGWMEAEIGDFFNDGGLMGFDEVEVSIVDVTSPNLK 291
Query: 117 GGLIVEGIEVRPK 129
G+++EGIE RPK
Sbjct: 292 CGVMIEGIEFRPK 304
>At1g80110 unknown protein
Length = 163
Score = 62.0 bits (149), Expect = 8e-11
Identities = 46/128 (35%), Positives = 66/128 (50%), Gaps = 20/128 (15%)
Query: 5 ILTPNTTYVVYLITKVSHRVYGLDSAPAEVSVAMANKVQNGMAYLYNKYEDTMFYENHRK 64
+L+ +T Y Y + K +H YG P E S+ +A DT +N +
Sbjct: 50 LLSDDTLYAAYFVFKWNHSPYGFRQ-PVETSLVLA---------------DTESTDNVVQ 93
Query: 65 MERNKLMEDN--KEIRVPSKRDDGWMEIELGEFFCGEVDM-EVKMSVMEV-GYRLKGGLI 120
LM+D+ +E + P R DGW E+ELG+FF D+ E++MS+ E G K GLI
Sbjct: 94 PSMISLMQDSGGEEGQSPVLRRDGWYEVELGQFFKRRGDLGEIEMSLKETKGPYEKKGLI 153
Query: 121 VEGIEVRP 128
V GIE+RP
Sbjct: 154 VYGIEIRP 161
>At2g02250 lectin-like protein
Length = 305
Score = 61.6 bits (148), Expect = 1e-10
Identities = 39/129 (30%), Positives = 64/129 (49%), Gaps = 13/129 (10%)
Query: 1 MRTHILTPNTTYVVYLITKVSHRVYGLDSAPAEVSVAMANKVQNGMAYLYNKYEDTMFYE 60
M T L+P T Y Y++ K + L P E +V + + ++ ++Y +
Sbjct: 182 MNTKELSPGTRYSAYIVFKTKNGCPNLGDVPVEATVGLVGQ-ESSQRHIY--------FV 232
Query: 61 NHRKMERNKLMEDNKEIRVPSKRDDGWMEIELGEFFCGEVDMEVKMSVMEVGYRL-KGGL 119
R++ + +++ P+KR DGWME ELG+FF V S++E+ K GL
Sbjct: 233 GPSDQRRDR---ETRDVTRPTKRKDGWMEAELGQFFNESGCDVVDTSILEIKTPYWKRGL 289
Query: 120 IVEGIEVRP 128
I++GIE RP
Sbjct: 290 IIQGIEFRP 298
>At2g02350 SKP1 interacting partner 3 (SKIP3)
Length = 294
Score = 58.2 bits (139), Expect = 1e-09
Identities = 40/132 (30%), Positives = 63/132 (47%), Gaps = 17/132 (12%)
Query: 1 MRTHILTPNTTYVVYLITKVSHRVYGLDSAPAEVSVAMANKVQNGMAYLYNKYEDTMFYE 60
M T +L+ T Y VY++ K + G E V + + + + ++
Sbjct: 164 MNTRVLSLRTRYSVYIVFKKADNWCGFKGVSIEAVVGIVGE---------ESFRSFICFD 214
Query: 61 NHRKMERNKLMEDNKEIRVPSKRDDGWMEIELGEFFCGEVDM---EVKMSVMEVGY-RLK 116
H K + K K + P R+DGWME E+GEF+ M EV++S +E Y + K
Sbjct: 215 THGKGQARK----RKVVAKPELREDGWMETEIGEFYNEGGLMSSDEVEISTVEGKYAQQK 270
Query: 117 GGLIVEGIEVRP 128
GL++ GIE+RP
Sbjct: 271 RGLVILGIEIRP 282
>At2g02270 putative phloem-specific lectin
Length = 265
Score = 53.9 bits (128), Expect = 2e-08
Identities = 37/128 (28%), Positives = 56/128 (42%), Gaps = 26/128 (20%)
Query: 1 MRTHILTPNTTYVVYLITKVSHRVYGLDSAPAEVSVAMANKVQNGMAYLYNKYEDTMFYE 60
+ T L+P T Y VY++ K + G+ P F
Sbjct: 141 LSTKYLSPRTRYSVYIVFKTNDLYPGVTLEP--------------------------FPR 174
Query: 61 NHRKMERNKLMEDNKEIRVPSKRDDGWMEIELGEFFCGEVDMEVKMSVMEVGYRLKGGLI 120
R + L + + + P KR+D WME ELGEFF +V++SV++ K GL+
Sbjct: 175 FVRFVGPTDLKYEREYVTRPEKREDKWMEAELGEFFNETSCGDVEVSVIDENSYWKSGLV 234
Query: 121 VEGIEVRP 128
++GIE RP
Sbjct: 235 IQGIEFRP 242
>At2g02300 putative phloem-specific lectin
Length = 284
Score = 53.1 bits (126), Expect = 4e-08
Identities = 39/137 (28%), Positives = 66/137 (47%), Gaps = 37/137 (27%)
Query: 1 MRTHILTPNTTYVVYLI-TKVSHRVYGLDSAPAEVSVAMANKVQNGMAYLYNKYEDTMFY 59
+ T I++P T Y Y++ TK SH G ++P + V ++GM+ + +++
Sbjct: 174 LNTRIISPGTHYSAYIVYTKTSH-FNGFQTSPIQAGVGFQ---RHGMSKTFIRFDS---- 225
Query: 60 ENHRKMERNKLMEDNKEIRVPSKRDDGWMEIELGEFF--CGEVDME-VKMSVMEVG---- 112
KR DGWME ++G+F+ G + +++SV++V
Sbjct: 226 ---------------------KKRQDGWMEAKIGDFYNEGGLIGFNLIEVSVVDVARYPH 264
Query: 113 YRLKGGLIVEGIEVRPK 129
+K GLI+EGIE RPK
Sbjct: 265 MNMKSGLIIEGIEFRPK 281
>At2g02280 putative phloem-specific lectin
Length = 144
Score = 46.6 bits (109), Expect = 3e-06
Identities = 39/144 (27%), Positives = 60/144 (41%), Gaps = 20/144 (13%)
Query: 1 MRTHILTPNTTYVVYLITKVSHRVYGLDSAPAEVSVAMANKVQN-----------GMAYL 49
M T IL+ T Y Y++ K +R +G K + G
Sbjct: 1 MNTQILSQKTRYSAYIVYKTIYRFHGFKHIGVGFIGHGTPKAKRWERKDLGNDWLGCKKK 60
Query: 50 YNKYEDTMFYENHRKMERNKLMEDNKEIRVPSKRDDGWMEIELGEFFC---GEVDM-EVK 105
+ + FY N + K + +DGWM E GEFF G +D E+
Sbjct: 61 FKASKKQKFYSNKSTFTDKPITHLIKL----EEGEDGWMATEFGEFFAEGGGLLDCDEIV 116
Query: 106 MSVMEVGYRL-KGGLIVEGIEVRP 128
+SV+++ Y K GLI++GI++RP
Sbjct: 117 LSVIDIDYAYWKCGLIIQGIDIRP 140
>At4g19850 lectin like protein
Length = 194
Score = 38.5 bits (88), Expect = 0.001
Identities = 37/130 (28%), Positives = 55/130 (41%), Gaps = 33/130 (25%)
Query: 3 THILTPNTTYVVYLITKVSHRVYGLDSAPAEVSVAMANKVQNGMAYLYNKYEDTMFYENH 62
T LTPN+ Y V + K+ +DSA G + N E
Sbjct: 94 TEKLTPNSLYEVVFVVKL------IDSA-------------KGWDFRVNFKLVLPTGETK 134
Query: 63 RKMERNKLMEDNKEIRVPSKRDDGWMEIELGEFFCGEVDM--EVKMSVMEV-GYRLKGGL 119
+ E L+E NK W+EI GEF + +++ S++EV + K GL
Sbjct: 135 ERRENVNLLERNK-----------WVEIPAGEFMISPEHLSGKIEFSMLEVKSDQWKSGL 183
Query: 120 IVEGIEVRPK 129
IV+G+ +RPK
Sbjct: 184 IVKGVAIRPK 193
>At1g65400 hypothetical protein
Length = 172
Score = 33.1 bits (74), Expect = 0.040
Identities = 17/47 (36%), Positives = 30/47 (63%), Gaps = 2/47 (4%)
Query: 85 DGWMEIELGEFFCGEVDM-EVKMSVMEVGYRL-KGGLIVEGIEVRPK 129
D W++I +GEF + ++ E+ ++ E +L K GL V+G+ +RPK
Sbjct: 125 DQWLDIPVGEFTTSKKNVGEISFAMYEHECQLWKSGLFVKGVTIRPK 171
>At2g26820 similar to avrRpt2-induced protein 1
Length = 463
Score = 30.4 bits (67), Expect = 0.26
Identities = 17/46 (36%), Positives = 28/46 (59%), Gaps = 4/46 (8%)
Query: 86 GWMEIELGEFFCGEVDMEV---KMSVMEVGYRLKGGLIVEGIEVRP 128
GW+ I GEF ++ + +MS ++ G +GGLIV+G+ +RP
Sbjct: 417 GWVTIHAGEFITTPENVGLIGFRMSEVDSGDN-RGGLIVKGVLIRP 461
>At3g54000 unknown protein
Length = 352
Score = 28.9 bits (63), Expect = 0.75
Identities = 18/80 (22%), Positives = 35/80 (43%), Gaps = 12/80 (15%)
Query: 22 HRVYGLDSAPAEVSVAMANKVQNGMAYLYNKYEDTMFY-----------ENHRKMERNKL 70
H GL +PA++S + N NG Y YN ++ + + R +L
Sbjct: 154 HSGRGLLGSPAKLSATVKNHSNNGTGY-YNNHQSLQYQKLQAIQFQQLKQQQLMKHRRQL 212
Query: 71 MEDNKEIRVPSKRDDGWMEI 90
+ N+ +RV ++ G +++
Sbjct: 213 VRQNRGVRVNGNKNVGPVDL 232
>At5g44650 unknown protein
Length = 280
Score = 28.5 bits (62), Expect = 0.97
Identities = 26/106 (24%), Positives = 42/106 (39%), Gaps = 17/106 (16%)
Query: 1 MRTHILTPNTTYVVYLITKVSHRVYGLDSAPAEVSVAMANKVQNGMAYLYNKYEDTMFYE 60
M T I Y + R YG S+P+ + + +N + +G+ ++ + + F
Sbjct: 1 MTTQIFQLPLKYCASSFSSTGQRNYGASSSPSPIVICKSNGISDGL-WVKRRKNNRRFGS 59
Query: 61 NHRKMERNKLMEDNKEIRVP------------SKRDDGWMEIELGE 94
K E+ D EIRVP RDD EI+ G+
Sbjct: 60 LIVKQEKG----DVTEIRVPVPLTLEQQEKEKQNRDDEEDEIDEGD 101
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.136 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,812,462
Number of Sequences: 26719
Number of extensions: 110236
Number of successful extensions: 371
Number of sequences better than 10.0: 35
Number of HSP's better than 10.0 without gapping: 22
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 316
Number of HSP's gapped (non-prelim): 42
length of query: 129
length of database: 11,318,596
effective HSP length: 88
effective length of query: 41
effective length of database: 8,967,324
effective search space: 367660284
effective search space used: 367660284
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 54 (25.4 bits)
Medicago: description of AC147963.2


