

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147961.4 - phase: 0
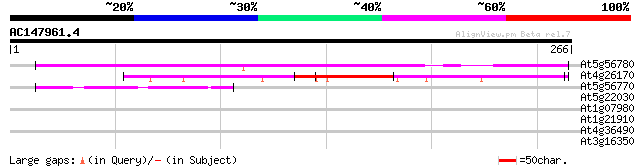
(266 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g56780 putative protein 167 7e-42
At4g26170 putative transcription factor 84 6e-17
At5g56770 putative protein 68 6e-12
At5g22030 ubiquitin-specific protease 8 (UBP8) 30 1.7
At1g07980 unknown protein 29 2.3
At1g21910 TINY like protein 28 3.9
At4g36490 hypothetical protein 28 6.6
At3g16350 putative MYB family transcription factor 27 8.6
>At5g56780 putative protein
Length = 488
Score = 167 bits (422), Expect = 7e-42
Identities = 97/256 (37%), Positives = 141/256 (54%), Gaps = 26/256 (10%)
Query: 13 LFQEIFSQGFPIVYRWAPMQNEGDALRTESQLLSTFDYAWNTISNGTRRPDDIHQMLNKL 72
LF++IFS+G I+YRWAPM ++ +A TE LLSTFDYAWN SNG RR D+ + L
Sbjct: 142 LFEDIFSKGGSILYRWAPMGSKREAEATEGMLLSTFDYAWNKGSNGERRQLDLLKKLGDR 201
Query: 73 ASGTRTFSDVAKLILPFTQKKVGIPIKSSKLHLTDDK---LDEADSGSYNVLSRVFKFNR 129
++ S +++++ PF + +VGI IK K L +++ D + S N L+ + K R
Sbjct: 202 EFMSKRKSGISRMLFPFLRNQVGIRIKGEKHVLKEERKLTCDVDEEKSNNFLTSILKLTR 261
Query: 130 SRPRIVQDTTVGSATNENAKICGVMLTDGSICKRPPVEKRVRCPEHKGMRINASTTKAIR 189
SRP+ V D + + +CGV+L DG C R PV+ R RC EHKG R+ + +
Sbjct: 262 SRPQPVSDRFDEVDGSCSDIVCGVLLEDGGCCIRSPVKGRKRCIEHKGQRVCRVSPEKQT 321
Query: 190 SPKSELESIVRNAYRYQNASHDVEDPPQRTVKCHVEEGITKTIICGIILDDGSTCRRQPV 249
PKSE+ + H+ +D ++CG+IL D C ++PV
Sbjct: 322 PPKSEIFT--------GQDHHNHKD---------------SDVVCGVILPDMEPCNKRPV 358
Query: 250 KGRKRCQEHKGRRIRA 265
GRKRC++HKG RI A
Sbjct: 359 PGRKRCEDHKGMRINA 374
>At4g26170 putative transcription factor
Length = 349
Score = 84.3 bits (207), Expect = 6e-17
Identities = 67/229 (29%), Positives = 105/229 (45%), Gaps = 46/229 (20%)
Query: 55 ISNGTRRPDDI--------HQMLNKLASGTRTFSD-------VAKLILPFTQKKVGIPIK 99
+S+ R DD+ + ++L S +R+F+ +++ ILP TQ K +
Sbjct: 35 LSHDHGRKDDVLVANLGQPESIRSRLRSYSRSFAHHDLLKQGLSQTILPTTQNKSDNQTE 94
Query: 100 SSKLHLTDDKLDEADSGSY--NVLSRVFKFNRSRPRIVQDTTVGSAT-NENAKICGVMLT 156
K +++ +D+ N L + + +RSRP+ V + +++A CGV+L
Sbjct: 95 EKKSDSEEEREVSSDAAEKESNSLPSILRLSRSRPQPVSEKHDDIVDESDSASACGVLLE 154
Query: 157 DGSICKRPPVEKRVRCPEHKGMRINASTTKAIRSPKSELESIVRNAYRYQNASHDVEDPP 216
DG+ C PV+ R RC EHKG R++ + I P E P
Sbjct: 155 DGTTCTTTPVKGRKRCTEHKGKRLSR-VSPGIHIP--------------------CEVPT 193
Query: 217 QRTVKCHVEEGITKTIICGIILDDGSTCRRQPVKGRKRCQEHKGRRIRA 265
R +C E I CG+IL D CR +PV RKRC++HKG R+ A
Sbjct: 194 VR--ECEETENI-----CGVILPDMIRCRSKPVSRRKRCEDHKGMRVNA 235
Score = 82.0 bits (201), Expect = 3e-16
Identities = 51/149 (34%), Positives = 69/149 (46%), Gaps = 31/149 (20%)
Query: 146 ENAKICGVMLTDGSICKRPPVEKRVRCPEHKGMRINA--------STTKAIRSPKSELE- 196
E ICGV+L D C+ PV +R RC +HKGMR+NA KA+ KS+ E
Sbjct: 199 ETENICGVILPDMIRCRSKPVSRRKRCEDHKGMRVNAFFFLLNPTERDKAVNEDKSKPET 258
Query: 197 --------SIVRNAYRYQNASHDVEDPPQRTVKC--------------HVEEGITKTIIC 234
S + +N P+ + +C +V+ +IC
Sbjct: 259 STGMNQEGSGLLCEATTKNGLPCTRSAPEGSKRCWQHKDKTLNHGSSENVQSATASQVIC 318
Query: 235 GIILDDGSTCRRQPVKGRKRCQEHKGRRI 263
G L +GS C + PVKGRKRC+EHKG RI
Sbjct: 319 GFKLYNGSVCEKSPVKGRKRCEEHKGMRI 347
Score = 49.3 bits (116), Expect = 2e-06
Identities = 23/54 (42%), Positives = 34/54 (62%), Gaps = 7/54 (12%)
Query: 136 QDTTVGSATNENAK-------ICGVMLTDGSICKRPPVEKRVRCPEHKGMRINA 182
+D T+ ++EN + ICG L +GS+C++ PV+ R RC EHKGMRI +
Sbjct: 296 KDKTLNHGSSENVQSATASQVICGFKLYNGSVCEKSPVKGRKRCEEHKGMRITS 349
>At5g56770 putative protein
Length = 254
Score = 67.8 bits (164), Expect = 6e-12
Identities = 39/94 (41%), Positives = 55/94 (58%), Gaps = 10/94 (10%)
Query: 13 LFQEIFSQGFPIVYRWAPMQNEGDALRTESQLLSTFDYAWNTISNGTRRPDDIHQMLNKL 72
L+++IFS+G+ + YRWAP +A TE LLSTFDYAWNT SNG RR H L KL
Sbjct: 158 LYEDIFSEGYSVFYRWAP-----EAAATEGMLLSTFDYAWNTCSNGERR----HLELQKL 208
Query: 73 ASGTRTFSDVAKLILPFTQKKVGIPIKSSKLHLT 106
+++++P + +V + IK K + T
Sbjct: 209 GDPEFMSKRKSQVLVPSIRDQV-VTIKVEKSNYT 241
>At5g22030 ubiquitin-specific protease 8 (UBP8)
Length = 901
Score = 29.6 bits (65), Expect = 1.7
Identities = 21/89 (23%), Positives = 36/89 (39%), Gaps = 2/89 (2%)
Query: 21 GFPIVYRWAPMQNEGDALRTESQLLSTFDYAWNTISNGTRRPDDIHQMLNKLASGTRTFS 80
G P+V R ++N D +LLS+F + P + GT +
Sbjct: 609 GIPLVSRLCDVENGSDVENLYLKLLSSFKMPTEFFTENLENPTEEEATDKTDTDGTTSVE 668
Query: 81 DVAKLILPFTQKKVGIPIKSSKLHLTDDK 109
D + T + + P+ +L+LTDD+
Sbjct: 669 DTNSTDVKETTESLPDPV--LRLYLTDDR 695
>At1g07980 unknown protein
Length = 206
Score = 29.3 bits (64), Expect = 2.3
Identities = 18/48 (37%), Positives = 25/48 (51%), Gaps = 2/48 (4%)
Query: 92 KKVGIPIKSSKLHLTDD--KLDEADSGSYNVLSRVFKFNRSRPRIVQD 137
K VGIP K+SK DD D N + R+ + + S P+I+QD
Sbjct: 84 KSVGIPTKTSKNREEDDGGAEDAKIKFPMNRIRRIMRSDNSAPQIMQD 131
>At1g21910 TINY like protein
Length = 230
Score = 28.5 bits (62), Expect = 3.9
Identities = 18/44 (40%), Positives = 21/44 (46%)
Query: 81 DVAKLILPFTQKKVGIPIKSSKLHLTDDKLDEADSGSYNVLSRV 124
DVA L L Q + P SS HL D+ LDE S + RV
Sbjct: 90 DVALLCLKGPQANLNFPTSSSSHHLLDNLLDENTLLSPKSIQRV 133
>At4g36490 hypothetical protein
Length = 543
Score = 27.7 bits (60), Expect = 6.6
Identities = 18/65 (27%), Positives = 29/65 (43%)
Query: 152 GVMLTDGSICKRPPVEKRVRCPEHKGMRINASTTKAIRSPKSELESIVRNAYRYQNASHD 211
G M +D K P + KRV +HK + + + ++ E +S A + AS +
Sbjct: 297 GCMRSDKGPWKNPEIMKRVHNGDHKCSKGSQAENSGEKTIPEEDDSTTEPASEEEKASKE 356
Query: 212 VEDPP 216
VE P
Sbjct: 357 VEIVP 361
>At3g16350 putative MYB family transcription factor
Length = 387
Score = 27.3 bits (59), Expect = 8.6
Identities = 12/23 (52%), Positives = 16/23 (69%)
Query: 141 GSATNENAKICGVMLTDGSICKR 163
GS ++ K+ GV LTDGSI K+
Sbjct: 40 GSGSSSAVKLFGVRLTDGSIIKK 62
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.133 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,037,193
Number of Sequences: 26719
Number of extensions: 243582
Number of successful extensions: 582
Number of sequences better than 10.0: 8
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 2
Number of HSP's that attempted gapping in prelim test: 561
Number of HSP's gapped (non-prelim): 14
length of query: 266
length of database: 11,318,596
effective HSP length: 98
effective length of query: 168
effective length of database: 8,700,134
effective search space: 1461622512
effective search space used: 1461622512
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC147961.4


