

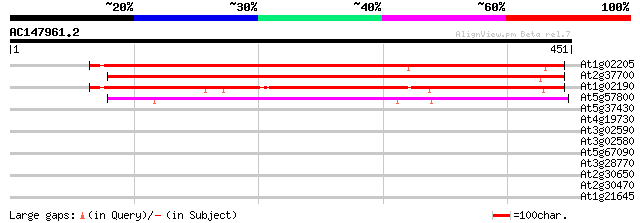
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147961.2 - phase: 0 /pseudo
(451 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g02205 CER1 protein 420 e-118
At2g37700 CER1-like protein 403 e-113
At1g02190 CER1-like protein 391 e-109
At5g57800 cuticle protein (WAX2) 204 6e-53
At5g37430 contains similarity to unknown protein (pir||C71432) 30 2.1
At4g19730 putative protein 30 2.7
At3g02590 putative sterol-C5-desaturase 30 2.7
At3g02580 sterol-C5-desaturase 30 2.7
At5g67090 subtilisin-type protease-like 29 4.6
At3g28770 hypothetical protein 29 6.1
At2g30650 3-hydroxyisobutyryl-coenzyme A hydrolase 28 7.9
At2g30470 putative VP1/ABI3 family regulatory protein 28 7.9
At1g21645 putative SecA-type chloroplast protein transport factor 28 7.9
>At1g02205 CER1 protein
Length = 625
Score = 420 bits (1080), Expect = e-118
Identities = 203/388 (52%), Positives = 266/388 (68%), Gaps = 8/388 (2%)
Query: 65 FSPSFFNCH*ACHLFHSLHHTKFRRNYSLFMPMYDYIYGTVDKSTDVIYETSLMRPKESP 124
F P F C+ +HSLHHT+FR NYSLFMP+YDYIYGT+D+STD +YE +L R +
Sbjct: 235 FPPLKFLCY--TPSYHSLHHTQFRTNYSLFMPLYDYIYGTMDESTDTLYEKTLERGDDIV 292
Query: 125 DVVHLTHLTTFNSIYQLRLGFASLASNPQTSKWYLHLMWPFTMFSMLMTWICGRAFVLES 184
DVVHLTHLTT SIY LR+G AS AS P +W++ L+WPFT SM+ T R FV E
Sbjct: 293 DVVHLTHLTTPESIYHLRIGLASFASYPFAYRWFMRLLWPFTSLSMIFTLFYARLFVAER 352
Query: 185 NSFKNLKLQCWLIPRFKRQYFSKWQSKTFNNLIEEAIVEAELNGAKVISLGLFNKNHQLN 244
NSF L LQ W+IPR+ QY KW+ + NN+IE+AI+EA+ G KV+SLGL N+ +LN
Sbjct: 353 NSFNKLNLQSWVIPRYNLQYLLKWRKEAINNMIEKAILEADKKGVKVLSLGLMNQGEELN 412
Query: 245 ERHEHYIGRLPQLKIKVVDGSSLAAATVLNNIPKGTNQVLLRGKFNKVAFVIANALCKKN 304
E YI P +K+++VDGS LAAA V+N++PK T V++ G KVA+ IA+ALC++
Sbjct: 413 RNGEVYIHNHPDMKVRLVDGSRLAAAVVINSVPKATTSVVMTGNLTKVAYTIASALCQRG 472
Query: 305 VQVVVLYKDELKELE----QRINTSKGNLALSPFNTPKIWLVGDEWDEYEQMEAPKGSLF 360
VQV L DE +++ Q L ++ K+WLVG+ EQ +A KG+LF
Sbjct: 473 VQVSTLRLDEYEKIRSCVPQECRDHLVYLTSEALSSNKVWLVGEGTTREEQEKATKGTLF 532
Query: 361 IPFSHFPPKKMRKDCFYHYTPAMITPTTFMNSHSCENWLPRRVMSAWRIAGIIHALEGWN 420
IPFS FP K++R+DC YH TPA+I P + +N HSCENWLPR+ MSA R+AGI+HALEGW
Sbjct: 533 IPFSQFPLKQLRRDCIYHTTPALIVPKSLVNVHSCENWLPRKAMSATRVAGILHALEGWE 592
Query: 421 VHECGDTIL--STEKVWEASIRHGFQPL 446
+HECG ++L ++VWEA + HGFQPL
Sbjct: 593 MHECGTSLLLSDLDQVWEACLSHGFQPL 620
>At2g37700 CER1-like protein
Length = 635
Score = 403 bits (1036), Expect = e-113
Identities = 202/373 (54%), Positives = 263/373 (70%), Gaps = 5/373 (1%)
Query: 79 FHSLHHTKFRRNYSLFMPMYDYIYGTVDKSTDVIYETSLMRPKESPDVVHLTHLTTFNSI 138
FHSLHHT+FR NYSLFMPMYDYIYGT D+ +D +YETSL + +E PD +HLTHLT+ +SI
Sbjct: 256 FHSLHHTQFRTNYSLFMPMYDYIYGTTDECSDSLYETSLEKEEEKPDAIHLTHLTSLDSI 315
Query: 139 YQLRLGFASLASNPQTSKWYLHLMWPFTMF-SMLMTWICGRAFVLESNSFKNLKLQCWLI 197
Y LRLGFASL+S+P +S+ YL LM PF + S ++ + FV+E N F++L L L+
Sbjct: 316 YHLRLGFASLSSHPLSSRCYLFLMKPFALILSFILRSFSFQTFVVERNRFRDLTLHSHLL 375
Query: 198 PRFKRQYFSKWQSKTFNNLIEEAIVEAELNGAKVISLGLFNKNHQLNERHEHYIGRLPQL 257
P+F Y S Q + N +IE AI+EA+ G KV+SLGL N+ +LN E Y+ R P+L
Sbjct: 376 PKFSSHYMSHQQKECINKMIEAAILEADKKGVKVMSLGLLNQGEELNGYGEMYVRRHPKL 435
Query: 258 KIKVVDGSSLAAATVLNNIPKGTNQVLLRGKFNKVAFVIANALCKKNVQVVVLYKDELKE 317
KI++VDG SLAA VL++IP GT +VL RG+ KVA I +LC+ ++V+VL K+E
Sbjct: 436 KIRIVDGGSLAAEVVLHSIPVGTKEVLFRGQITKVARAIVFSLCQNAIKVMVLRKEEHSM 495
Query: 318 LEQRINTS-KGNLALSPFNTPKIWLVGDEWDEYEQMEAPKGSLFIPFSHFPPKKMRKDCF 376
L + ++ K NL L+ P IWLVGD EQ A G+LF+PFS FPPK +RKDCF
Sbjct: 496 LAEFLDDKCKENLVLTTNYYPMIWLVGDGLSTKEQKMAKDGTLFLPFSQFPPKTLRKDCF 555
Query: 377 YHYTPAMITPTTFMNSHSCENWLPRRVMSAWRIAGIIHALEGWNVHECG---DTILSTEK 433
YH TPAMI P + N SCENWL RRVMSAWR+ GI+HALEGW HECG ++I++ +
Sbjct: 556 YHTTPAMIIPHSAQNIDSCENWLGRRVMSAWRVGGIVHALEGWKEHECGLDDNSIINPPR 615
Query: 434 VWEASIRHGFQPL 446
VWEA++R+GFQPL
Sbjct: 616 VWEAALRNGFQPL 628
>At1g02190 CER1-like protein
Length = 623
Score = 391 bits (1005), Expect = e-109
Identities = 207/392 (52%), Positives = 267/392 (67%), Gaps = 18/392 (4%)
Query: 65 FSPSFFNCH*ACHLFHSLHHTKFRRNYSLFMPMYDYIYGTVDKSTDVIYETSLMRPKESP 124
F P F C+ FHSLHHT+FR NYSLFMP+YD+IYGT D TD +YE SL +ESP
Sbjct: 235 FPPLKFLCY--TPSFHSLHHTQFRTNYSLFMPIYDFIYGTTDNLTDSLYERSLEIEEESP 292
Query: 125 DVVHLTHLTTFNSIYQLRLGFASLASNPQTSK--WYLH-LMWPFTMFSM--LMTWICGRA 179
DV+HLTHLTT NSIYQ+RLGF SL+S P S+ WYL MWPFT+ L + I R
Sbjct: 293 DVIHLTHLTTHNSIYQMRLGFPSLSSCPLWSRPPWYLTCFMWPFTLLCSFALTSAIPLRT 352
Query: 180 FVLESNSFKNLKLQCWLIPRFKRQYFSKWQSKTFNNLIEEAIVEAELNGAKVISLGLFNK 239
FV E N ++L + L+P+F F + ++ N +IEEAI+EA+ G KV+SLGL N
Sbjct: 353 FVFERNRLRDLTVHSHLLPKFS---FHR-HHESINTIIEEAILEADEKGVKVMSLGLMNN 408
Query: 240 NHQLNERHEHYIGRLPQLKIKVVDGSSLAAATVLNNIPKGTNQVLLRGKFNKVAFVIANA 299
+LN E Y+ + P+LKI++VDGSS+AA V+NNIPK +++ RG KVA + A
Sbjct: 409 REELNGSGEMYVQKYPKLKIRLVDGSSMAATVVINNIPKEATEIVFRGNLTKVASAVVFA 468
Query: 300 LCKKNVQVVVLYKDELKELEQRINTSKGNLALSPFNT---PKIWLVGDEWDEYEQMEAPK 356
LC+K V+VVVL ++E +L + + NL LS N+ PK+WLVGD + EQM+A +
Sbjct: 469 LCQKGVKVVVLREEEHSKLIK--SGVDKNLVLSTSNSYYSPKVWLVGDGIENEEQMKAKE 526
Query: 357 GSLFIPFSHFPPKKMRKDCFYHYTPAMITPTTFMNSHSCENWLPRRVMSAWRIAGIIHAL 416
G+LF+PFSHFPP K+RKDCFY TPAM P + N SCENWL RRVMSAW+I GI+HAL
Sbjct: 527 GTLFVPFSHFPPNKLRKDCFYQSTPAMRVPKSAQNIDSCENWLGRRVMSAWKIGGIVHAL 586
Query: 417 EGWNVHECGDT--ILSTEKVWEASIRHGFQPL 446
EGW H+CG+T +L +WEA++RH FQPL
Sbjct: 587 EGWEEHDCGNTCNVLRLHAIWEAALRHDFQPL 618
>At5g57800 cuticle protein (WAX2)
Length = 632
Score = 204 bits (520), Expect = 6e-53
Identities = 126/382 (32%), Positives = 194/382 (49%), Gaps = 11/382 (2%)
Query: 79 FHSLHHTKFRRNYSLFMPMYDYIYGTVDKSTDVIYET---SLMRPKESPDVVHLTHLTTF 135
+HSLHH + N+ LFMP++D + T + ++ + + S K P+ V L H
Sbjct: 249 YHSLHHQEMGTNFCLFMPLFDVLGDTQNPNSWELQKKIRLSAGERKRVPEFVFLAHGVDV 308
Query: 136 NSIYQLRLGFASLASNPQTSKWYLHLMWPFTMFSMLMTWICGRAFVLESNSFKNLKLQCW 195
S F S AS P T++ +L MWPFT ML W + F+ + +N Q W
Sbjct: 309 MSAMHAPFVFRSFASMPYTTRIFLLPMWPFTFCVMLGMWAWSKTFLFSFYTLRNNLCQTW 368
Query: 196 LIPRFKRQYFSKWQSKTFNNLIEEAIVEAELNGAKVISLGLFNKNHQLNERHEHYIGRLP 255
+PRF QYF + +K N+ IE AI+ A+ G KVISL NKN LN ++ + P
Sbjct: 369 GVPRFGFQYFLPFATKGINDQIEAAILRADKIGVKVISLAALNKNEALNGGGTLFVNKHP 428
Query: 256 QLKIKVVDGSSLAAATVLNNIPKGTNQVLLRGKFNKVAFVIANALCKKNVQVVVL----Y 311
L+++VV G++L AA +L IPK N+V L G +K+ IA LC++ V+V++L
Sbjct: 429 DLRVRVVHGNTLTAAVILYEIPKDVNEVFLTGATSKLGRAIALYLCRRGVRVLMLTLSME 488
Query: 312 KDELKELEQRINTSKGNLALSPFNTP---KIWLVGDEWDEYEQMEAPKGSLFIPFSHFPP 368
+ + + E + + ++ +N K W+VG EQ AP G+ F F P
Sbjct: 489 RFQKIQKEAPVEFQNNLVQVTKYNAAQHCKTWIVGKWLTPREQSWAPAGTHFHQFVVPPI 548
Query: 369 KKMRKDCFYHYTPAMITPTTFMNSHSCENWLPRRVMSAWRIAGIIHALEGWNVHECGD-T 427
K R++C Y AM P +CE + R V+ A G++H LEGW HE G
Sbjct: 549 LKFRRNCTYGDLAAMKLPKDVEGLGTCEYTMERGVVHACHAGGVVHMLEGWKHHEVGAID 608
Query: 428 ILSTEKVWEASIRHGFQPLKNI 449
+ + VWEA++++G + ++
Sbjct: 609 VDRIDLVWEAAMKYGLSAVSSL 630
>At5g37430 contains similarity to unknown protein (pir||C71432)
Length = 607
Score = 30.4 bits (67), Expect = 2.1
Identities = 18/62 (29%), Positives = 29/62 (46%), Gaps = 2/62 (3%)
Query: 369 KKMRKDCFYHYTPAMITPTTFMNSHSCENWLPRRVMSAWRIAGIIHALEGWNVHECGDTI 428
+K+ DC Y +I T + C+ + RR++S +A + L + E GD I
Sbjct: 92 RKLSLDCLYEIKTLLIACLTMQETKECDIKILRRIVSC--VAYNVMDLHKYKWDELGDCI 149
Query: 429 LS 430
LS
Sbjct: 150 LS 151
>At4g19730 putative protein
Length = 332
Score = 30.0 bits (66), Expect = 2.7
Identities = 28/83 (33%), Positives = 37/83 (43%), Gaps = 25/83 (30%)
Query: 145 FASLASNPQTSKWYLHLMWPFTMFSMLMTWICGRAFVLESNSFKNLKLQCWLIPRFKRQY 204
FAS+ASN Q+ K ++ +WI F+ SN F L L W P
Sbjct: 97 FASMASNHQSRKTFID------------SWI----FIARSNGFHGLDL-AWEYP------ 133
Query: 205 FSKWQSKTFNNLIEE--AIVEAE 225
+S + F NL+ E A VEAE
Sbjct: 134 YSDHEMTDFGNLVGELRAAVEAE 156
>At3g02590 putative sterol-C5-desaturase
Length = 279
Score = 30.0 bits (66), Expect = 2.7
Identities = 8/27 (29%), Positives = 19/27 (69%)
Query: 79 FHSLHHTKFRRNYSLFMPMYDYIYGTV 105
+H++HHT ++ NY + D+++G++
Sbjct: 238 YHTIHHTTYKHNYGHYTIWMDWMFGSL 264
>At3g02580 sterol-C5-desaturase
Length = 281
Score = 30.0 bits (66), Expect = 2.7
Identities = 8/27 (29%), Positives = 19/27 (69%)
Query: 79 FHSLHHTKFRRNYSLFMPMYDYIYGTV 105
+H++HHT ++ NY + D+++G++
Sbjct: 237 YHTIHHTTYKHNYGHYTIWMDWMFGSL 263
>At5g67090 subtilisin-type protease-like
Length = 736
Score = 29.3 bits (64), Expect = 4.6
Identities = 30/127 (23%), Positives = 53/127 (41%), Gaps = 5/127 (3%)
Query: 206 SKWQSKTFNNLIEEAIVEAELNGAKVISLGLFNKNHQLNERHEHYIGRLPQLKIKVVDGS 265
SKW+ N ++ +L GAKV + GLF N L E IG+ + G+
Sbjct: 153 SKWKGACEFN--SSSLCNKKLIGAKVFNKGLFANNPDLRETK---IGQYSSPYDTIGHGT 207
Query: 266 SLAAATVLNNIPKGTNQVLLRGKFNKVAFVIANALCKKNVQVVVLYKDELKELEQRINTS 325
+AA N++ + +G + +A A+ K + + D + ++Q I
Sbjct: 208 HVAAIAAGNHVKNASYFSYAQGTASGIAPHAHLAIYKAAWEEGIYSSDVIAAIDQAIRDG 267
Query: 326 KGNLALS 332
++LS
Sbjct: 268 VHVISLS 274
>At3g28770 hypothetical protein
Length = 2081
Score = 28.9 bits (63), Expect = 6.1
Identities = 26/97 (26%), Positives = 45/97 (45%), Gaps = 9/97 (9%)
Query: 201 KRQYFSKWQSKT-FNNLIEEAIVEAELNGAKVISLGLFNKN--HQLNERHEHYIGRLPQL 257
+++ F +S T + I I+E + G + +SL L KN + E I R+ ++
Sbjct: 120 RKEMFEVMKSLTELHAAIGRVIIEKHIKGDEAMSLSLEQKNAVETSITQWEQTITRIVKI 179
Query: 258 KIKVVDGSSLAA------ATVLNNIPKGTNQVLLRGK 288
++V SS A +T NN+ G+N V G+
Sbjct: 180 VVEVKSKSSSEASSEESSSTEHNNVTTGSNMVETNGE 216
>At2g30650 3-hydroxyisobutyryl-coenzyme A hydrolase
Length = 410
Score = 28.5 bits (62), Expect = 7.9
Identities = 14/45 (31%), Positives = 25/45 (55%), Gaps = 3/45 (6%)
Query: 213 FNNLIEEAIVEAELNGAKVISLGLFNKNHQLNERHEHYIGRLPQL 257
F+NL+ ++ E + ++++ FN+ QLN H + RL QL
Sbjct: 43 FSNLVRSQVLVEEKSSVRILT---FNRPKQLNALSFHMVSRLLQL 84
>At2g30470 putative VP1/ABI3 family regulatory protein
Length = 790
Score = 28.5 bits (62), Expect = 7.9
Identities = 19/78 (24%), Positives = 39/78 (49%), Gaps = 1/78 (1%)
Query: 271 TVLNNIPKGTNQVLLRGKFNKVAFVIANALCKKNVQVVVLYKDELKELEQRINTSKGNLA 330
TV + + + +++R K ++ + A KK + + D+ KE E+ +NT++ +L
Sbjct: 673 TVCSTVKRRFKTLMMRRKKKQLERDVTAAEDKKKKDMELAESDKSKE-EKEVNTARIDLN 731
Query: 331 LSPFNTPKIWLVGDEWDE 348
P+N + V E +E
Sbjct: 732 SDPYNKEDVEAVAVEKEE 749
>At1g21645 putative SecA-type chloroplast protein transport factor
Length = 882
Score = 28.5 bits (62), Expect = 7.9
Identities = 23/80 (28%), Positives = 34/80 (41%), Gaps = 11/80 (13%)
Query: 276 IPKGTNQVLLRGKFNK------VAFVIANALCKKNVQVVVLYKDELKELEQRINTSKGNL 329
I +G N +L+ G+ N+ VA +A L K + YK ELKE + +L
Sbjct: 235 IDEGRNPLLISGEANENAARYPVAAKVAELLVKDSH-----YKVELKENSVELTEEGISL 289
Query: 330 ALSPFNTPKIWLVGDEWDEY 349
A T +W D W +
Sbjct: 290 AEMALETGDLWDENDPWARF 309
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.328 0.139 0.455
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,471,495
Number of Sequences: 26719
Number of extensions: 449884
Number of successful extensions: 1243
Number of sequences better than 10.0: 13
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 1224
Number of HSP's gapped (non-prelim): 15
length of query: 451
length of database: 11,318,596
effective HSP length: 103
effective length of query: 348
effective length of database: 8,566,539
effective search space: 2981155572
effective search space used: 2981155572
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC147961.2


