

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147960.7 + phase: 0
(248 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
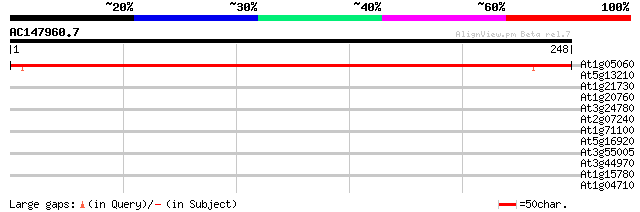
At1g05060 unknown protein 288 2e-78
At5g13210 unknown protein 32 0.32
At1g21730 kinesin-related protein (MKRP1) 31 0.70
At1g20760 unknown protein 29 2.0
At3g24780 hypothetical protein 28 4.6
At2g07240 hypothetical protein 28 6.0
At1g71100 ribose 5-phosphate isomerase like protein 28 6.0
At5g16920 unknown protein 27 7.8
At3g55005 tonneau 1b (TON1b) 27 7.8
At3g44970 cytochrome P450 - like protein 27 7.8
At1g15780 unknown protein 27 7.8
At1g04710 acetyl-CoA acyltransferase like protein 27 7.8
>At1g05060 unknown protein
Length = 253
Score = 288 bits (736), Expect = 2e-78
Identities = 146/252 (57%), Positives = 184/252 (72%), Gaps = 4/252 (1%)
Query: 1 MSFL-AGRLAGKEAAYFFQESKQAVTKLAQKNNPISKTNVVDQRHVVQDNADVLPEVLRH 59
MSFL AGRLAGKEAAYFFQESK AV +LA+K+ K + DVLPE+LRH
Sbjct: 1 MSFLGAGRLAGKEAAYFFQESKHAVNRLAEKSPATGKKLPSSPPDPPEIQPDVLPEILRH 60
Query: 60 SLPSKLFRDETASSSSFSASKWVLQSDPKLRSSVSPDAINPLRAFVSLPQVTFGARRWEL 119
SLPSK++ SS SKW L+SDP S+SPD +NPLR +VSLPQVTFG RRW+L
Sbjct: 61 SLPSKIYGRPPDPSSLSQFSKWALESDPNATVSISPDVLNPLRGYVSLPQVTFGRRRWDL 120
Query: 120 PEAKHGVSASTANELRQDRYDVNVNPEKLKAASEGLANLGKAFAIATAVVFGGAAMVIGM 179
PE+++ V ASTANELR+DRY VNPEKLKAA EGL ++GKAFA AT ++FG A +V G
Sbjct: 121 PESENSVLASTANELRRDRYGTPVNPEKLKAAGEGLQHIGKAFAAATIIIFGSATLVFGT 180
Query: 180 VASKLELHNMGDLKTKGKDVVEPQLENIKNYFVPMKVWAENMSRKWHLERE---DVKQKA 236
ASKL++ N D++TKGKD+ +P+LE++K P++ WAENMS+KWH+E E +K+K
Sbjct: 181 AASKLDMRNADDIRTKGKDLFQPKLESMKEQVEPLRTWAENMSKKWHIENEGGNTIKEKP 240
Query: 237 IVKDLSKILGSK 248
I+K+LSKILG K
Sbjct: 241 ILKELSKILGPK 252
>At5g13210 unknown protein
Length = 673
Score = 32.0 bits (71), Expect = 0.32
Identities = 20/66 (30%), Positives = 35/66 (52%), Gaps = 7/66 (10%)
Query: 56 VLRHSLPSKLFRDETASSSSFSASKWVLQSDP--KLRSSVSPDAINPLRAFVSLPQVTFG 113
+L S+ K+F E SF + V+++ ++R + D + PLR + LP+V G
Sbjct: 311 LLCESIARKIFTRE-----SFPEYEGVVEAHYAYRVRDRLRKDVLVPLRKTLQLPEVYMG 365
Query: 114 ARRWEL 119
AR W++
Sbjct: 366 ARNWDI 371
>At1g21730 kinesin-related protein (MKRP1)
Length = 890
Score = 30.8 bits (68), Expect = 0.70
Identities = 21/36 (58%), Positives = 23/36 (63%), Gaps = 3/36 (8%)
Query: 60 SLPSKLFRDETASSSSFSASKWVLQSDPKLRSSVSP 95
++P K R ET SSS FSAS V S P LRSS SP
Sbjct: 23 TIPMK--RPETPSSSHFSASP-VTSSSPLLRSSPSP 55
>At1g20760 unknown protein
Length = 1019
Score = 29.3 bits (64), Expect = 2.0
Identities = 27/110 (24%), Positives = 48/110 (43%), Gaps = 18/110 (16%)
Query: 6 GRLAGKEAAYFFQESKQAVTKLAQ--KNNPISKTNVVDQRH---------VVQDNADVLP 54
GR++G EA FFQ S + LAQ + S + +D+++ V Q D+ P
Sbjct: 24 GRISGAEAVGFFQGSGLSKQVLAQIWSLSDRSHSGFLDRQNFYNSLRLVTVAQSKRDLTP 83
Query: 55 EVLRHSLPSKLFRDETASSSSFSASKWVLQSDPKLRSSVSPDAINPLRAF 104
E++ +L T +++ K L + P R + + + P+ F
Sbjct: 84 EIVNAAL-------NTPAAAKIPPPKINLSAIPAPRPNPAATTVGPVSGF 126
>At3g24780 hypothetical protein
Length = 715
Score = 28.1 bits (61), Expect = 4.6
Identities = 11/31 (35%), Positives = 18/31 (57%)
Query: 88 KLRSSVSPDAINPLRAFVSLPQVTFGARRWE 118
++R + + PLR + LP+V GAR W+
Sbjct: 392 RVRDRLRKQVLVPLRKTLQLPEVYMGARAWQ 422
>At2g07240 hypothetical protein
Length = 928
Score = 27.7 bits (60), Expect = 6.0
Identities = 15/39 (38%), Positives = 20/39 (50%)
Query: 72 SSSSFSASKWVLQSDPKLRSSVSPDAINPLRAFVSLPQV 110
SSS+ S K V + L V+PD +NP+ LP V
Sbjct: 487 SSSTVSPPKSVTTENTALGPDVAPDPLNPVAPLDPLPSV 525
>At1g71100 ribose 5-phosphate isomerase like protein
Length = 267
Score = 27.7 bits (60), Expect = 6.0
Identities = 15/45 (33%), Positives = 23/45 (50%)
Query: 164 IATAVVFGGAAMVIGMVASKLELHNMGDLKTKGKDVVEPQLENIK 208
I T+ A+ +G+ S L+ H + DL G D V+P L +K
Sbjct: 80 IPTSTTTHEQAVSLGIPLSDLDSHPVVDLSIDGADEVDPALNLVK 124
>At5g16920 unknown protein
Length = 256
Score = 27.3 bits (59), Expect = 7.8
Identities = 15/42 (35%), Positives = 25/42 (58%), Gaps = 6/42 (14%)
Query: 24 VTKLAQKNNPISKTNVVDQRHVVQDNADVLPEVLRHSLPSKL 65
VT L K+ +S +N+++Q+ V + +LRHS+PS L
Sbjct: 69 VTFLMPKDKTLSTSNIINQQDSVTEF------LLRHSIPSSL 104
>At3g55005 tonneau 1b (TON1b)
Length = 257
Score = 27.3 bits (59), Expect = 7.8
Identities = 19/73 (26%), Positives = 30/73 (41%)
Query: 86 DPKLRSSVSPDAINPLRAFVSLPQVTFGARRWELPEAKHGVSASTANELRQDRYDVNVNP 145
+ + SS+S D NP R + + R +A ++ + RYD P
Sbjct: 157 ESETESSLSLDTRNPPRRSSASDSLPHQRRSVSASQASGAATSGYRKDESNWRYDTEDMP 216
Query: 146 EKLKAASEGLANL 158
E++ AS L NL
Sbjct: 217 EEVMRASTALENL 229
>At3g44970 cytochrome P450 - like protein
Length = 455
Score = 27.3 bits (59), Expect = 7.8
Identities = 14/44 (31%), Positives = 23/44 (51%)
Query: 15 YFFQESKQAVTKLAQKNNPISKTNVVDQRHVVQDNADVLPEVLR 58
Y F E + K + P+ +TN++ + VV + DV E+LR
Sbjct: 55 YGFYEISPYLKKKMLRYGPLFRTNILGVKTVVSTDKDVNMEILR 98
>At1g15780 unknown protein
Length = 1335
Score = 27.3 bits (59), Expect = 7.8
Identities = 19/62 (30%), Positives = 28/62 (44%), Gaps = 9/62 (14%)
Query: 38 NVVDQRHVVQDNADVLPEVLRHSLPSKLFRDETASSSSFSASKW---VLQSDPKLRSSVS 94
NVVDQ+ + + LPE+ SL D TA + S + W V Q ++ +
Sbjct: 556 NVVDQQRQLYQSQRTLPEMPSSSL------DSTAQTESANGGDWQEEVYQKIKSMKETYL 609
Query: 95 PD 96
PD
Sbjct: 610 PD 611
>At1g04710 acetyl-CoA acyltransferase like protein
Length = 443
Score = 27.3 bits (59), Expect = 7.8
Identities = 10/35 (28%), Positives = 22/35 (62%)
Query: 203 QLENIKNYFVPMKVWAENMSRKWHLEREDVKQKAI 237
+ E N +PM + +EN++ ++++ RE+ Q A+
Sbjct: 177 KFEQAHNCLLPMGITSENVAHRFNVSREEQDQAAV 211
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.314 0.129 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,234,248
Number of Sequences: 26719
Number of extensions: 206724
Number of successful extensions: 500
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 489
Number of HSP's gapped (non-prelim): 12
length of query: 248
length of database: 11,318,596
effective HSP length: 97
effective length of query: 151
effective length of database: 8,726,853
effective search space: 1317754803
effective search space used: 1317754803
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 59 (27.3 bits)
Medicago: description of AC147960.7


