

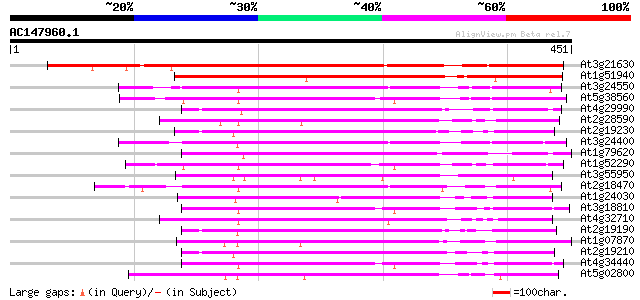
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147960.1 - phase: 1 /pseudo
(451 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g21630 receptor like protein kinase 398 e-111
At1g51940 protein kinase like protein 240 1e-63
At3g24550 protein kinase, putative 213 2e-55
At5g38560 putative protein 211 9e-55
At4g29990 serine/threonine-specific receptor protein kinase LRRPK 208 6e-54
At2g28590 putative protein kinase 206 2e-53
At2g19230 putative receptor-like protein kinase 205 5e-53
At3g24400 protein kinase, putative 204 8e-53
At1g79620 unknown protein 204 8e-53
At1g52290 protein kinase, putative 202 4e-52
At3g55950 receptor kinase - like protein 201 5e-52
At2g18470 putative protein kinase 201 5e-52
At1g24030 protein kinase like protein 201 7e-52
At3g18810 protein kinase, putative 201 9e-52
At4g32710 putative protein kinase 200 1e-51
At2g19190 putative receptor-like protein kinase 200 1e-51
At1g07870 putative protein kinase (At1g07870) 200 1e-51
At2g19210 putative receptor-like protein kinase 199 2e-51
At4g34440 serine/threonine protein kinase - like 199 3e-51
At5g02800 protein kinase - like 199 3e-51
>At3g21630 receptor like protein kinase
Length = 617
Score = 398 bits (1022), Expect = e-111
Identities = 227/457 (49%), Positives = 291/457 (63%), Gaps = 61/457 (13%)
Query: 31 REFLYFLYLPFCP*ATLESIAKDTMIEIELLKRYN--VNFSQGSGLVYIRGKDGNGLYVP 88
++F F+ P P +L SIA+ + + ++L+RYN VNF+ G+G+VY+ G+D NG + P
Sbjct: 162 KDFGLFVTYPLRPEDSLSSIARSSGVSADILQRYNPGVNFNSGNGIVYVPGRDPNGAFPP 221
Query: 89 LPPK-----------------------------YFRKKNGEEESKFPPEDSMTPSTK-DV 118
Y +KN + F S+ STK D
Sbjct: 222 FKSSKQDGVGAGVIAGIVIGVIVALLLILFIVYYAYRKNKSKGDSF--SSSIPLSTKADH 279
Query: 119 DKDTNDDNGS----------KYIWVDKSPEFSYEELANATDNFSLAKKIGQGGFGEVYYG 168
T+ +G I VDKS EFS EELA ATDNF+L+ KIGQGGFG VYY
Sbjct: 280 ASSTSLQSGGLGGAGVSPGIAAISVDKSVEFSLEELAKATDNFNLSFKIGQGGFGAVYYA 339
Query: 169 ELRGQKIAIKKMKMQATREFLSELKVLTSVHHWNLVHLIGYCVEGFLFLVYEYMENGNLS 228
ELRG+K AIKKM M+A+++FL+ELKVLT VHH NLV LIGYCVEG LFLVYEY+ENGNL
Sbjct: 340 ELRGEKAAIKKMDMEASKQFLAELKVLTRVHHVNLVRLIGYCVEGSLFLVYEYVENGNLG 399
Query: 229 QHLHNSEKEPMTLSTRMKIALDVARGLEYIHDHSVPVYIHRDIKSDNILLNENFTGKVAD 288
QHLH S +EP+ + R++IALD ARGLEYIH+H+VPVY+HRDIKS NIL+++ F KVAD
Sbjct: 400 QHLHGSGREPLPWTKRVQIALDSARGLEYIHEHTVPVYVHRDIKSANILIDQKFRAKVAD 459
Query: 289 FGLTKLTDAANSADNTVHVAGTFGYMPPENAYGRISRKIDVYAFGVVLYELISAKAAVIK 348
FGLTKLT+ SA T GTFGYM PE YG +S K+DVYAFGVVLYELISAK AV+K
Sbjct: 460 FGLTKLTEVGGSA--TRGAMGTFGYMAPETVYGEVSAKVDVYAFGVVLYELISAKGAVVK 517
Query: 349 IDKTEFEFKSLEIKTNESIDEYKSLVALFDEVMNQTGDCIDDLRKLVDPRLGYNYSIDSI 408
+ E++ E++ LV +F+E +T D + LRK++DPRLG +Y DS+
Sbjct: 518 M--------------TEAVGEFRGLVGVFEESFKET-DKEEALRKIIDPRLGDSYPFDSV 562
Query: 409 SKMAKLAKACINRDPKQRPTMRDVVVSLMELNSSIDN 445
KMA+L KAC + + RP+MR +VV+L L SS N
Sbjct: 563 YKMAELGKACTQENAQLRPSMRYIVVALSTLFSSTGN 599
>At1g51940 protein kinase like protein
Length = 601
Score = 240 bits (612), Expect = 1e-63
Identities = 139/321 (43%), Positives = 200/321 (62%), Gaps = 20/321 (6%)
Query: 133 VDKSPEFSYEELANATDNFSLAKKIGQGGFGEVYYGELRGQKIAIKKMKMQATREFLSEL 192
++K F+YEE+ ATD FS + +G G +G VY+G LR Q++A+K+M T+EF +E+
Sbjct: 273 IEKPMVFTYEEIRAATDEFSDSNLLGHGNYGSVYFGLLREQEVAVKRMTATKTKEFAAEM 332
Query: 193 KVLTSVHHWNLVHLIGYCVE-GFLFLVYEYMENGNLSQHLHNSEKE---PMTLSTRMKIA 248
KVL VHH NLV LIGY LF+VYEY+ G L HLH+ + + P++ R +IA
Sbjct: 333 KVLCKVHHSNLVELIGYAATVDELFVVYEYVRKGMLKSHLHDPQSKGNTPLSWIMRNQIA 392
Query: 249 LDVARGLEYIHDHSVPVYIHRDIKSDNILLNENFTGKVADFGLTKLTDAANSADNTV-HV 307
LD ARGLEYIH+H+ Y+HRDIK+ NILL+E F K++DFGL KL + + +V V
Sbjct: 393 LDAARGLEYIHEHTKTHYVHRDIKTSNILLDEAFRAKISDFGLAKLVEKTGEGEISVTKV 452
Query: 308 AGTFGYMPPEN-AYGRISRKIDVYAFGVVLYELISAKAAVIKIDKTEFEFKSLEIKTNES 366
GT+GY+ PE + G + K D+YAFGVVL+E+IS + AVI+ + I T
Sbjct: 453 VGTYGYLAPEYLSDGLATSKSDIYAFGVVLFEIISGREAVIRTE---------AIGTKN- 502
Query: 367 IDEYKSLVALFDEVMNQTGDCID--DLRKLVDPRLGYNYSIDSISKMAKLAKACINRDPK 424
E + L ++ V+ + D ++ L++ VDP + Y D + K+A LAK C++ DP
Sbjct: 503 -PERRPLASIMLAVLKNSPDSMNMSSLKEFVDPNMMDLYPHDCLFKIATLAKQCVDDDPI 561
Query: 425 QRPTMRDVVVSLME-LNSSID 444
RP M+ VV+SL + L SSI+
Sbjct: 562 LRPNMKQVVISLSQILLSSIE 582
>At3g24550 protein kinase, putative
Length = 652
Score = 213 bits (541), Expect = 2e-55
Identities = 138/368 (37%), Positives = 200/368 (53%), Gaps = 45/368 (12%)
Query: 88 PLPPKYFRKKNGEEESKFPPEDSMTPSTKDVDKDTNDDNGSKYIWVDKSPEFSYEELANA 147
P PP + G + S P +P + KS F+YEEL+ A
Sbjct: 232 PPPPAFMSSSGGSDYSDLPVLPPPSPGL--------------VLGFSKST-FTYEELSRA 276
Query: 148 TDNFSLAKKIGQGGFGEVYYGEL-RGQKIAIKKMKM---QATREFLSELKVLTSVHHWNL 203
T+ FS A +GQGGFG V+ G L G+++A+K++K Q REF +E+++++ VHH +L
Sbjct: 277 TNGFSEANLLGQGGFGYVHKGILPSGKEVAVKQLKAGSGQGEREFQAEVEIISRVHHRHL 336
Query: 204 VHLIGYCVEGFL-FLVYEYMENGNLSQHLHNSEKEPMTLSTRMKIALDVARGLEYIHDHS 262
V LIGYC+ G LVYE++ N NL HLH + M STR+KIAL A+GL Y+H+
Sbjct: 337 VSLIGYCMAGVQRLLVYEFVPNNNLEFHLHGKGRPTMEWSTRLKIALGSAKGLSYLHEDC 396
Query: 263 VPVYIHRDIKSDNILLNENFTGKVADFGLTKLTDAANSADNTVHVAGTFGYMPPE-NAYG 321
P IHRDIK+ NIL++ F KVADFGL K+ N+ +T V GTFGY+ PE A G
Sbjct: 397 NPKIIHRDIKASNILIDFKFEAKVADFGLAKIASDTNTHVST-RVMGTFGYLAPEYAASG 455
Query: 322 RISRKIDVYAFGVVLYELISAKAAVIKIDKTEFEFKSLEIKTNESIDEYKSLVALFDEVM 381
+++ K DV++FGVVL ELI+ + V N +D+ SLV ++
Sbjct: 456 KLTEKSDVFSFGVVLLELITGRRPV--------------DANNVYVDD--SLVDWARPLL 499
Query: 382 NQTGDCIDDLRKLVDPRLGYNYSIDSISKMAKLAKACINRDPKQRPTMRDVV------VS 435
N+ + D L D ++G Y + +++M A AC+ ++RP M +V VS
Sbjct: 500 NRASE-EGDFEGLADSKMGNEYDREEMARMVACAAACVRHSARRRPRMSQIVRALEGNVS 558
Query: 436 LMELNSSI 443
L +LN +
Sbjct: 559 LSDLNEGM 566
>At5g38560 putative protein
Length = 681
Score = 211 bits (536), Expect = 9e-55
Identities = 136/377 (36%), Positives = 202/377 (53%), Gaps = 45/377 (11%)
Query: 89 LPPKYFRKKNGEEESKFPPEDSMTPSTKDVDKDTNDDNGSKYIWVDKSPE--------FS 140
+PP + G + F S P + +GS Y++ FS
Sbjct: 276 MPPSAYSSPQGSDVVLFNSRSSAPPKMRS-------HSGSDYMYASSDSGMVSNQRSWFS 328
Query: 141 YEELANATDNFSLAKKIGQGGFGEVYYGELR-GQKIAIKKMKM---QATREFLSELKVLT 196
Y+EL+ T FS +G+GGFG VY G L G+++A+K++K+ Q REF +E+++++
Sbjct: 329 YDELSQVTSGFSEKNLLGEGGFGCVYKGVLSDGREVAVKQLKIGGSQGEREFKAEVEIIS 388
Query: 197 SVHHWNLVHLIGYCV-EGFLFLVYEYMENGNLSQHLHNSEKEPMTLSTRMKIALDVARGL 255
VHH +LV L+GYC+ E LVY+Y+ N L HLH + MT TR+++A ARG+
Sbjct: 389 RVHHRHLVTLVGYCISEQHRLLVYDYVPNNTLHYHLHAPGRPVMTWETRVRVAAGAARGI 448
Query: 256 EYIHDHSVPVYIHRDIKSDNILLNENFTGKVADFGLTKLTDAANSADNTVHVA----GTF 311
Y+H+ P IHRDIKS NILL+ +F VADFGL K+ A D HV+ GTF
Sbjct: 449 AYLHEDCHPRIIHRDIKSSNILLDNSFEALVADFGLAKI---AQELDLNTHVSTRVMGTF 505
Query: 312 GYMPPENA-YGRISRKIDVYAFGVVLYELISAKAAVIKIDKTEFEFKSLEIKTNESIDEY 370
GYM PE A G++S K DVY++GV+L ELI+ + V T++ + +
Sbjct: 506 GYMAPEYATSGKLSEKADVYSYGVILLELITGRKPV---------------DTSQPLGD- 549
Query: 371 KSLVALFDEVMNQTGDCIDDLRKLVDPRLGYNYSIDSISKMAKLAKACINRDPKQRPTMR 430
+SLV ++ Q + ++ +LVDPRLG N+ + +M + A AC+ +RP M
Sbjct: 550 ESLVEWARPLLGQAIE-NEEFDELVDPRLGKNFIPGEMFRMVEAAAACVRHSAAKRPKMS 608
Query: 431 DVVVSLMELNSSIDNKN 447
VV +L L + D N
Sbjct: 609 QVVRALDTLEEATDITN 625
>At4g29990 serine/threonine-specific receptor protein kinase LRRPK
Length = 876
Score = 208 bits (529), Expect = 6e-54
Identities = 126/310 (40%), Positives = 182/310 (58%), Gaps = 32/310 (10%)
Query: 139 FSYEELANATDNFSLAKKIGQGGFGEVYYGELRGQKIAIKKMKMQAT---REFLSELKVL 195
F Y E+ N T+NF + +G+GGFG+VY+G L G ++A+K + ++T +EF +E+++L
Sbjct: 564 FIYSEVVNITNNFE--RVLGKGGFGKVYHGFLNGDQVAVKILSEESTQGYKEFRAEVELL 621
Query: 196 TSVHHWNLVHLIGYCVE-GFLFLVYEYMENGNLSQHLHNSEKEPMTLSTRMKIALDVARG 254
VHH NL LIGYC E + L+YEYM NGNL +L ++ R++I+LD A+G
Sbjct: 622 MRVHHTNLTSLIGYCNEDNHMALIYEYMANGNLGDYLSGKSSLILSWEERLQISLDAAQG 681
Query: 255 LEYIHDHSVPVYIHRDIKSDNILLNENFTGKVADFGLTKLTDAANSADNTVHVAGTFGYM 314
LEY+H P +HRD+K NILLNEN K+ADFGL++ S+ + VAGT GY+
Sbjct: 682 LEYLHYGCKPPIVHRDVKPANILLNENLQAKIADFGLSRSFPVEGSSQVSTVVAGTIGYL 741
Query: 315 PPE-NAYGRISRKIDVYAFGVVLYELISAKAAVIKIDKTEFEFKSLEIKTNESIDEYKSL 373
PE A +++ K DVY+FGVVL E+I+ K A+ +TE
Sbjct: 742 DPEYYATRQMNEKSDVYSFGVVLLEVITGKPAIWH-SRTE-------------------S 781
Query: 374 VALFDEVMNQTGDCIDDLRKLVDPRLGYNYSIDSISKMAKLAKACINRDPKQRPTMRDVV 433
V L D+V + + D++ +VD RLG + + S K+ +LA AC + +QRPTM VV
Sbjct: 782 VHLSDQVGSMLAN--GDIKGIVDQRLGDRFEVGSAWKITELALACASESSEQRPTMSQVV 839
Query: 434 VSLMELNSSI 443
MEL SI
Sbjct: 840 ---MELKQSI 846
>At2g28590 putative protein kinase
Length = 424
Score = 206 bits (525), Expect = 2e-53
Identities = 128/331 (38%), Positives = 187/331 (55%), Gaps = 29/331 (8%)
Query: 121 DTNDDNGSKYIWVDKSPEFSYEELANATDNFSLAKKIGQGGFGEVYYG--ELRGQKIAIK 178
D D N + V K+ F++EEL+ +T NF +G+GGFG+VY G E Q +AIK
Sbjct: 68 DAKDTNVEDEVIVKKAQTFTFEELSVSTGNFKSDCFLGEGGFGKVYKGFIEKINQVVAIK 127
Query: 179 KMKM---QATREFLSELKVLTSVHHWNLVHLIGYCVEGFL-FLVYEYMENGNLSQHLHN- 233
++ Q REF+ E+ L+ H NLV LIG+C EG LVYEYM G+L HLH+
Sbjct: 128 QLDRNGAQGIREFVVEVLTLSLADHPNLVKLIGFCAEGVQRLLVYEYMPLGSLDNHLHDL 187
Query: 234 -SEKEPMTLSTRMKIALDVARGLEYIHDHSVPVYIHRDIKSDNILLNENFTGKVADFGLT 292
S K P+ +TRMKIA ARGLEY+HD P I+RD+K NIL++E + K++DFGL
Sbjct: 188 PSGKNPLAWNTRMKIAAGAARGLEYLHDTMKPPVIYRDLKCSNILIDEGYHAKLSDFGLA 247
Query: 293 KLTDAANSADNTVHVAGTFGYMPPENAY-GRISRKIDVYAFGVVLYELISAKAAVIKIDK 351
K+ + + V GT+GY P+ A G+++ K DVY+FGVVL ELI+ + A
Sbjct: 248 KVGPRGSETHVSTRVMGTYGYCAPDYALTGQLTFKSDVYSFGVVLLELITGRKA------ 301
Query: 352 TEFEFKSLEIKTNESIDEYKSLVALFDEVMNQTGDCIDDLRKLVDPRLGYNYSIDSISKM 411
+ + + ++S+ E+ + LF + N +K+VDP L +Y + + +
Sbjct: 302 ----YDNTRTRNHQSLVEWAN--PLFKDRKN--------FKKMVDPLLEGDYPVRGLYQA 347
Query: 412 AKLAKACINRDPKQRPTMRDVVVSLMELNSS 442
+A C+ P RP + DVV++L L SS
Sbjct: 348 LAIAAMCVQEQPSMRPVIADVVMALDHLASS 378
>At2g19230 putative receptor-like protein kinase
Length = 877
Score = 205 bits (521), Expect = 5e-53
Identities = 122/311 (39%), Positives = 178/311 (57%), Gaps = 28/311 (9%)
Query: 133 VDKSPEFSYEELANATDNFSLAKKIGQGGFGEVYYGELRGQKIAIK---KMKMQATREFL 189
+D + Y E+ T+NF + +GQGGFG+VYYG LRG+++AIK K Q +EF
Sbjct: 553 LDTKRYYKYSEIVEITNNFE--RVLGQGGFGKVYYGVLRGEQVAIKMLSKSSAQGYKEFR 610
Query: 190 SELKVLTSVHHWNLVHLIGYCVEGF-LFLVYEYMENGNLSQHLHNSEKEPMTLSTRMKIA 248
+E+++L VHH NL+ LIGYC EG + L+YEY+ NG L +L ++ R++I+
Sbjct: 611 AEVELLLRVHHKNLIALIGYCHEGDQMALIYEYIGNGTLGDYLSGKNSSILSWEERLQIS 670
Query: 249 LDVARGLEYIHDHSVPVYIHRDIKSDNILLNENFTGKVADFGLTKLTDAANSADNTVHVA 308
LD A+GLEY+H+ P +HRD+K NIL+NE K+ADFGL++ + + VA
Sbjct: 671 LDAAQGLEYLHNGCKPPIVHRDVKPTNILINEKLQAKIADFGLSRSFTLEGDSQVSTEVA 730
Query: 309 GTFGYMPPEN-AYGRISRKIDVYAFGVVLYELISAKAAVIKIDKTEFEFKSLEIKTNESI 367
GT GY+ PE+ + + S K DVY+FGVVL E+I+ + VI +TE N I
Sbjct: 731 GTIGYLDPEHYSMQQFSEKSDVYSFGVVLLEVITGQ-PVISRSRTE---------ENRHI 780
Query: 368 DEYKSLVALFDEVMNQTGDCIDDLRKLVDPRLGYNYSIDSISKMAKLAKACINRDPKQRP 427
+ SL M G D++ +VDP+LG ++ K+ ++A AC + K R
Sbjct: 781 SDRVSL-------MLSKG----DIKSIVDPKLGERFNAGLAWKITEVALACASESTKTRL 829
Query: 428 TMRDVVVSLME 438
TM VV L E
Sbjct: 830 TMSQVVAELKE 840
>At3g24400 protein kinase, putative
Length = 694
Score = 204 bits (519), Expect = 8e-53
Identities = 129/367 (35%), Positives = 202/367 (54%), Gaps = 36/367 (9%)
Query: 88 PLPPKYFRK-KNGEEESKFPPEDSMTPSTKDVDKDTNDDNGSKYIWVDKSPEFSYEELAN 146
P PP + +G+ +S + + + P + + G+ F+YEEL+
Sbjct: 277 PRPPHFMSSGSSGDYDSNYSDQSVLPPPSPGLALGLGIYQGT----------FNYEELSR 326
Query: 147 ATDNFSLAKKIGQGGFGEVYYGELR-GQKIAIKKMK---MQATREFLSELKVLTSVHHWN 202
AT+ FS A +GQGGFG V+ G LR G+++A+K++K Q REF +E+ +++ VHH +
Sbjct: 327 ATNGFSEANLLGQGGFGYVFKGMLRNGKEVAVKQLKEGSSQGEREFQAEVGIISRVHHRH 386
Query: 203 LVHLIGYCV-EGFLFLVYEYMENGNLSQHLHNSEKEPMTLSTRMKIALDVARGLEYIHDH 261
LV L+GYC+ + LVYE++ N L HLH + M S+R+KIA+ A+GL Y+H++
Sbjct: 387 LVALVGYCIADAQRLLVYEFVPNNTLEFHLHGKGRPTMEWSSRLKIAVGSAKGLSYLHEN 446
Query: 262 SVPVYIHRDIKSDNILLNENFTGKVADFGLTKLTDAANSADNTVHVAGTFGYMPPENA-Y 320
P IHRDIK+ NIL++ F KVADFGL K+ N+ +T V GTFGY+ PE A
Sbjct: 447 CNPKIIHRDIKASNILIDFKFEAKVADFGLAKIASDTNTHVST-RVMGTFGYLAPEYASS 505
Query: 321 GRISRKIDVYAFGVVLYELISAKAAVIKIDKTEFEFKSLEIKTNESIDEYKSLVALFDEV 380
G+++ K DV++FGVVL ELI+ + + ++ SLV +
Sbjct: 506 GKLTEKSDVFSFGVVLLELITGRRPI----------------DVNNVHADNSLVDWARPL 549
Query: 381 MNQTGDCIDDLRKLVDPRLGYNYSIDSISKMAKLAKACINRDPKQRPTMRDVVVSLMELN 440
+NQ + + + +VD +L Y + +++M A AC+ +RP M D V ++E N
Sbjct: 550 LNQVSE-LGNFEVVVDKKLNNEYDKEEMARMVACAAACVRSTAPRRPRM-DQVARVLEGN 607
Query: 441 SSIDNKN 447
S + N
Sbjct: 608 ISPSDLN 614
>At1g79620 unknown protein
Length = 971
Score = 204 bits (519), Expect = 8e-53
Identities = 121/319 (37%), Positives = 183/319 (56%), Gaps = 30/319 (9%)
Query: 139 FSYEELANATDNFSLAKKIGQGGFGEVYYGELR-GQKIAIKKMKMQATR---EFLSELKV 194
FSYEEL T+NFS++ ++G GG+G+VY G L+ G +AIK+ + +T+ EF +E+++
Sbjct: 626 FSYEELKKITNNFSVSSELGYGGYGKVYKGMLQDGHMVAIKRAQQGSTQGGLEFKTEIEL 685
Query: 195 LTSVHHWNLVHLIGYCVE-GFLFLVYEYMENGNLSQHLHNSEKEPMTLSTRMKIALDVAR 253
L+ VHH NLV L+G+C E G LVYEYM NG+L L + R+++AL AR
Sbjct: 686 LSRVHHKNLVGLVGFCFEQGEQILVYEYMSNGSLKDSLTGRSGITLDWKRRLRVALGSAR 745
Query: 254 GLEYIHDHSVPVYIHRDIKSDNILLNENFTGKVADFGLTKLTDAANSADNTVHVAGTFGY 313
GL Y+H+ + P IHRD+KS NILL+EN T KVADFGL+KL + V GT GY
Sbjct: 746 GLAYLHELADPPIIHRDVKSTNILLDENLTAKVADFGLSKLVSDCTKGHVSTQVKGTLGY 805
Query: 314 MPPE-NAYGRISRKIDVYAFGVVLYELISAKAAVIKIDKTEFEFKSLEIKTNESIDEYKS 372
+ PE +++ K DVY+FGVV+ ELI+AK I+K ++ + +++ N+S D++
Sbjct: 806 LDPEYYTTQKLTEKSDVYSFGVVMMELITAKQ---PIEKGKYIVREIKLVMNKSDDDFYG 862
Query: 373 LVALFDEVMNQTGDCIDDLRKLVDPRLGYNYSIDSISKMAKLAKACINRDPKQRPTMRDV 432
L D + G ++ + + +LA C++ +RPTM +V
Sbjct: 863 LRDKMDRSLRDVG------------------TLPELGRYMELALKCVDETADERPTMSEV 904
Query: 433 VVSLMELNSSIDNKNSTGS 451
V E+ I N ++ S
Sbjct: 905 V---KEIEIIIQNSGASSS 920
>At1g52290 protein kinase, putative
Length = 509
Score = 202 bits (513), Expect = 4e-52
Identities = 137/365 (37%), Positives = 199/365 (53%), Gaps = 36/365 (9%)
Query: 94 FRKKNGEEESKFPPEDSMTPSTKDVDKDTNDDNGSKYIWVDKSPE---FSYEELANATDN 150
F K+ + K ED +D D DD+ + W F+YE+L+ AT N
Sbjct: 84 FYKRKKRKLKKKKKEDIEASINRD-SLDPKDDSNNLQQWSSSEIGQNLFTYEDLSKATSN 142
Query: 151 FSLAKKIGQGGFGEVYYGEL-RGQKIAIKKMKM---QATREFLSELKVLTSVHHWNLVHL 206
FS +GQGGFG V+ G L G +AIK++K Q REF +E++ ++ VHH +LV L
Sbjct: 143 FSNTNLLGQGGFGYVHRGVLVDGTLVAIKQLKSGSGQGEREFQAEIQTISRVHHRHLVSL 202
Query: 207 IGYCVEGFL-FLVYEYMENGNLSQHLHNSEKEPMTLSTRMKIALDVARGLEYIHDHSVPV 265
+GYC+ G LVYE++ N L HLH E+ M S RMKIAL A+GL Y+H+ P
Sbjct: 203 LGYCITGAQRLLVYEFVPNKTLEFHLHEKERPVMEWSKRMKIALGAAKGLAYLHEDCNPK 262
Query: 266 YIHRDIKSDNILLNENFTGKVADFGLTKLTDAANSADNTVHVA----GTFGYMPPENA-Y 320
IHRD+K+ NIL+++++ K+ADFGL A +S D HV+ GTFGY+ PE A
Sbjct: 263 TIHRDVKAANILIDDSYEAKLADFGL-----ARSSLDTDTHVSTRIMGTFGYLAPEYASS 317
Query: 321 GRISRKIDVYAFGVVLYELISAKAAVIKIDKTEFEFKSLEIKTNESIDEYKSLVALFDEV 380
G+++ K DV++ GVVL ELI+ + V KS ++SI ++ L +
Sbjct: 318 GKLTEKSDVFSIGVVLLELITGRRPV---------DKSQPFADDDSIVDWAK--PLMIQA 366
Query: 381 MNQTGDCIDDLRKLVDPRLGYNYSIDSISKMAKLAKACINRDPKQRPTMRDVVVSLMELN 440
+N + LVDPRL ++ I+ +++M A A + K+RP M +V E N
Sbjct: 367 LND-----GNFDGLVDPRLENDFDINEMTRMVACAAASVRHSAKRRPKMSQ-IVRAFEGN 420
Query: 441 SSIDN 445
SID+
Sbjct: 421 ISIDD 425
>At3g55950 receptor kinase - like protein
Length = 814
Score = 201 bits (512), Expect = 5e-52
Identities = 131/326 (40%), Positives = 186/326 (56%), Gaps = 37/326 (11%)
Query: 134 DKSPEFSYEELANATDNFSLAKKIGQGGFGEVYYGELR-GQKIAIK------KMKMQATR 186
DK+ EFS+ ELA+AT NFSL KIG G FG VY G+L G+++AIK KMK +
Sbjct: 479 DKAEEFSFSELASATGNFSLENKIGSGSFGVVYRGKLNDGREVAIKRGEVNAKMKKFQEK 538
Query: 187 E--FLSELKVLTSVHHWNLVHLIGYCVEGF-LFLVYEYMENGNLSQHLH---NSEKEPMT 240
E F SE+ L+ +HH +LV L+GYC E LVY+YM+NG L HLH N EK
Sbjct: 539 ETAFDSEIAFLSRLHHKHLVRLVGYCEEREEKLLVYDYMKNGALYDHLHDKNNVEKHSSL 598
Query: 241 LST---RMKIALDVARGLEYIHDHSVPVYIHRDIKSDNILLNENFTGKVADFGLTKLTDA 297
+++ R+KIALD ARG+EY+H+++VP IHRDIKS NILL+ N+ +V+DFGL+ +
Sbjct: 599 INSWKMRIKIALDAARGIEYLHNYAVPPIIHRDIKSSNILLDSNWVARVSDFGLSLMGPV 658
Query: 298 A----NSADNTVHVAGTFGYMPPE-NAYGRISRKIDVYAFGVVLYELISAKAAVIKIDKT 352
N AGT GY+ PE + ++ K DVY GVVL EL++ K A+
Sbjct: 659 LGKDHNPYQRPTKAAGTVGYIDPEYYSLNVLTDKSDVYGLGVVLLELLTGKRAI------ 712
Query: 353 EFEFKSLEIKTNESIDEYKSLVALFDEVMNQTGDCIDDLRKLVDPRLGYNY--SIDSISK 410
+ N ++E + V + + D+L ++DPR+G D++
Sbjct: 713 --------FRNNGDVEEEEGCVPVHLVDYSVPAITADELSTILDPRVGSPELGEGDAVEL 764
Query: 411 MAKLAKACINRDPKQRPTMRDVVVSL 436
+A A C+N + + RPTM D+V +L
Sbjct: 765 VAYTAMHCVNAEGRNRPTMTDIVGNL 790
>At2g18470 putative protein kinase
Length = 633
Score = 201 bits (512), Expect = 5e-52
Identities = 146/392 (37%), Positives = 203/392 (51%), Gaps = 53/392 (13%)
Query: 69 SQGSGLVYIRGKDGNGLYVPLPPKYFRKKNGEEESKF--PPEDSMTPSTKDVDKDTNDDN 126
SQG G G G P PP+ +GE+ S + P + P + + N
Sbjct: 214 SQGQNQQSTGGWGGGGPSPPPPPRM--PTSGEDSSMYSGPSRPVLPPPSPALALGFNKST 271
Query: 127 GSKYIWVDKSPEFSYEELANATDNFSLAKKIGQGGFGEVYYGEL-RGQKIAIKKMKM--- 182
F+Y+ELA AT F+ A +GQGGFG V+ G L G+++A+K +K
Sbjct: 272 ------------FTYQELAAATGGFTDANLLGQGGFGYVHKGVLPSGKEVAVKSLKAGSG 319
Query: 183 QATREFLSELKVLTSVHHWNLVHLIGYCV-EGFLFLVYEYMENGNLSQHLHNSEKEPMTL 241
Q REF +E+ +++ VHH LV L+GYC+ +G LVYE++ N L HLH M
Sbjct: 320 QGEREFQAEVDIISRVHHRYLVSLVGYCIADGQRMLVYEFVPNKTLEYHLHGKNLPVMEF 379
Query: 242 STRMKIALDVARGLEYIHDHSVPVYIHRDIKSDNILLNENFTGKVADFGLTKLTDAANSA 301
STR++IAL A+GL Y+H+ P IHRDIKS NILL+ NF VADFGL KLT N+
Sbjct: 380 STRLRIALGAAKGLAYLHEDCHPRIIHRDIKSANILLDFNFDAMVADFGLAKLTSDNNTH 439
Query: 302 DNTVHVAGTFGYMPPENA-YGRISRKIDVYAFGVVLYELISAKAAV---IKIDKTEFEFK 357
+T V GTFGY+ PE A G+++ K DV+++GV+L ELI+ K V I +D T
Sbjct: 440 VST-RVMGTFGYLAPEYASSGKLTEKSDVFSYGVMLLELITGKRPVDNSITMDDT----- 493
Query: 358 SLEIKTNESIDEYKSLVALFDEVMNQTGDCIDDLRKLVDPRLGYNYSIDSISKMAKLAKA 417
+D + L+A E N +L D RL NY+ +++M A A
Sbjct: 494 --------LVDWARPLMARALEDGN--------FNELADARLEGNYNPQEMARMVTCAAA 537
Query: 418 CINRDPKQRPTMRDVV------VSLMELNSSI 443
I ++RP M +V VSL LN +
Sbjct: 538 SIRHSGRKRPKMSQIVRALEGEVSLDALNEGV 569
>At1g24030 protein kinase like protein
Length = 375
Score = 201 bits (511), Expect = 7e-52
Identities = 125/314 (39%), Positives = 181/314 (56%), Gaps = 34/314 (10%)
Query: 136 SPEFSYEELANATDNFSLAKKIGQGGFGEVYYGELR-GQKIAIKKM------KMQATREF 188
S ++ +E+ AT +FS +G+GGFG VY G L+ G+ +AIKKM K REF
Sbjct: 61 SSVYTLKEMEEATSSFSDENLLGKGGFGRVYQGTLKTGEVVAIKKMDLPTFKKADGEREF 120
Query: 189 LSELKVLTSVHHWNLVHLIGYCVEG-FLFLVYEYMENGNLSQHLHNSEKEPMTLSTRMKI 247
E+ +L+ + H NLV LIGYC +G FLVYEYM+NGNL HL+ ++ ++ R++I
Sbjct: 121 RVEVDILSRLDHPNLVSLIGYCADGKHRFLVYEYMQNGNLQDHLNGIKEAKISWPIRLRI 180
Query: 248 ALDVARGLEYIHDHS---VPVYIHRDIKSDNILLNENFTGKVADFGLTKLTDAANSADNT 304
AL A+GL Y+H S +P+ +HRD KS N+LL+ N+ K++DFGL KL T
Sbjct: 181 ALGAAKGLAYLHSSSSVGIPI-VHRDFKSTNVLLDSNYNAKISDFGLAKLMPEGKDTCVT 239
Query: 305 VHVAGTFGYMPPE-NAYGRISRKIDVYAFGVVLYELISAKAAVIKIDKTEFEFKSLEIKT 363
V GTFGY PE + G+++ + D+YAFGVVL EL++ + AV L
Sbjct: 240 ARVLGTFGYFDPEYTSTGKLTLQSDIYAFGVVLLELLTGRRAV-----------DLTQGP 288
Query: 364 NESIDEYKSLVALFDEVMNQTGDCIDDLRKLVDPRLGYN-YSIDSISKMAKLAKACINRD 422
NE ++LV ++N LRK++D L N YS+++I+ A LA CI +
Sbjct: 289 NE-----QNLVLQVRNILNDR----KKLRKVIDVELPRNSYSMEAITMFADLASRCIRIE 339
Query: 423 PKQRPTMRDVVVSL 436
K+RP++ D V L
Sbjct: 340 SKERPSVMDCVKEL 353
>At3g18810 protein kinase, putative
Length = 700
Score = 201 bits (510), Expect = 9e-52
Identities = 127/322 (39%), Positives = 188/322 (57%), Gaps = 34/322 (10%)
Query: 139 FSYEELANATDNFSLAKKIGQGGFGEVYYGEL-RGQKIAIKKMKM---QATREFLSELKV 194
F+Y+ELA AT FS ++ +GQGGFG V+ G L G++IA+K +K Q REF +E+ +
Sbjct: 325 FTYDELAAATQGFSQSRLLGQGGFGYVHKGILPNGKEIAVKSLKAGSGQGEREFQAEVDI 384
Query: 195 LTSVHHWNLVHLIGYCVEGFL-FLVYEYMENGNLSQHLHNSEKEPMTLSTRMKIALDVAR 253
++ VHH LV L+GYC+ G LVYE++ N L HLH + + TR+KIAL A+
Sbjct: 385 ISRVHHRFLVSLVGYCIAGGQRMLVYEFLPNDTLEFHLHGKSGKVLDWPTRLKIALGSAK 444
Query: 254 GLEYIHDHSVPVYIHRDIKSDNILLNENFTGKVADFGLTKLTDAANSADNTVHVA----G 309
GL Y+H+ P IHRDIK+ NILL+E+F KVADFGL KL S DN HV+ G
Sbjct: 445 GLAYLHEDCHPRIIHRDIKASNILLDESFEAKVADFGLAKL-----SQDNVTHVSTRIMG 499
Query: 310 TFGYMPPENA-YGRISRKIDVYAFGVVLYELISAKAAVIKIDKTEFEFKSLEIKTNESID 368
TFGY+ PE A G+++ + DV++FGV+L EL++ + V L + +S+
Sbjct: 500 TFGYLAPEYASSGKLTDRSDVFSFGVMLLELVTGRRPV-----------DLTGEMEDSLV 548
Query: 369 EYKSLVALFDEVMNQTGDCIDDLRKLVDPRLGYNYSIDSISKMAKLAKACINRDPKQRPT 428
++ + L N D D +LVDPRL Y +++M A A + ++RP
Sbjct: 549 DWARPICL-----NAAQD--GDYSELVDPRLENQYEPHEMAQMVACAAAAVRHSARRRPK 601
Query: 429 MRDVVVSLMELNSSIDNKNSTG 450
M +V +L E ++++D+ + G
Sbjct: 602 MSQIVRAL-EGDATLDDLSEGG 622
>At4g32710 putative protein kinase
Length = 731
Score = 200 bits (509), Expect = 1e-51
Identities = 128/324 (39%), Positives = 183/324 (55%), Gaps = 25/324 (7%)
Query: 121 DTNDDNGSKYIWVDKSPEFSYEELANATDNFSLAKKIGQGGFGEVYYGELR-GQKIAIKK 179
DT ++N S FSYEEL+ AT FS +G+GGFG V+ G L+ G ++A+K+
Sbjct: 359 DTKENNSVAKNISMPSGMFSYEELSKATGGFSEENLLGEGGFGYVHKGVLKNGTEVAVKQ 418
Query: 180 MKM---QATREFLSELKVLTSVHHWNLVHLIGYCVEGFL-FLVYEYMENGNLSQHLHNSE 235
+K+ Q REF +E+ ++ VHH +LV L+GYCV G LVYE++ L HLH +
Sbjct: 419 LKIGSYQGEREFQAEVDTISRVHHKHLVSLVGYCVNGDKRLLVYEFVPKDTLEFHLHENR 478
Query: 236 KEPMTLSTRMKIALDVARGLEYIHDHSVPVYIHRDIKSDNILLNENFTGKVADFGLTKLT 295
+ R++IA+ A+GL Y+H+ P IHRDIK+ NILL+ F KV+DFGL K
Sbjct: 479 GSVLEWEMRLRIAVGAAKGLAYLHEDCSPTIIHRDIKAANILLDSKFEAKVSDFGLAKFF 538
Query: 296 DAANSADN--TVHVAGTFGYMPPENA-YGRISRKIDVYAFGVVLYELISAKAAVIKIDKT 352
NS+ + V GTFGYM PE A G+++ K DVY+FGVVL ELI+ + ++ D +
Sbjct: 539 SDTNSSFTHISTRVVGTFGYMAPEYASSGKVTDKSDVYSFGVVLLELITGRPSIFAKDSS 598
Query: 353 EFEFKSLEIKTNESIDEYKSLVALFDEVMNQTGDCIDDLRKLVDPRLGYNYSIDSISKMA 412
TN+S+ ++ L + + +G+ D LVD RL NY ++ MA
Sbjct: 599 ----------TNQSLVDWAR--PLLTKAI--SGESFD---FLVDSRLEKNYDTTQMANMA 641
Query: 413 KLAKACINRDPKQRPTMRDVVVSL 436
A ACI + RP M VV +L
Sbjct: 642 ACAAACIRQSAWLRPRMSQVVRAL 665
>At2g19190 putative receptor-like protein kinase
Length = 876
Score = 200 bits (509), Expect = 1e-51
Identities = 120/306 (39%), Positives = 175/306 (56%), Gaps = 29/306 (9%)
Query: 139 FSYEELANATDNFSLAKKIGQGGFGEVYYGELRGQKIAIKKMK---MQATREFLSELKVL 195
F Y E+ N T+NF + IG+GGFG+VY+G + G+++A+K + Q +EF +E+ +L
Sbjct: 564 FKYSEVVNITNNFE--RVIGKGGFGKVYHGVINGEQVAVKVLSEESAQGYKEFRAEVDLL 621
Query: 196 TSVHHWNLVHLIGYCVE-GFLFLVYEYMENGNLSQHLHNSEKEPMTLSTRMKIALDVARG 254
VHH NL L+GYC E + L+YEYM N NL +L ++ R+KI+LD A+G
Sbjct: 622 MRVHHTNLTSLVGYCNEINHMVLIYEYMANENLGDYLAGKRSFILSWEERLKISLDAAQG 681
Query: 255 LEYIHDHSVPVYIHRDIKSDNILLNENFTGKVADFGLTKLTDAANSADNTVHVAGTFGYM 314
LEY+H+ P +HRD+K NILLNE K+ADFGL++ S + VAG+ GY+
Sbjct: 682 LEYLHNGCKPPIVHRDVKPTNILLNEKLQAKMADFGLSRSFSVEGSGQISTVVAGSIGYL 741
Query: 315 PPENAYGR-ISRKIDVYAFGVVLYELISAKAAVIKIDKTEFEFKSLEIKTNESIDEYKSL 373
PE R ++ K DVY+ GVVL E+I+ + A+ KTE K + S D +S+
Sbjct: 742 DPEYYSTRQMNEKSDVYSLGVVLLEVITGQPAIAS-SKTE--------KVHIS-DHVRSI 791
Query: 374 VALFDEVMNQTGDCIDDLRKLVDPRLGYNYSIDSISKMAKLAKACINRDPKQRPTMRDVV 433
+A D+R +VD RL Y + S KM+++A AC QRPTM VV
Sbjct: 792 LA------------NGDIRGIVDQRLRERYDVGSAWKMSEIALACTEHTSAQRPTMSQVV 839
Query: 434 VSLMEL 439
+ L ++
Sbjct: 840 MELKQI 845
>At1g07870 putative protein kinase (At1g07870)
Length = 423
Score = 200 bits (509), Expect = 1e-51
Identities = 128/326 (39%), Positives = 183/326 (55%), Gaps = 29/326 (8%)
Query: 135 KSPEFSYEELANATDNFSLAKKIGQGGFGEVYYGELR--GQKIAIKKMK---MQATREFL 189
K+ F+++ELA AT NF +G+GGFG+V+ G + Q +AIK++ +Q REF+
Sbjct: 87 KAQTFTFQELAEATGNFRSDCFLGEGGFGKVFKGTIEKLDQVVAIKQLDRNGVQGIREFV 146
Query: 190 SELKVLTSVHHWNLVHLIGYCVEGFL-FLVYEYMENGNLSQHLH--NSEKEPMTLSTRMK 246
E+ L+ H NLV LIG+C EG LVYEYM G+L HLH S K+P+ +TRMK
Sbjct: 147 VEVLTLSLADHPNLVKLIGFCAEGDQRLLVYEYMPQGSLEDHLHVLPSGKKPLDWNTRMK 206
Query: 247 IALDVARGLEYIHDHSVPVYIHRDIKSDNILLNENFTGKVADFGLTKLTDAANSADNTVH 306
IA ARGLEY+HD P I+RD+K NILL E++ K++DFGL K+ + + +
Sbjct: 207 IAAGAARGLEYLHDRMTPPVIYRDLKCSNILLGEDYQPKLSDFGLAKVGPSGDKTHVSTR 266
Query: 307 VAGTFGYMPPENAY-GRISRKIDVYAFGVVLYELISAKAAVIKIDKTEFEFKSLEIKTNE 365
V GT+GY P+ A G+++ K D+Y+FGVVL ELI+ + A ID T KT +
Sbjct: 267 VMGTYGYCAPDYAMTGQLTFKSDIYSFGVVLLELITGRKA---IDNT---------KTRK 314
Query: 366 SIDEYKSLVALFDEVMNQTGDCIDDLRKLVDPRLGYNYSIDSISKMAKLAKACINRDPKQ 425
+ LF + N K+VDP L Y + + + ++ C+ P
Sbjct: 315 DQNLVGWARPLFKDRRN--------FPKMVDPLLQGQYPVRGLYQALAISAMCVQEQPTM 366
Query: 426 RPTMRDVVVSLMELNSSIDNKNSTGS 451
RP + DVV++L L SS + NS S
Sbjct: 367 RPVVSDVVLALNFLASSKYDPNSPSS 392
>At2g19210 putative receptor-like protein kinase
Length = 881
Score = 199 bits (507), Expect = 2e-51
Identities = 114/305 (37%), Positives = 174/305 (56%), Gaps = 27/305 (8%)
Query: 139 FSYEELANATDNFSLAKKIGQGGFGEVYYGELRGQKIAIK---KMKMQATREFLSELKVL 195
+ Y E+ T+NF + +GQGGFG+VY+G L ++A+K + Q +EF +E+++L
Sbjct: 566 YKYSEVVKVTNNFE--RVLGQGGFGKVYHGVLNDDQVAVKILSESSAQGYKEFRAEVELL 623
Query: 196 TSVHHWNLVHLIGYCVEGF-LFLVYEYMENGNLSQHLHNSEKEPMTLSTRMKIALDVARG 254
VHH NL LIGYC EG + L+YE+M NG L +L + ++ R++I+LD A+G
Sbjct: 624 LRVHHKNLTALIGYCHEGKKMALIYEFMANGTLGDYLSGEKSYVLSWEERLQISLDAAQG 683
Query: 255 LEYIHDHSVPVYIHRDIKSDNILLNENFTGKVADFGLTKLTDAANSADNTVHVAGTFGYM 314
LEY+H+ P + RD+K NIL+NE K+ADFGL++ + +T VAGT GY+
Sbjct: 684 LEYLHNGCKPPIVQRDVKPANILINEKLQAKIADFGLSRSVALDGNNQDTTAVAGTIGYL 743
Query: 315 PPE-NAYGRISRKIDVYAFGVVLYELISAKAAVIKIDKTEFEFKSLEIKTNESIDEYKSL 373
PE + ++S K D+Y+FGVVL E++S + + + T + I + +D
Sbjct: 744 DPEYHLTQKLSEKSDIYSFGVVLLEVVSGQPVIARSRTT-----AENIHITDRVD----- 793
Query: 374 VALFDEVMNQTGDCIDDLRKLVDPRLGYNYSIDSISKMAKLAKACINRDPKQRPTMRDVV 433
+M TG D+R +VDP+LG + S K+ ++A AC + K RPTM VV
Sbjct: 794 ------LMLSTG----DIRGIVDPKLGERFDAGSAWKITEVAMACASSSSKNRPTMSHVV 843
Query: 434 VSLME 438
L E
Sbjct: 844 AELKE 848
>At4g34440 serine/threonine protein kinase - like
Length = 670
Score = 199 bits (506), Expect = 3e-51
Identities = 129/317 (40%), Positives = 187/317 (58%), Gaps = 34/317 (10%)
Query: 139 FSYEELANATDNFSLAKKIGQGGFGEVYYGEL-RGQKIAIKKMKM---QATREFLSELKV 194
F+Y+EL+ AT+ F+ + +GQGGFG V+ G L G+++A+K +K+ Q REF +E+ +
Sbjct: 300 FTYDELSIATEGFAQSNLLGQGGFGYVHKGVLPSGKEVAVKSLKLGSGQGEREFQAEVDI 359
Query: 195 LTSVHHWNLVHLIGYCVEGFL-FLVYEYMENGNLSQHLHNSEKEPMTLSTRMKIALDVAR 253
++ VHH +LV L+GYC+ G LVYE++ N L HLH + + TR+KIAL AR
Sbjct: 360 ISRVHHRHLVSLVGYCISGGQRLLVYEFIPNNTLEFHLHGKGRPVLDWPTRVKIALGSAR 419
Query: 254 GLEYIHDHSVPVYIHRDIKSDNILLNENFTGKVADFGLTKLTDAANSADNTVHVA----G 309
GL Y+H+ P IHRDIK+ NILL+ +F KVADFGL KL S DN HV+ G
Sbjct: 420 GLAYLHEDCHPRIIHRDIKAANILLDFSFETKVADFGLAKL-----SQDNYTHVSTRVMG 474
Query: 310 TFGYMPPENA-YGRISRKIDVYAFGVVLYELISAKAAVIKIDKTEFEFKSLEIKTNESID 368
TFGY+ PE A G++S K DV++FGV+L ELI+ + + L + +S+
Sbjct: 475 TFGYLAPEYASSGKLSDKSDVFSFGVMLLELITGRPPL-----------DLTGEMEDSLV 523
Query: 369 EYKSLVALFDEVMNQTGDCIDDLRKLVDPRLGYNYSIDSISKMAKLAKACINRDPKQRPT 428
++ + L Q G D +L DPRL NYS + +MA A A I ++RP
Sbjct: 524 DWARPLCL---KAAQDG----DYNQLADPRLELNYSHQEMVQMASCAAAAIRHSARRRPK 576
Query: 429 MRDVVVSLMELNSSIDN 445
M +V +L E + S+D+
Sbjct: 577 MSQIVRAL-EGDMSMDD 592
>At5g02800 protein kinase - like
Length = 395
Score = 199 bits (505), Expect = 3e-51
Identities = 136/359 (37%), Positives = 198/359 (54%), Gaps = 36/359 (10%)
Query: 96 KKNGEEESKFPPED-SMTPSTKDVDKDTNDDNGSKYIWVDKSPEFSYEELANATDNFSLA 154
++NG+ + D S++ S K K + ++ SK + F++ ELA AT NF
Sbjct: 17 RRNGDHKLDRKSSDCSVSTSEKSRAKSSLSESKSKGSDHIVAQTFTFSELATATRNFRKE 76
Query: 155 KKIGQGGFGEVYYGELRG--QKIAIKKMK---MQATREFLSELKVLTSVHHWNLVHLIGY 209
IG+GGFG VY G L Q AIK++ +Q REFL E+ +L+ +HH NLV+LIGY
Sbjct: 77 CLIGEGGFGRVYKGYLASTSQTAAIKQLDHNGLQGNREFLVEVLMLSLLHHPNLVNLIGY 136
Query: 210 CVEGFL-FLVYEYMENGNLSQHLHNSE--KEPMTLSTRMKIALDVARGLEYIHDHSVPVY 266
C +G LVYEYM G+L HLH+ K+P+ +TRMKIA A+GLEY+HD ++P
Sbjct: 137 CADGDQRLLVYEYMPLGSLEDHLHDISPGKQPLDWNTRMKIAAGAAKGLEYLHDKTMPPV 196
Query: 267 IHRDIKSDNILLNENFTGKVADFGLTKLTDAANSADNTVHVAGTFGYMPPENAY-GRISR 325
I+RD+K NILL++++ K++DFGL KL + + + V GT+GY PE A G+++
Sbjct: 197 IYRDLKCSNILLDDDYFPKLSDFGLAKLGPVGDKSHVSTRVMGTYGYCAPEYAMTGQLTL 256
Query: 326 KIDVYAFGVVLYELISAKAAVIKIDKTEFEFKSLEIKTNESIDEYKSLVALFDEVMNQTG 385
K DVY+FGVVL E+I+ + A I ++ S E ++LVA +
Sbjct: 257 KSDVYSFGVVLLEIITGRKA---------------IDSSRSTGE-QNLVAWARPLFK--- 297
Query: 386 DCIDDLRK---LVDPRLGYNYSIDSISKMAKLAKACINRDPKQRPTMRDVVVSLMELNS 441
D RK + DP L Y + + +A C+ P RP + DVV +L L S
Sbjct: 298 ----DRRKFSQMADPMLQGQYPPRGLYQALAVAAMCVQEQPNLRPLIADVVTALSYLAS 352
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.140 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,249,499
Number of Sequences: 26719
Number of extensions: 481016
Number of successful extensions: 5639
Number of sequences better than 10.0: 1013
Number of HSP's better than 10.0 without gapping: 838
Number of HSP's successfully gapped in prelim test: 175
Number of HSP's that attempted gapping in prelim test: 1449
Number of HSP's gapped (non-prelim): 1322
length of query: 451
length of database: 11,318,596
effective HSP length: 103
effective length of query: 348
effective length of database: 8,566,539
effective search space: 2981155572
effective search space used: 2981155572
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC147960.1


