

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147875.5 - phase: 0
(459 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
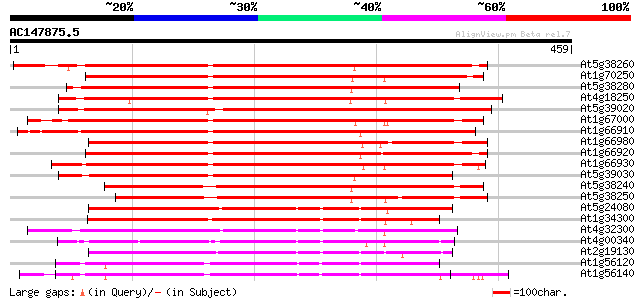
At5g38260 receptor serine/threonine protein kinase - like 384 e-107
At1g70250 hypothetical protein 380 e-105
At5g38280 receptor serine/threonine kinase PR5K 378 e-105
At4g18250 receptor serine/threonine kinase-like protein 378 e-105
At5g39020 receptor protein kinase - like protein 377 e-105
At1g67000 hypothetical protein 377 e-105
At1g66910 receptor serine/threonine kinase PR5K, putative 375 e-104
At1g66980 putative kinase 375 e-104
At1g66920 receptor serine/threonine kinase PR5K, putative 374 e-104
At1g66930 receptor serine/threonine kinase PR5K, putative 373 e-103
At5g39030 receptor protein kinase - like protein 361 e-100
At5g38240 receptor serine/threonine protein kinase -like 343 9e-95
At5g38250 receptor serine/threonine protein kinase - like 332 2e-91
At5g24080 receptor-like protein kinase 242 4e-64
At1g34300 putative protein 240 1e-63
At4g32300 S-receptor kinase -like protein 238 7e-63
At4g00340 receptor-like protein kinase 221 5e-58
At2g19130 putative receptor-like protein kinase 219 3e-57
At1g56120 receptor-like protein kinase, putative 216 2e-56
At1g56140 receptor-like protein kinase, putative 215 5e-56
>At5g38260 receptor serine/threonine protein kinase - like
Length = 638
Score = 384 bits (987), Expect = e-107
Identities = 198/394 (50%), Positives = 269/394 (68%), Gaps = 31/394 (7%)
Query: 4 INSGASVAGFGVTMFFIIFISCYFKKGIRRPQMTIFRKRRKHV---DNNVEVFMQSYNLS 60
I G G T+ + + +F+K R+ H+ DNN++ +Q
Sbjct: 263 IGIGLGCGFLGATLITVCLLCFFFQK----------RRTSHHLRPRDNNLKGLVQ----- 307
Query: 61 IARRYSYAEVKRITNSFRDKLGHGGYGVVYKASLTDGRQVAVKVINESKGNGEEFINEVA 120
++YSYAEV++IT F LG GG+G VY +L DGR+VAVK++ + K NGE+FINEVA
Sbjct: 308 -LKQYSYAEVRKITKLFSHTLGKGGFGTVYGGNLCDGRKVAVKILKDFKSNGEDFINEVA 366
Query: 121 SISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIA 180
S+S+TSH+NIVSLLG+CYE +KRA++YEF+ GSLD+F+ + + D +TL++IA
Sbjct: 367 SMSQTSHVNIVSLLGFCYEGSKRAIVYEFLENGSLDQFLSEK----KSLNLDVSTLYRIA 422
Query: 181 IGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTR 240
+G+ARGL+YLH GC +RI+H DIKPQNILLD+ FCPK+SDFGLAK+C+ +SI+S+ R
Sbjct: 423 LGVARGLDYLHHGCKTRIVHFDIKPQNILLDDTFCPKVSDFGLAKLCEKRESILSLLDAR 482
Query: 241 GTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKN--YQTGGSCTSEMYFPDWIY 298
GTIGY+APEVFS +G VS+KSDVYSYGML+LEMIG + +T S +S YFPDWIY
Sbjct: 483 GTIGYIAPEVFSGMYGRVSHKSDVYSYGMLVLEMIGAKNKEIEETAASNSSSAYFPDWIY 542
Query: 299 KDLEQGNDLLN-SLTISEEENDMVKKITMVSLWCIQTNPLDRPPMNKVIEMLQGPLSSVT 357
K+LE G D IS E+ ++ KK+T+V LWCIQ +PL+RPPMN+++EM++G L +
Sbjct: 543 KNLENGEDTWKFGDEISREDKEVAKKMTLVGLWCIQPSPLNRPPMNRIVEMMEGSLDVLE 602
Query: 358 FPPKPVLFSPKRPPLQLSNMSSSDWQETNSITTE 391
PPKP + P QLS+ S E NSI TE
Sbjct: 603 VPPKPSIHYSAEPLPQLSSFS-----EENSIYTE 631
>At1g70250 hypothetical protein
Length = 676
Score = 380 bits (975), Expect = e-105
Identities = 192/331 (58%), Positives = 242/331 (73%), Gaps = 14/331 (4%)
Query: 63 RRYSYAEVKRITNSFRDKLGHGGYGVVYKASLTDG-RQVAVKVINESKGNGEEFINEVAS 121
+R+SY +VK++T SF + LG GG+G VYK L DG R VAVK++ ES +GE+FINE+AS
Sbjct: 324 KRFSYVQVKKMTKSFENVLGKGGFGTVYKGKLPDGSRDVAVKILKESNEDGEDFINEIAS 383
Query: 122 ISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIAI 181
+SRTSH NIVSLLG+CYE K+A+IYE MP GSLDKFI K+ + TL+ IA+
Sbjct: 384 MSRTSHANIVSLLGFCYEGRKKAIIYELMPNGSLDKFISKN----MSAKMEWKTLYNIAV 439
Query: 182 GIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTRG 241
G++ GLEYLH C SRI+H DIKPQNIL+D + CPKISDFGLAK+C+ N+SI+S+ RG
Sbjct: 440 GVSHGLEYLHSHCVSRIVHFDIKPQNILIDGDLCPKISDFGLAKLCKNNESIISMLHARG 499
Query: 242 TIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRK--NYQTGGSCTSEMYFPDWIYK 299
TIGY+APEVFS+ FGGVS+KSDVYSYGM++LEMIG R Q GS + MYFPDWIYK
Sbjct: 500 TIGYIAPEVFSQNFGGVSHKSDVYSYGMVVLEMIGARNIGRAQNAGSSNTSMYFPDWIYK 559
Query: 300 DLEQGN--DLLNSLTISEEENDMVKKITMVSLWCIQTNPLDRPPMNKVIEMLQGPLSSVT 357
DLE+G L EE+ +VKK+ +V LWCIQTNP DRPPM+KV+EML+G L ++
Sbjct: 560 DLEKGEIMSFLADQITEEEDEKIVKKMVLVGLWCIQTNPYDRPPMSKVVEMLEGSLEALQ 619
Query: 358 FPPKPVLFSPK-RPPLQLSNMSSSDWQETNS 387
PPKP+L P P+ + D QET+S
Sbjct: 620 IPPKPLLCLPAITAPITV----DEDIQETSS 646
>At5g38280 receptor serine/threonine kinase PR5K
Length = 665
Score = 378 bits (970), Expect = e-105
Identities = 188/326 (57%), Positives = 242/326 (73%), Gaps = 14/326 (4%)
Query: 47 DNNVEVFMQSYNLSIARRYSYAEVKRITNSFRDKLGHGGYGVVYKASLTD-GRQVAVKVI 105
D NVE +++ +RYSY VK++TNSF LG GG+G VYK L D GR VAVK++
Sbjct: 309 DQNVEA------VAMLKRYSYTRVKKMTNSFAHVLGKGGFGTVYKGKLADSGRDVAVKIL 362
Query: 106 NESKGNGEEFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFP 165
S+GNGEEFINEVAS+SRTSH+NIVSLLG+CYE NKRA+IYEFMP GSLDK+I +
Sbjct: 363 KVSEGNGEEFINEVASMSRTSHVNIVSLLGFCYEKNKRAIIYEFMPNGSLDKYISAN--- 419
Query: 166 DAVCDCDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAK 225
+ L+ +A+GI+RGLEYLH C +RI+H DIKPQNIL+DEN CPKISDFGLAK
Sbjct: 420 -MSTKMEWERLYDVAVGISRGLEYLHNRCVTRIVHFDIKPQNILMDENLCPKISDFGLAK 478
Query: 226 ICQMNDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGR--KNYQT 283
+C+ +SI+S+ RGT GY+APE+FS+ FG VS+KSDVYSYGM++LEMIG + + +
Sbjct: 479 LCKNKESIISMLHMRGTFGYIAPEMFSKNFGAVSHKSDVYSYGMVVLEMIGAKNIEKVEY 538
Query: 284 GGSCTSEMYFPDWIYKDLEQGN-DLLNSLTISEEENDMVKKITMVSLWCIQTNPLDRPPM 342
GS MYFP+W+YKD E+G + +I++EE + KK+ +V+LWCIQ NP DRPPM
Sbjct: 539 SGSNNGSMYFPEWVYKDFEKGEITRIFGDSITDEEEKIAKKLVLVALWCIQMNPSDRPPM 598
Query: 343 NKVIEMLQGPLSSVTFPPKPVLFSPK 368
KVIEML+G L ++ PP P+LFSP+
Sbjct: 599 IKVIEMLEGNLEALQVPPNPLLFSPE 624
>At4g18250 receptor serine/threonine kinase-like protein
Length = 687
Score = 378 bits (970), Expect = e-105
Identities = 195/372 (52%), Positives = 266/372 (71%), Gaps = 23/372 (6%)
Query: 41 KRRKHV-DNNVEVFMQSYNLSIARRYSYAEVKRITNSFRDKLGHGGYGVVYKASLTD--G 97
KR+ + D N+E + + +RYS+ +VK++TNSF +G GG+G VYK L D G
Sbjct: 324 KRKSELNDENIEAVV------MLKRYSFEKVKKMTNSFDHVIGKGGFGTVYKGKLPDASG 377
Query: 98 RQVAVKVINESKGNGEEFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDK 157
R +A+K++ ESKGNGEEFINE+ S+SR SH+NIVSL G+CYE ++RA+IYEFMP GSLDK
Sbjct: 378 RDIALKILKESKGNGEEFINELVSMSRASHVNIVSLFGFCYEGSQRAIIYEFMPNGSLDK 437
Query: 158 FIYKSGFPDAVCDCDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPK 217
FI ++ + TL+ IA+G+ARGLEYLH C S+I+H DIKPQNIL+DE+ CPK
Sbjct: 438 FISEN----MSTKIEWKTLYNIAVGVARGLEYLHNSCVSKIVHFDIKPQNILIDEDLCPK 493
Query: 218 ISDFGLAKICQMNDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGG 277
ISDFGLAK+C+ +SI+S+ RGT+GY+APE+FS+ +GGVS+KSDVYSYGM++LEMIG
Sbjct: 494 ISDFGLAKLCKKKESIISMLDARGTVGYIAPEMFSKNYGGVSHKSDVYSYGMVVLEMIGA 553
Query: 278 --RKNYQTGGSCTSEMYFPDWIYKDLEQGND--LLNSLTISEEEND-MVKKITMVSLWCI 332
R+ +T + S MYFPDW+Y+DLE+ LL I EEE + +VK++T+V LWCI
Sbjct: 554 TKREEVETSATDKSSMYFPDWVYEDLERKETMRLLEDHIIEEEEEEKIVKRMTLVGLWCI 613
Query: 333 QTNPLDRPPMNKVIEMLQGP-LSSVTFPPKPVLFSPKRPPLQLSNMSSSDWQETNSITTE 391
QTNP DRPPM KV+EML+G L ++ PPKP+L + +S D Q+T+ ++T+
Sbjct: 614 QTNPSDRPPMRKVVEMLEGSRLEALQVPPKPLL----NLHVVTDWETSEDSQQTSRLSTQ 669
Query: 392 TELEEEPIEGYQ 403
+ LE + YQ
Sbjct: 670 SLLERKTRSNYQ 681
>At5g39020 receptor protein kinase - like protein
Length = 813
Score = 377 bits (968), Expect = e-105
Identities = 190/357 (53%), Positives = 257/357 (71%), Gaps = 13/357 (3%)
Query: 41 KRRKHVDNNVEVFMQSYNLSIARRYSYAEVKRITNSFRDKLGHGGYGVVYKASLTDGRQV 100
KR+K+ N + + + ++Y YAE+K+IT SF +G GG+G VY+ +L++GR V
Sbjct: 466 KRKKNKKENSVIMFKL----LLKQYIYAELKKITKSFSHTVGKGGFGTVYRGNLSNGRTV 521
Query: 101 AVKVINESKGNGEEFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIY 160
AVKV+ + KGNG++FINEV S+S+TSH+NIVSLLG+CYE +KRA+I EF+ GSLD+FI
Sbjct: 522 AVKVLKDLKGNGDDFINEVTSMSQTSHVNIVSLLGFCYEGSKRAIISEFLEHGSLDQFIS 581
Query: 161 --KSGFPDAVCDCDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKI 218
KS P+ TL+ IA+GIARGLEYLH GC +RI+H DIKPQNILLD+NFCPK+
Sbjct: 582 RNKSLTPNVT------TLYGIALGIARGLEYLHYGCKTRIVHFDIKPQNILLDDNFCPKV 635
Query: 219 SDFGLAKICQMNDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGR 278
+DFGLAK+C+ +SI+S+ TRGTIGY+APEV SR +GG+S+KSDVYSYGML+L+MIG R
Sbjct: 636 ADFGLAKLCEKRESILSLIDTRGTIGYIAPEVVSRMYGGISHKSDVYSYGMLVLDMIGAR 695
Query: 279 KNYQTGGSCTSEMYFPDWIYKDLEQGNDL-LNSLTISEEENDMVKKITMVSLWCIQTNPL 337
+T S YFPDWIYKDLE G+ + I+EE+N +VKK+ +VSLWCI+ P
Sbjct: 696 NKVETTTCNGSTAYFPDWIYKDLENGDQTWIIGDEINEEDNKIVKKMILVSLWCIRPCPS 755
Query: 338 DRPPMNKVIEMLQGPLSSVTFPPKPVLFSPKRPPLQLSNMSSSDWQETNSITTETEL 394
DRPPMNKV+EM++G L ++ PPKP L+ S++S E + T ++ +
Sbjct: 756 DRPPMNKVVEMIEGSLDALELPPKPSRHISTELVLESSSLSDGQEAEKQTQTLDSTI 812
>At1g67000 hypothetical protein
Length = 717
Score = 377 bits (967), Expect = e-105
Identities = 198/387 (51%), Positives = 269/387 (69%), Gaps = 35/387 (9%)
Query: 15 VTMFFIIFISCYFKKGIRRPQMTIFRKRRKHVDNNVEVFMQSYNLSIA-RRYSYAEVKRI 73
V +F ++ C+ ++ IFRKR+ + EV +Q I + Y+YAEVK++
Sbjct: 332 VIVFLVLLCPCF--------RVQIFRKRK----TSDEVRLQKLKALIPLKHYTYAEVKKM 379
Query: 74 TNSFRDKLGHGGYGVVYKASLTDGRQVAVKVINESKG-NGEEFINEVASISRTSHMNIVS 132
T SF + +G GG+G+VY +L+D VAVKV+ +SKG +GE+FINEVAS+S+TSH+NIVS
Sbjct: 380 TKSFTEVVGRGGFGIVYSGTLSDSSMVAVKVLKDSKGTDGEDFINEVASMSQTSHVNIVS 439
Query: 133 LLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIAIGIARGLEYLHQ 192
LLG+C E ++RA+IYEF+ GSLDKFI + + D TL+ IA+G+ARGLEYLH
Sbjct: 440 LLGFCCEGSRRAIIYEFLGNGSLDKFISDK----SSVNLDLKTLYGIALGVARGLEYLHY 495
Query: 193 GCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTRGTIGYMAPEVFS 252
GC +RI+H DIKPQN+LLD+N CPK+SDFGLAK+C+ +SI+S+ TRGTIGY+APE+ S
Sbjct: 496 GCKTRIVHFDIKPQNVLLDDNLCPKVSDFGLAKLCEKKESILSLLDTRGTIGYIAPEMIS 555
Query: 253 RAFGGVSYKSDVYSYGMLILEMIGGRKNY---QTGGSCTSEMYFPDWIYKDLEQGN--DL 307
R +G VS+KSDVYSYGML+LEMIG RK Q S S +YFP+WIYKDLE+ N D+
Sbjct: 556 RLYGSVSHKSDVYSYGMLVLEMIGARKKERFDQNSRSDGSSIYFPEWIYKDLEKANIKDI 615
Query: 308 -------LNSLTISEEENDMVKKITMVSLWCIQTNPLDRPPMNKVIEMLQGPLSSVTFPP 360
L IS EE ++ +K+T+V LWCIQ++P DRPPMNKV+EM++G L ++ PP
Sbjct: 616 EKTENGGLIENGISSEEEEIARKMTLVGLWCIQSSPSDRPPMNKVVEMMEGSLDALEVPP 675
Query: 361 KPVLFSPKRPPLQLSNMSSSDWQETNS 387
+PVL + S++S S W S
Sbjct: 676 RPVL-----QQISASSVSDSFWNSEES 697
>At1g66910 receptor serine/threonine kinase PR5K, putative
Length = 655
Score = 375 bits (964), Expect = e-104
Identities = 199/378 (52%), Positives = 263/378 (68%), Gaps = 11/378 (2%)
Query: 7 GASVAGFGVTMFFIIFISCYFKKGIRRPQMTIFRKRRKHVDNNVEVFMQSYNLSIARRYS 66
G A F + M I+ ++C IRR + T+ R + D++ + +++ L + YS
Sbjct: 273 GIGAASFAM-MGVILVVTC-LNCLIRRQRKTLNDPRMRTSDDSRQQNLKA--LIPLKHYS 328
Query: 67 YAEVKRITNSFRDKLGHGGYGVVYKASLTDGRQVAVKVINESKGNGEEFINEVASISRTS 126
YA+V IT SF + +G GG+G VY+ +L DGR VAVKV+ ES+GNGE+FINEVAS+S+TS
Sbjct: 329 YAQVTSITKSFAEVIGKGGFGTVYRGTLYDGRSVAVKVLKESQGNGEDFINEVASMSQTS 388
Query: 127 HMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIAIGIARG 186
H+NIV+LLG+C E KRA+IYEFM GSLDKFI D L+ IA+G+ARG
Sbjct: 389 HVNIVTLLGFCSEGYKRAIIYEFMENGSLDKFISSK----KSSTMDWRELYGIALGVARG 444
Query: 187 LEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTRGTIGYM 246
LEYLH GC +RI+H DIKPQN+LLD+N PK+SDFGLAK+C+ +SI+S+ TRGTIGY+
Sbjct: 445 LEYLHHGCRTRIVHFDIKPQNVLLDDNLSPKVSDFGLAKLCERKESILSLMDTRGTIGYI 504
Query: 247 APEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGG--SCTSEMYFPDWIYKDLEQG 304
APEVFSR +G VS+KSDVYSYGML+L++IG R T S TS MYFP+WIY+DLE+
Sbjct: 505 APEVFSRVYGRVSHKSDVYSYGMLVLDIIGARNKTSTEDTTSSTSSMYFPEWIYRDLEKA 564
Query: 305 -NDLLNSLTISEEENDMVKKITMVSLWCIQTNPLDRPPMNKVIEMLQGPLSSVTFPPKPV 363
N IS EE+++ KK+T+V LWCIQ PLDRP MN+V+EM++G L ++ PP+PV
Sbjct: 565 HNGKSIETAISNEEDEIAKKMTLVGLWCIQPWPLDRPAMNRVVEMMEGNLDALEVPPRPV 624
Query: 364 LFSPKRPPLQLSNMSSSD 381
L LQ S+ S D
Sbjct: 625 LQQIPTATLQESSTFSED 642
>At1g66980 putative kinase
Length = 1109
Score = 375 bits (962), Expect = e-104
Identities = 184/334 (55%), Positives = 251/334 (75%), Gaps = 18/334 (5%)
Query: 65 YSYAEVKRITNSFRDKLGHGGYGVVYKASLTDGRQVAVKVINESKGNGEEFINEVASISR 124
Y+YA+VKRIT SF + +G GG+G+VYK +L+DGR VAVKV+ ++KGNGE+FINEVA++SR
Sbjct: 786 YTYAQVKRITKSFAEVVGRGGFGIVYKGTLSDGRVVAVKVLKDTKGNGEDFINEVATMSR 845
Query: 125 TSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIAIGIA 184
TSH+NIVSLLG+C E +KRA+IYEF+ GSLDKFI + D L++IA+G+A
Sbjct: 846 TSHLNIVSLLGFCSEGSKRAIIYEFLENGSLDKFILGK----TSVNMDWTALYRIALGVA 901
Query: 185 RGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTRGTIG 244
GLEYLH C +RI+H DIKPQN+LLD++FCPK+SDFGLAK+C+ +SI+S+ TRGTIG
Sbjct: 902 HGLEYLHHSCKTRIVHFDIKPQNVLLDDSFCPKVSDFGLAKLCEKKESILSMLDTRGTIG 961
Query: 245 YMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSC---TSEMYFPDWIYKDL 301
Y+APE+ SR +G VS+KSDVYSYGML+LE+IG R + +C TS MYFP+W+Y+DL
Sbjct: 962 YIAPEMISRVYGNVSHKSDVYSYGMLVLEIIGARNKEKANQACASNTSSMYFPEWVYRDL 1021
Query: 302 E---QGNDLLNSLTISEEENDMVKKITMVSLWCIQTNPLDRPPMNKVIEMLQGPLSSVTF 358
E G + + I+ EE+++ KK+T+V LWCIQ +P+DRP MN+V+EM++G L ++
Sbjct: 1022 ESCKSGRHIEDG--INSEEDELAKKMTLVGLWCIQPSPVDRPAMNRVVEMMEGSLEALEV 1079
Query: 359 PPKPVLFSPKRPPLQLSNM-SSSDWQETNSITTE 391
PP+PVL + +SN+ SS E S+ TE
Sbjct: 1080 PPRPVL-----QQIPISNLHESSILSEDVSVYTE 1108
>At1g66920 receptor serine/threonine kinase PR5K, putative
Length = 609
Score = 374 bits (960), Expect = e-104
Identities = 191/332 (57%), Positives = 247/332 (73%), Gaps = 13/332 (3%)
Query: 63 RRYSYAEVKRITNSFRDKLGHGGYGVVYKASLTDGRQVAVKVINESKGN-GEEFINEVAS 121
++YSY +VKRITNSF + +G GG+G+VY+ +L+DGR VAVKV+ + KGN GE+FINEVAS
Sbjct: 287 KQYSYEQVKRITNSFAEVVGRGGFGIVYRGTLSDGRMVAVKVLKDLKGNNGEDFINEVAS 346
Query: 122 ISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIAI 181
+S+TSH+NIV+LLG+C E KRA+IYEFM GSLDKFI D L+ IA+
Sbjct: 347 MSQTSHVNIVTLLGFCSEGYKRAIIYEFMENGSLDKFISSK----KSSTMDWRELYGIAL 402
Query: 182 GIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTRG 241
G+ARGLEYLH GC +RI+H DIKPQN+LLD+N PK+SDFGLAK+C+ +SI+S+ TRG
Sbjct: 403 GVARGLEYLHHGCRTRIVHFDIKPQNVLLDDNLSPKVSDFGLAKLCERKESILSLMDTRG 462
Query: 242 TIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGG--SCTSEMYFPDWIYK 299
TIGY+APEVFSR +G VS+KSDVYSYGML+L++IG R T S TS MYFP+WIYK
Sbjct: 463 TIGYIAPEVFSRVYGSVSHKSDVYSYGMLVLDIIGARNKTSTEDTTSSTSSMYFPEWIYK 522
Query: 300 DLEQGNDLLNSLTISEEENDMVKKITMVSLWCIQTNPLDRPPMNKVIEMLQGPLSSVTFP 359
DLE+G D + EE+++ KK+T+V LWCIQ PLDRP MN+V+EM++G L ++ P
Sbjct: 523 DLEKG-DNGRLIVNRSEEDEIAKKMTLVGLWCIQPWPLDRPAMNRVVEMMEGNLDALEVP 581
Query: 360 PKPVLFSPKRPPLQLSNMSSSDWQETNSITTE 391
P+PVL P L S +S E NSI++E
Sbjct: 582 PRPVLQCSVVPHLDSSWIS-----EENSISSE 608
>At1g66930 receptor serine/threonine kinase PR5K, putative
Length = 876
Score = 373 bits (958), Expect = e-103
Identities = 191/363 (52%), Positives = 261/363 (71%), Gaps = 20/363 (5%)
Query: 35 QMTIFRKRRKHVDNNVEVFMQSYNLSIARRYSYAEVKRITNSFRDKLGHGGYGVVYKASL 94
++ IFR R+ D E L + Y+YA+VKR+T SF + +G GG+G+VY+ +L
Sbjct: 511 KVRIFRNRKTSDDRRQEKLKALIPL---KHYTYAQVKRMTKSFAEVVGRGGFGIVYRGTL 567
Query: 95 TDGRQVAVKVINESKGNG-EEFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKG 153
DGR VAVKV+ ESKGN E+FINEV+S+S+TSH+NIVSLLG+C E ++RA+IYEF+ G
Sbjct: 568 CDGRMVAVKVLKESKGNNSEDFINEVSSMSQTSHVNIVSLLGFCSEGSRRAIIYEFLENG 627
Query: 154 SLDKFIYKSGFPDAVCDCDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDEN 213
SLDKFI + D L+ IA+G+ARGLEYLH GC +RI+H DIKPQN+LLD+N
Sbjct: 628 SLDKFISEK----TSVILDLTALYGIALGVARGLEYLHYGCKTRIVHFDIKPQNVLLDDN 683
Query: 214 FCPKISDFGLAKICQMNDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILE 273
PK+SDFGLAK+C+ +S++S+ TRGTIGY+APE+ SR +G VS+KSDVYSYGML+ E
Sbjct: 684 LSPKVSDFGLAKLCEKKESVMSLMDTRGTIGYIAPEMISRVYGSVSHKSDVYSYGMLVFE 743
Query: 274 MIGGRKNYQTGGSCT--SEMYFPDWIYKDLEQGN--DLLN-SLTISEEENDMVKKITMVS 328
MIG RK + G + S MYFP+WIYKDLE+ + DL + + IS EE ++ KK+T+V
Sbjct: 744 MIGARKKERFGQNSANGSSMYFPEWIYKDLEKADNGDLEHIEIGISSEEEEIAKKMTLVG 803
Query: 329 LWCIQTNPLDRPPMNKVIEMLQGPLSSVTFPPKPVLFSPKRPPLQLSNMSSSDW--QETN 386
LWCIQ++P DRPPMNKV+EM++G L ++ PP+PVL + + + S W +E++
Sbjct: 804 LWCIQSSPSDRPPMNKVVEMMEGSLDALEVPPRPVL-----QQIHVGPLLESSWITEESS 858
Query: 387 SIT 389
SI+
Sbjct: 859 SIS 861
>At5g39030 receptor protein kinase - like protein
Length = 806
Score = 361 bits (926), Expect = e-100
Identities = 177/325 (54%), Positives = 240/325 (73%), Gaps = 12/325 (3%)
Query: 41 KRRKHVDNNVEVFMQSYNLSIARRYSYAEVKRITNSFRDKLGHGGYGVVYKASLTDGRQV 100
KR+ + V +F + N+ Y+YAE+K+IT SF +G GG+G VY +L++GR+V
Sbjct: 469 KRKNRKEERVVMFKKLLNM-----YTYAELKKITKSFSYIIGKGGFGTVYGGNLSNGRKV 523
Query: 101 AVKVINESKGNGEEFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIY 160
AVKV+ + KG+ E+FINEVAS+S+TSH+NIVSLLG+C+E +KRA++YEF+ GSLD+F+
Sbjct: 524 AVKVLKDLKGSAEDFINEVASMSQTSHVNIVSLLGFCFEGSKRAIVYEFLENGSLDQFMS 583
Query: 161 KSGFPDAVCDCDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISD 220
++ D TL+ IA+GIARGLEYLH GC +RI+H DIKPQNILLD N CPK+SD
Sbjct: 584 RN----KSLTQDVTTLYGIALGIARGLEYLHYGCKTRIVHFDIKPQNILLDGNLCPKVSD 639
Query: 221 FGLAKICQMNDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKN 280
FGLAK+C+ +S++S+ TRGTIGY+APEVFSR +G VS+KSDVYS+GML+++MIG R
Sbjct: 640 FGLAKLCEKRESVLSLMDTRGTIGYIAPEVFSRMYGRVSHKSDVYSFGMLVIDMIGARSK 699
Query: 281 --YQTGGSCTSEMYFPDWIYKDLEQGNDL-LNSLTISEEENDMVKKITMVSLWCIQTNPL 337
+T S S YFPDWIYKDLE G + I++EE ++ KK+ +V LWCIQ P
Sbjct: 700 EIVETVDSAASSTYFPDWIYKDLEDGEQTWIFGDEITKEEKEIAKKMIVVGLWCIQPCPS 759
Query: 338 DRPPMNKVIEMLQGPLSSVTFPPKP 362
DRP MN+V+EM++G L ++ PPKP
Sbjct: 760 DRPSMNRVVEMMEGSLDALEIPPKP 784
>At5g38240 receptor serine/threonine protein kinase -like
Length = 588
Score = 343 bits (881), Expect = 9e-95
Identities = 172/313 (54%), Positives = 229/313 (72%), Gaps = 17/313 (5%)
Query: 78 RDKLGHGGYGVVYKASLTDGRQVAVKVINESKGNGEEFINEVASISRTSHMNIVSLLGYC 137
++ +G GG+G VYK +L DGR+VAVK++ +S GN E+FINEVASIS+TSH+NIVSLLG+C
Sbjct: 284 QEVVGRGGFGTVYKGNLRDGRKVAVKILKDSNGNCEDFINEVASISQTSHVNIVSLLGFC 343
Query: 138 YEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIAIGIARGLEYLHQGCSSR 197
+E +KRA++YEF+ GSLD+ + D +TL+ IA+G+ARG+EYLH GC R
Sbjct: 344 FEKSKRAIVYEFLENGSLDQS----------SNLDVSTLYGIALGVARGIEYLHFGCKKR 393
Query: 198 ILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTRGTIGYMAPEVFSRAFGG 257
I+H DIKPQN+LLDEN PK++DFGLAK+C+ +SI+S+ TRGTIGY+APE+FSR +G
Sbjct: 394 IVHFDIKPQNVLLDENLKPKVADFGLAKLCEKQESILSLLDTRGTIGYIAPELFSRVYGN 453
Query: 258 VSYKSDVYSYGMLILEMIGGR--KNYQTGGSCTSEMYFPDWIYKDLEQGNDL-LNSLTIS 314
VS+KSDVYSYGML+LEM G R + Q S S YFPDWI+KDLE G+ + L + ++
Sbjct: 454 VSHKSDVYSYGMLVLEMTGARNKERVQNADSNNSSAYFPDWIFKDLENGDYVKLLADGLT 513
Query: 315 EEENDMVKKITMVSLWCIQTNPLDRPPMNKVIEMLQGPLSSVTFPPKPVLFSPKRPPLQL 374
EE D+ KK+ +V LWCIQ P DRP MNKV+ M++G L S+ PPKP+L P+Q
Sbjct: 514 REEEDIAKKMILVGLWCIQFRPSDRPSMNKVVGMMEGNLDSLDPPPKPLL----HMPMQN 569
Query: 375 SNMSSSDWQETNS 387
+N SS E +S
Sbjct: 570 NNAESSQPSEEDS 582
>At5g38250 receptor serine/threonine protein kinase - like
Length = 579
Score = 332 bits (852), Expect = 2e-91
Identities = 173/310 (55%), Positives = 224/310 (71%), Gaps = 21/310 (6%)
Query: 87 GVVYKASLTDGRQVAVKVINESKGNGEEFINEVASISRTSHMNIVSLLGYCYEANKRALI 146
G + L DGR+VAVKV+ +SKGN E+FINEVAS+S+TSH+NIV+LLG+CYE +KRA+I
Sbjct: 285 GTLRGGRLRDGRKVAVKVLKDSKGNCEDFINEVASMSQTSHVNIVTLLGFCYEGSKRAII 344
Query: 147 YEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQ 206
YEF+ GSLD+ + + D +TL+ IA+G+ARGLEYLH GC +RI+H DIKPQ
Sbjct: 345 YEFLENGSLDQSL----------NLDVSTLYGIALGVARGLEYLHYGCKTRIVHFDIKPQ 394
Query: 207 NILLDENFCPKISDFGLAKICQMNDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYS 266
N+LLDEN PK++DFGLAK+C+ +SI+S+ TRGTIGY+APE+FSR +G VS+KSDVYS
Sbjct: 395 NVLLDENLRPKVADFGLAKLCEKQESILSLLDTRGTIGYIAPELFSRMYGSVSHKSDVYS 454
Query: 267 YGMLILEMIGGR--KNYQTGGSCTSEMYFPDWIYKDLEQGND---LLNSLTISEEENDMV 321
YGML+LEMIG R + Q S YFPDWIYKDLE ++ L + LT EE+N
Sbjct: 455 YGMLVLEMIGARNKERVQNADPNNSSAYFPDWIYKDLENFDNTRLLGDGLTREEEKN--A 512
Query: 322 KKITMVSLWCIQTNPLDRPPMNKVIEMLQGPLSSVTFPPKPVLFSPKRPPLQLSNMSSSD 381
KK+ +V LWCIQ P DRP MNKV+EM++G L S+ PPKP+L P+Q +N SS
Sbjct: 513 KKMILVGLWCIQFRPSDRPSMNKVVEMMEGSLDSLDPPPKPLL----HMPMQNNNAESSQ 568
Query: 382 WQETNSITTE 391
+SI +E
Sbjct: 569 LSVESSIYSE 578
>At5g24080 receptor-like protein kinase
Length = 872
Score = 242 bits (617), Expect = 4e-64
Identities = 129/301 (42%), Positives = 184/301 (60%), Gaps = 9/301 (2%)
Query: 65 YSYAEVKRITNSFRDKLGHGGYGVVYKASLTDGRQVAVKVINESKGNGE-EFINEVASIS 123
++Y +++ TN+F LG GG+G VYK ++ VAVK ++ + +GE EFI EV +I
Sbjct: 520 FTYRDLQNCTNNFSQLLGSGGFGTVYKGTVAGETLVAVKRLDRALSHGEREFITEVNTIG 579
Query: 124 RTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIAIGI 183
HMN+V L GYC E + R L+YE+M GSLDK+I+ S + D T F+IA+
Sbjct: 580 SMHHMNLVRLCGYCSEDSHRLLVYEYMINGSLDKWIFSSEQTANLLDW--RTRFEIAVAT 637
Query: 184 ARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTRGTI 243
A+G+ Y H+ C +RI+H DIKP+NILLD+NFCPK+SDFGLAK+ S V + RGT
Sbjct: 638 AQGIAYFHEQCRNRIIHCDIKPENILLDDNFCPKVSDFGLAKMMGREHSHV-VTMIRGTR 696
Query: 244 GYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSCTSEMYFPDWIYKDLEQ 303
GY+APE S ++ K+DVYSYGML+LE++GGR+N + ++P W YK+L
Sbjct: 697 GYLAPEWVSNR--PITVKADVYSYGMLLLEIVGGRRNLDMSYD-AEDFFYPGWAYKELTN 753
Query: 304 GNDL--LNSLTISEEENDMVKKITMVSLWCIQTNPLDRPPMNKVIEMLQGPLSSVTFPPK 361
G L ++ E + V K V+ WCIQ RP M +V+++L+G + PP
Sbjct: 754 GTSLKAVDKRLQGVAEEEEVVKALKVAFWCIQDEVSMRPSMGEVVKLLEGTSDEINLPPM 813
Query: 362 P 362
P
Sbjct: 814 P 814
>At1g34300 putative protein
Length = 829
Score = 240 bits (613), Expect = 1e-63
Identities = 124/292 (42%), Positives = 187/292 (63%), Gaps = 10/292 (3%)
Query: 64 RYSYAEVKRITNSFRDKLGHGGYGVVYKASLTDGRQVAVKVINESKGNGEEFINEVASIS 123
+++Y E++R T SF++KLG GG+G VY+ LT+ VAVK + + ++F EVA+IS
Sbjct: 473 QFTYKELQRCTKSFKEKLGAGGFGTVYRGVLTNRTVVAVKQLEGIEQGEKQFRMEVATIS 532
Query: 124 RTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIAIGI 183
T H+N+V L+G+C + R L+YEFM GSLD F++ + D+ F IA+G
Sbjct: 533 STHHLNLVRLIGFCSQGRHRLLVYEFMRNGSLDNFLFTT---DSAKFLTWEYRFNIALGT 589
Query: 184 ARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTRGTI 243
A+G+ YLH+ C I+H DIKP+NIL+D+NF K+SDFGLAK+ D+ ++ RGT
Sbjct: 590 AKGITYLHEECRDCIVHCDIKPENILVDDNFAAKVSDFGLAKLLNPKDNRYNMSSVRGTR 649
Query: 244 GYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSCTSEMYFPDWIYKDLEQ 303
GY+APE + ++ KSDVYSYGM++LE++ G++N+ T+ F W Y++ E+
Sbjct: 650 GYLAPEWLANL--PITSKSDVYSYGMVLLELVSGKRNFDVSEK-TNHKKFSIWAYEEFEK 706
Query: 304 GND--LLNSLTISEEENDMVKKITMV--SLWCIQTNPLDRPPMNKVIEMLQG 351
GN +L++ ++ DM + + MV S WCIQ PL RP M KV++ML+G
Sbjct: 707 GNTKAILDTRLSEDQTVDMEQVMRMVKTSFWCIQEQPLQRPTMGKVVQMLEG 758
>At4g32300 S-receptor kinase -like protein
Length = 821
Score = 238 bits (606), Expect = 7e-63
Identities = 143/357 (40%), Positives = 212/357 (59%), Gaps = 17/357 (4%)
Query: 15 VTMFFI-IFISCYFKKGIRRPQMTIFRKRRKHVDNNVEVFMQSYNLS-IARRYSYAEVKR 72
VT+F I + I F+ R+ + + DN +E NLS + R++Y +++
Sbjct: 437 VTVFIIAVLIFVAFRIHKRKKMILEAPQESSEEDNFLE------NLSGMPIRFAYKDLQS 490
Query: 73 ITNSFRDKLGHGGYGVVYKASLTDGRQVAVKVINESKGNGEEFINEVASISRTSHMNIVS 132
TN+F KLG GG+G VY+ +L DG ++AVK + +EF EV+ I H+++V
Sbjct: 491 ATNNFSVKLGQGGFGSVYEGTLPDGSRLAVKKLEGIGQGKKEFRAEVSIIGSIHHLHLVR 550
Query: 133 LLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIAIGIARGLEYLHQ 192
L G+C E R L YEF+ KGSL+++I++ D + D D T F IA+G A+GL YLH+
Sbjct: 551 LRGFCAEGAHRLLAYEFLSKGSLERWIFRKKDGDVLLDWD--TRFNIALGTAKGLAYLHE 608
Query: 193 GCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTRGTIGYMAPEVFS 252
C +RI+H DIKP+NILLD+NF K+SDFGLAK+ S V RGT GY+APE +
Sbjct: 609 DCDARIVHCDIKPENILLDDNFNAKVSDFGLAKLMTREQSHV-FTTMRGTRGYLAPEWIT 667
Query: 253 RAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSCTSEMYFPDWIYKDLEQGN--DLLNS 310
+S KSDVYSYGM++LE+IGGRKNY + + +FP + +K +E+G D+++
Sbjct: 668 NY--AISEKSDVYSYGMVLLELIGGRKNYDP-SETSEKCHFPSFAFKKMEEGKLMDIVDG 724
Query: 311 LTISEEEND-MVKKITMVSLWCIQTNPLDRPPMNKVIEMLQGPLSSVTFPPKPVLFS 366
+ + D V++ +LWCIQ + RP M+KV++ML+G V P + S
Sbjct: 725 KMKNVDVTDERVQRAMKTALWCIQEDMQTRPSMSKVVQMLEGVFPVVQPPSSSTMGS 781
>At4g00340 receptor-like protein kinase
Length = 797
Score = 221 bits (564), Expect = 5e-58
Identities = 136/336 (40%), Positives = 200/336 (59%), Gaps = 24/336 (7%)
Query: 40 RKRRKHVDNNVEVFMQSYNLSIARRYSYAEVKRITNSFRDKLGHGGYGVVYKASLTDGRQ 99
RKR+K + + F NL + +S+ E++ TN F DK+GHGG+G V+K +L
Sbjct: 430 RKRKKTRKQDEDGFAV-LNLKV---FSFKELQSATNGFSDKVGHGGFGAVFKGTLPGSST 485
Query: 100 -VAVKVINESKGNGE-EFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDK 157
VAVK + E G+GE EF EV +I H+N+V L G+C E R L+Y++MP+GSL
Sbjct: 486 FVAVKRL-ERPGSGESEFRAEVCTIGNIQHVNLVRLRGFCSENLHRLLVYDYMPQGSLSS 544
Query: 158 FIYKSGFPDAVCDCDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPK 217
++ ++ P + T F+IA+G A+G+ YLH+GC I+H DIKP+NILLD ++ K
Sbjct: 545 YLSRTS-PKLL---SWETRFRIALGTAKGIAYLHEGCRDCIIHCDIKPENILLDSDYNAK 600
Query: 218 ISDFGLAKICQMNDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGG 277
+SDFGLAK+ + S V + RGT GY+APE S ++ K+DVYS+GM +LE+IGG
Sbjct: 601 VSDFGLAKLLGRDFSRV-LATMRGTWGYVAPEWISGL--PITTKADVYSFGMTLLELIGG 657
Query: 278 RKNYQTGGSCTSE-------MYFPDWIYKDLEQGN--DLLNSLTISEEENDMVKKITMVS 328
R+N E +FP W +++ QGN +++S E + V ++ V+
Sbjct: 658 RRNVIVNSDTLGEKETEPEKWFFPPWAAREIIQGNVDSVVDSRLNGEYNTEEVTRMATVA 717
Query: 329 LWCIQTNPLDRPPMNKVIEMLQGPLSSVTFPPKPVL 364
+WCIQ N RP M V++ML+G + VT PP P L
Sbjct: 718 IWCIQDNEEIRPAMGTVVKMLEG-VVEVTVPPPPKL 752
>At2g19130 putative receptor-like protein kinase
Length = 828
Score = 219 bits (557), Expect = 3e-57
Identities = 122/303 (40%), Positives = 177/303 (58%), Gaps = 12/303 (3%)
Query: 65 YSYAEVKRITNSFRDKLGHGGYGVVYKASLTDGRQVAVKVINESKGNGEEFINEVASISR 124
+SY E++ T +F DKLG GG+G V+K +L D +AVK + ++F EV +I
Sbjct: 483 FSYRELQNATKNFSDKLGGGGFGSVFKGALPDSSDIAVKRLEGISQGEKQFRTEVVTIGT 542
Query: 125 TSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIAIGIA 184
H+N+V L G+C E +K+ L+Y++MP GSLD ++ + + + FQIA+G A
Sbjct: 543 IQHVNLVRLRGFCSEGSKKLLVYDYMPNGSLDSHLFLNQVEEKIV-LGWKLRFQIALGTA 601
Query: 185 RGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTRGTIG 244
RGL YLH C I+H DIKP+NILLD FCPK++DFGLAK+ + S V + RGT G
Sbjct: 602 RGLAYLHDECRDCIIHCDIKPENILLDSQFCPKVADFGLAKLVGRDFSRV-LTTMRGTRG 660
Query: 245 YMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSCTSEMYFPDWIYKDLEQG 304
Y+APE S ++ K+DVYSYGM++ E++ GR+N + + +FP W L +
Sbjct: 661 YLAPEWISGV--AITAKADVYSYGMMLFELVSGRRNTEQSEN-EKVRFFPSWAATILTKD 717
Query: 305 NDLLNSLTISEEENDM-----VKKITMVSLWCIQTNPLDRPPMNKVIEMLQGPLSSVTFP 359
D + SL E D V + V+ WCIQ RP M++V+++L+G L V P
Sbjct: 718 GD-IRSLVDPRLEGDAVDIEEVTRACKVACWCIQDEESHRPAMSQVVQILEGVL-EVNPP 775
Query: 360 PKP 362
P P
Sbjct: 776 PFP 778
>At1g56120 receptor-like protein kinase, putative
Length = 1045
Score = 216 bits (551), Expect = 2e-56
Identities = 124/318 (38%), Positives = 189/318 (58%), Gaps = 15/318 (4%)
Query: 38 IFRKRRKHVDNNVEVFMQSYNLSIARRYSYAEVKRITNSF--RDKLGHGGYGVVYKASLT 95
+ RKRRK ++ E+ ++Y+E+K T F +KLG GG+G VYK +L
Sbjct: 672 VIRKRRKPYTDDEEILSMDVK---PYTFTYSELKNATQDFDLSNKLGEGGFGAVYKGNLN 728
Query: 96 DGRQVAVKVINESKGNGE-EFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGS 154
DGR+VAVK ++ G+ +F+ E+ +IS H N+V L G C+E + R L+YE++P GS
Sbjct: 729 DGREVAVKQLSIGSRQGKGQFVAEIIAISSVLHRNLVKLYGCCFEGDHRLLVYEYLPNGS 788
Query: 155 LDKFIYKSGFPDAVCDCDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENF 214
LD+ + F D D +T ++I +G+ARGL YLH+ S RI+H D+K NILLD
Sbjct: 789 LDQAL----FGDKSLHLDWSTRYEICLGVARGLVYLHEEASVRIIHRDVKASNILLDSEL 844
Query: 215 CPKISDFGLAKICQMNDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEM 274
PK+SDFGLAK+ + +S GTIGY+APE R G ++ K+DVY++G++ LE+
Sbjct: 845 VPKVSDFGLAKLYDDKKTHIS-TRVAGTIGYLAPEYAMR--GHLTEKTDVYAFGVVALEL 901
Query: 275 IGGRKNYQTGGSCTSEMYFPDWIYKDLEQGNDL-LNSLTISEEENDMVKKITMVSLWCIQ 333
+ GRKN + Y +W + E+ D+ L +SE + VK++ ++L C Q
Sbjct: 902 VSGRKNSDENLE-EGKKYLLEWAWNLHEKNRDVELIDDELSEYNMEEVKRMIGIALLCTQ 960
Query: 334 TNPLDRPPMNKVIEMLQG 351
++ RPPM++V+ ML G
Sbjct: 961 SSYALRPPMSRVVAMLSG 978
>At1g56140 receptor-like protein kinase, putative
Length = 2083
Score = 215 bits (547), Expect = 5e-56
Identities = 126/329 (38%), Positives = 191/329 (57%), Gaps = 17/329 (5%)
Query: 38 IFRKRRKHVDNNVEVFMQSYNLSIARRYSYAEVKRITNSF--RDKLGHGGYGVVYKASLT 95
I RKRRK ++ E+ ++Y+E+K T F +KLG GG+G VYK L
Sbjct: 1707 IIRKRRKRYTDDEEILSMDVK---PYTFTYSELKSATQDFDPSNKLGEGGFGPVYKGKLN 1763
Query: 96 DGRQVAVKVINESKGNGE-EFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGS 154
DGR+VAVK+++ G+ +F+ E+ +IS H N+V L G CYE R L+YE++P GS
Sbjct: 1764 DGREVAVKLLSVGSRQGKGQFVAEIVAISAVQHRNLVKLYGCCYEGEHRLLVYEYLPNGS 1823
Query: 155 LDKFIYKSGFPDAVCDCDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENF 214
LD+ + F + D +T ++I +G+ARGL YLH+ RI+H D+K NILLD
Sbjct: 1824 LDQAL----FGEKTLHLDWSTRYEICLGVARGLVYLHEEARLRIVHRDVKASNILLDSKL 1879
Query: 215 CPKISDFGLAKICQMNDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEM 274
PK+SDFGLAK+ + +S GTIGY+APE R G ++ K+DVY++G++ LE+
Sbjct: 1880 VPKVSDFGLAKLYDDKKTHIS-TRVAGTIGYLAPEYAMR--GHLTEKTDVYAFGVVALEL 1936
Query: 275 IGGRKNYQTGGSCTSEMYFPDWIYKDLEQGNDL-LNSLTISEEENDMVKKITMVSLWCIQ 333
+ GR N + Y +W + E+G ++ L ++E + K++ ++L C Q
Sbjct: 1937 VSGRPNSDENLE-DEKRYLLEWAWNLHEKGREVELIDHQLTEFNMEEGKRMIGIALLCTQ 1995
Query: 334 TNPLDRPPMNKVIEMLQG--PLSSVTFPP 360
T+ RPPM++V+ ML G +S VT P
Sbjct: 1996 TSHALRPPMSRVVAMLSGDVEVSDVTSKP 2024
Score = 198 bits (504), Expect = 5e-51
Identities = 130/413 (31%), Positives = 222/413 (53%), Gaps = 38/413 (9%)
Query: 9 SVAGFGVTMFFIIFISCYFKKGIRRPQMTIFRKRRKHVDNNV--EVFMQSYNLSIARRYS 66
++ G G+ +I I + + RKR++ D V + ++ Y +S
Sbjct: 620 AIVGAGMLCILVIAILLFIR-----------RKRKRAADEEVLNSLHIRPYT------FS 662
Query: 67 YAEVKRITNSF--RDKLGHGGYGVVYKASLTDGRQVAVKVINESKGNGE-EFINEVASIS 123
Y+E++ T F +KLG GG+G V+K L DGR++AVK ++ + G+ +F+ E+A+IS
Sbjct: 663 YSELRTATQDFDPSNKLGEGGFGPVFKGKLNDGREIAVKQLSVASRQGKGQFVAEIATIS 722
Query: 124 RTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIAIGI 183
H N+V L G C E N+R L+YE++ SLD+ + F + + F+I +G+
Sbjct: 723 AVQHRNLVKLYGCCIEGNQRMLVYEYLSNKSLDQAL----FEEKSLQLGWSQRFEICLGV 778
Query: 184 ARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTRGTI 243
A+GL Y+H+ + RI+H D+K NILLD + PK+SDFGLAK+ + +S GTI
Sbjct: 779 AKGLAYMHEESNPRIVHRDVKASNILLDSDLVPKLSDFGLAKLYDDKKTHIS-TRVAGTI 837
Query: 244 GYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSCTSEMYFPDWIYKDLEQ 303
GY++PE G ++ K+DV+++G++ LE++ GR N + + Y +W + ++
Sbjct: 838 GYLSPEYV--MLGHLTEKTDVFAFGIVALEIVSGRPN-SSPELDDDKQYLLEWAWSLHQE 894
Query: 304 GNDL-LNSLTISEEENDMVKKITMVSLWCIQTNPLDRPPMNKVIEMLQGPLSSVTFPPKP 362
D+ + ++E + + VK++ V+ C QT+ RP M++V+ ML G + KP
Sbjct: 895 QRDMEVVDPDLTEFDKEEVKRVIGVAFLCTQTDHAIRPTMSRVVGMLTGDVEITEANAKP 954
Query: 363 VLFSPKRPPLQLSNMS---SSDW--QET--NSITTETELEEEPIEGYQSSKFE 408
S + +S MS SS W ET +S + L + P++ S+F+
Sbjct: 955 GYVSERTFENAMSFMSGSTSSSWILPETPKDSSKQQPPLLKSPVDNAAESRFK 1007
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.138 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,881,224
Number of Sequences: 26719
Number of extensions: 434331
Number of successful extensions: 4970
Number of sequences better than 10.0: 1014
Number of HSP's better than 10.0 without gapping: 834
Number of HSP's successfully gapped in prelim test: 180
Number of HSP's that attempted gapping in prelim test: 1302
Number of HSP's gapped (non-prelim): 1128
length of query: 459
length of database: 11,318,596
effective HSP length: 103
effective length of query: 356
effective length of database: 8,566,539
effective search space: 3049687884
effective search space used: 3049687884
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC147875.5


