

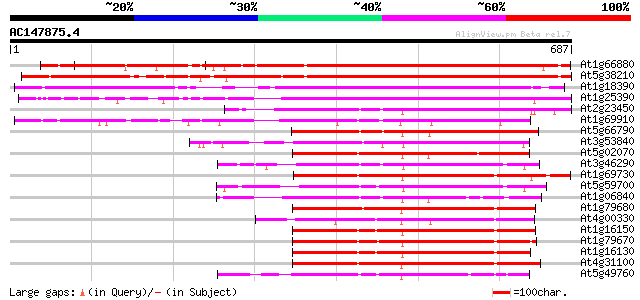
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147875.4 + phase: 0
(687 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g66880 wall-associated kinase like protein 538 e-153
At5g38210 protein kinase - like protein 533 e-151
At1g18390 wall-associated kinase, putative 468 e-132
At1g25390 putative protein 443 e-124
At2g23450 protein kinase like protein 288 5e-78
At1g69910 protein kinase like protein 278 7e-75
At5g66790 unknown protein 265 6e-71
At3g53840 protein kinase-like protein 254 9e-68
At5g02070 putative protein kinase 254 1e-67
At3g46290 receptor protein kinase -like 253 2e-67
At1g69730 putative protein kinase 253 2e-67
At5g59700 receptor-like protein kinase precursor - like 246 2e-65
At1g06840 receptor protein kinase, putative 244 9e-65
At1g79680 unknown protein 244 2e-64
At4g00330 unknown protein 241 1e-63
At1g16150 240 2e-63
At1g79670 putative protein 240 2e-63
At1g16130 unknown protein 238 1e-62
At4g31100 serine/threonine-specific protein kinase - like 234 1e-61
At5g49760 receptor protein kinase-like 234 2e-61
>At1g66880 wall-associated kinase like protein
Length = 848
Score = 538 bits (1386), Expect = e-153
Identities = 322/679 (47%), Positives = 412/679 (60%), Gaps = 45/679 (6%)
Query: 38 NTTFNCGHITKLSYPFTGGDRPSYCGPPQFHLN-CKNNVPELNISSVSYRVLQVNSVTHS 96
N TF+CG +L YPF R CG P F L+ C EL+ISSV +R+L ++
Sbjct: 184 NGTFSCGDQRELFYPFWTSGRED-CGHPDFKLDDCSGRFAELSISSVKFRILASVYGSNI 242
Query: 97 LTLARLDLWNETCTHHYVNSTFGGTSFSNGLGNSKITLFYGCQPTSLFTEKPHNL--FYC 154
+ L R + + C +N+ F + +T+FY C F ++ N F C
Sbjct: 243 IRLGRSEYIGDLCPQDPINAPFSESVLPFAPNTELLTIFYNCSRD--FPQQVTNFGDFAC 300
Query: 155 DSNGYKNNSYTLIGPFPLDPVLKF--------VQCDYGVGVPILEEQANRFAGNRSL--L 204
+ + SY + P+ + CD V +P N S L
Sbjct: 301 GDDSDDDRSYYVTRNLSFPPLSEINDLLYDFSQSCDRNVSIPASGSTLNILQSTPSNDNL 360
Query: 205 REVLMEGFNVNYNNPFENDCLECISSGGQQCGFDSDENEHICIC------GNGLCPSGNS 258
++ L GF + N DC CI S G CG+ + +C P+ N
Sbjct: 361 KKALEYGFELELNQ----DCRTCIDSKGA-CGYSQTSSRFVCYSIEEPQTPTPPNPTRNK 415
Query: 259 STN------GGLIGGVVGGVAALCLLGFVACFVVRRRRKNAKKPISNDLYMPPSSTTSGT 312
T+ G+ V G+A L L G C +RRRRK ++ +P +S +S
Sbjct: 416 DTSLSIGAKAGIAVASVSGLAILLLAGLFLC--IRRRRKTQDAQYTSKS-LPITSYSSRD 472
Query: 313 NTGTLTSTTNSSQSIPSYPSSKTSTMPKSFYFGVQVFTYEELEEATNNFHTSKELGEGGF 372
+ TSTT SS S S S ++ +S Y GVQVF+YEELEEAT NF S+ELG+GGF
Sbjct: 473 TSRNPTSTTISSSSNHSLLPSISNLANRSDYCGVQVFSYEELEEATENF--SRELGDGGF 530
Query: 373 GTVYKGDLKDGRVVAVKRHYESNFKRVAQFMNEVEILARLRHKNLVTLYGCTSKHSRELL 432
GTVY G LKDGR VAVKR YE + KRV QF NE+EIL L+H NLV LYGCTS+HSRELL
Sbjct: 531 GTVYYGVLKDGRAVAVKRLYERSLKRVEQFKNEIEILKSLKHPNLVILYGCTSRHSRELL 590
Query: 433 LVYEYISNGTVADHLHGDRSSSCLLPWSVRLDIALETAEALAYLHASDVMHRDVKSNNIL 492
LVYEYISNGT+A+HLHG+R+ + L WS RL+IA+ETA AL++LH ++HRD+K+ NIL
Sbjct: 591 LVYEYISNGTLAEHLHGNRAEARPLCWSTRLNIAIETASALSFLHIKGIIHRDIKTTNIL 650
Query: 493 LDEKFHVKVADFGLSRLFPNDVTHVSTAPQGTPGYVDPEYYQCYQLTDKSDVYSFGVVLV 552
LD+ + VKVADFGLSRLFP D TH+STAPQGTPGYVDPEYYQCYQL +KSDVYSFGVVL
Sbjct: 651 LDDNYQVKVADFGLSRLFPMDQTHISTAPQGTPGYVDPEYYQCYQLNEKSDVYSFGVVLT 710
Query: 553 ELISSLQAVDITRHRNDVNLANMAVNKIQSQELYDLVDPNLGYEKDNSVKRMTTAVAELA 612
ELISS +AVDITRHR+D+NLANMAV+KIQ+ L++LVD +LGY+ D V+R AVAELA
Sbjct: 711 ELISSKEAVDITRHRHDINLANMAVSKIQNNALHELVDSSLGYDNDPEVRRKMMAVAELA 770
Query: 613 FRCLQQQRDLRPSMDEIVEVLRAIKSDEPETQESKVLDVV----VRTDELVLLKKG-PYP 667
FRCLQQ+RD+RP+MDEIVE+LR IK DE + K DVV D++ LL+ P P
Sbjct: 771 FRCLQQERDVRPAMDEIVEILRGIKDDEKKRVLVKSPDVVDIECGGGDDVGLLRNSVPPP 830
Query: 668 TSPDSVAEKWVSGSSTSTS 686
SP++ +KW S S T+ S
Sbjct: 831 ISPET--DKWTSSSDTAAS 847
Score = 46.6 bits (109), Expect = 5e-05
Identities = 46/166 (27%), Positives = 63/166 (37%), Gaps = 26/166 (15%)
Query: 80 ISSVSYRVLQVNSVTHSLTLARLDLWNETCTHHYVNSTFGGTSFSNGLGNSKITLFYGCQ 139
IS Y VL ++ +++L LAR +L C Y +T F +T+FY C
Sbjct: 6 ISDHFYNVLHIDQTSNTLRLARAELEGSFCNATYTATTLPSKIFEISSTYKSLTVFYLCD 65
Query: 140 P----TSLFTEKPHNLFYCDSNGYKNNSYTLIGPFPLDPVLKFVQCDYGVGVP--ILEEQ 193
P S +T L N +NS Q + + VP + E+
Sbjct: 66 PKVSYRSSYTCPGRGLVSVSQNSDYHNS---------------CQDSFTINVPKSFVPEE 110
Query: 194 ANRFAGNRSLLREVLMEGFNVNYNNPFENDCLECISSGGQQCGFDS 239
N L L EGF V E C +C SSGG CGF +
Sbjct: 111 KELDVTN---LESALREGFEVKVKVD-EKTCQKCTSSGG-TCGFQN 151
>At5g38210 protein kinase - like protein
Length = 686
Score = 533 bits (1374), Expect = e-151
Identities = 326/694 (46%), Positives = 429/694 (60%), Gaps = 46/694 (6%)
Query: 15 IFFIFYLRHTTSLPSHASLSSCNNTTFNCGHITKLSYPFTGGDRPSYCGPPQFHLNCKN- 73
+ +F+L + LP S +T F CG +T +PF G RP CG P L+C+
Sbjct: 16 LVLLFFLSYIHFLPCAQSQREPCDTLFRCGDLTA-GFPFWGVARPQPCGHPSLGLHCQKQ 74
Query: 74 -NVPELNISSVSYRVLQVNSVTHSLTLARLDLWNETCTHHYVNSTFGGTSFSNGLGNSKI 132
N L ISS+ YRVL+VN+ T +L L R D C+ + +T F +
Sbjct: 75 TNSTSLIISSLMYRVLEVNTTTSTLKLVRQDFSGPFCSASFSGATLTPELFELLPDYKTL 134
Query: 133 TLFYGCQPTSLFTEKPHNLFYCDSNGYKNNSYTLIGPFPLDPVLKFVQCD--YGVGVPIL 190
+ +Y C P+ + K F C + G +G D L C + + VPI
Sbjct: 135 SAYYLCNPSLHYPAK----FICPNKG--------VGSIHQDD-LYHNHCGGIFNITVPI- 180
Query: 191 EEQANRFAGNRSLLREVLMEGFNVNYNNPFENDCLECISSGG--------QQCGFDSDEN 242
A N + L VL +GF V + E C EC ++GG C + +
Sbjct: 181 GYAPEEGALNVTNLESVLKKGFEVKLSID-ERPCQECKTNGGICAYHVATPVCCKTNSSS 239
Query: 243 EHICICGNGLCPSGNSSTNGGL-------IGGVVGGVAALCLLGFVACFVVRRRRKNAKK 295
E C + PSG SS + GL IG G + A + G + C +RRR+K A +
Sbjct: 240 EVNC---TPMMPSG-SSAHAGLSKKGKIGIGFASGFLGATLIGGCLLCIFIRRRKKLATQ 295
Query: 296 PISNDLYMPPSSTTSGTNTGTLTSTTNSSQSIPSYPSSKTSTMPKSFYFGVQVFTYEELE 355
+ L +++ T + T TSTT S + PS ++ S Y G+QVF+YEELE
Sbjct: 296 YTNKGLSTTTPYSSNYTMSNTPTSTTISGSNHSLVPSI-SNLGNGSVYSGIQVFSYEELE 354
Query: 356 EATNNFHTSKELGEGGFGTVYKGDLKDGRVVAVKRHYESNFKRVAQFMNEVEILARLRHK 415
EAT NF SKELG+GGFGTVY G LKDGR VAVKR +E + KRV QF NE++IL L+H
Sbjct: 355 EATENF--SKELGDGGFGTVYYGTLKDGRAVAVKRLFERSLKRVEQFKNEIDILKSLKHP 412
Query: 416 NLVTLYGCTSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDIALETAEALAY 475
NLV LYGCT++HSRELLLVYEYISNGT+A+HLHG+++ S + W RL IA+ETA AL+Y
Sbjct: 413 NLVILYGCTTRHSRELLLVYEYISNGTLAEHLHGNQAQSRPICWPARLQIAIETASALSY 472
Query: 476 LHASDVMHRDVKSNNILLDEKFHVKVADFGLSRLFPNDVTHVSTAPQGTPGYVDPEYYQC 535
LHAS ++HRDVK+ NILLD + VKVADFGLSRLFP D TH+STAPQGTPGYVDPEYYQC
Sbjct: 473 LHASGIIHRDVKTTNILLDSNYQVKVADFGLSRLFPMDQTHISTAPQGTPGYVDPEYYQC 532
Query: 536 YQLTDKSDVYSFGVVLVELISSLQAVDITRHRNDVNLANMAVNKIQSQELYDLVDPNLGY 595
Y+L +KSDVYSFGVVL ELISS +AVDITRHR+D+NLANMA++KIQ+ +++L D +LG+
Sbjct: 533 YRLNEKSDVYSFGVVLSELISSKEAVDITRHRHDINLANMAISKIQNDAVHELADLSLGF 592
Query: 596 EKDNSVKRMTTAVAELAFRCLQQQRDLRPSMDEIVEVLRAIKSDEPETQESKVLDVVVR- 654
+D SVK+M ++VAELAFRCLQQ+RD+RPSMDEIVEVLR I+ D + V+++ V
Sbjct: 593 ARDPSVKKMMSSVAELAFRCLQQERDVRPSMDEIVEVLRVIQKDGISDSKDVVVEIDVNG 652
Query: 655 TDELVLLKKG-PYPTSPDSVAEKWVSGSSTSTSS 687
D++ LLK G P P SP++ +K + SS +T+S
Sbjct: 653 GDDVGLLKHGVPPPLSPET--DKTTASSSNTTAS 684
>At1g18390 wall-associated kinase, putative
Length = 605
Score = 468 bits (1205), Expect = e-132
Identities = 294/682 (43%), Positives = 379/682 (55%), Gaps = 90/682 (13%)
Query: 7 SSPIITFI-IFFIFYLRHTTSLPSHASLSSCNNTTFNCGHITKLSYPF-TGGDRPSYCGP 64
S+P + + IFF F + T +L +C + CG ++SYPF G + S+CG
Sbjct: 4 STPSLLYTSIFFYFTIIATQTLSLDPKFKACEPKS--CGKGPQISYPFYLSGKQESFCGY 61
Query: 65 PQFHLNC--KNNVPELNISSVSYRVLQVNSVTHSLTLARLDLWNETCTHHYVNSTFGGTS 122
P F L C + +P L IS Y + ++ +T S + ++ C N T T
Sbjct: 62 PSFELTCDDEEKLPVLGISGEEYVIKNISYLTQSFQVVNSKASHDPCPRPLNNLTLHRTP 121
Query: 123 FSNGLGNSKITLFYGCQPTSLFTEKPHNLFYCDSNGYKNNSYTLIGPFPL--DPVLKFVQ 180
F + T+ Y C L + + L C N S+ + L + + +
Sbjct: 122 FFVNPSHINFTILYNCSDHLLEDFRTYPLT-CARNTSLLRSFGVFDRKKLGKEKQIASMS 180
Query: 181 CDYGVGVPILEEQANRFAGNRSLLREVLMEGFNVNYNNPFENDCLECISSGGQQCGFDSD 240
C V VP+L + G + E+L GF +N+ N C CI
Sbjct: 181 CQKLVDVPVLASNESDVMGMTYV--EILKRGFVLNWT---ANSCFRCI------------ 223
Query: 241 ENEHICICGNGLCPSGNSSTNGGLIGGVVGGVAALCLLGFVACFVVRRRRKNAKKPISND 300
T+GG G LC
Sbjct: 224 -------------------TSGGRCGTDQQEFVCLC------------------------ 240
Query: 301 LYMPPSSTTSGTNTGTLTSTTNSSQSIPSYPSSKTSTMPKS--FYFGVQVFTYEELEEAT 358
P T T L ++I S PS+K+ + K+ GV +F+YEELEEAT
Sbjct: 241 ---PDGPKLHDTCTNALLP-----RNISSDPSAKSFDIEKAEELLVGVHIFSYEELEEAT 292
Query: 359 NNFHTSKELGEGGFGTVYKGDLKDGRVVAVKRHYESNFKRVAQFMNEVEILARLRHKNLV 418
NNF SKELG+GGFGTVY G LKDGR VAVKR Y++NFKR QF NEVEIL LRH NLV
Sbjct: 293 NNFDPSKELGDGGFGTVYYGKLKDGRSVAVKRLYDNNFKRAEQFRNEVEILTGLRHPNLV 352
Query: 419 TLYGCTSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDIALETAEALAYLHA 478
L+GC+SK SR+LLLVYEY++NGT+ADHLHG +++ LPWS+RL IA+ETA AL YLHA
Sbjct: 353 ALFGCSSKQSRDLLLVYEYVANGTLADHLHGPQANPSSLPWSIRLKIAVETASALKYLHA 412
Query: 479 SDVMHRDVKSNNILLDEKFHVKVADFGLSRLFPNDVTHVSTAPQGTPGYVDPEYYQCYQL 538
S ++HRDVKSNNILLD+ F+VKVADFGLSRLFP D THVSTAPQGTPGYVDP+Y+ CYQL
Sbjct: 413 SKIIHRDVKSNNILLDQNFNVKVADFGLSRLFPMDKTHVSTAPQGTPGYVDPDYHLCYQL 472
Query: 539 TDKSDVYSFGVVLVELISSLQAVDITRHRNDVNLANMAVNKIQSQELYDLVDPNLGYEKD 598
++KSDVYSF VVL+ELISSL AVDITR R ++NL+NMAV KIQ+ EL D+VDP+LG++ D
Sbjct: 473 SNKSDVYSFAVVLMELISSLPAVDITRPRQEINLSNMAVVKIQNHELRDMVDPSLGFDTD 532
Query: 599 NSVKRMTTAVAELAFRCLQQQRDLRPSMDEIVEVLRAIKSDEPETQESKVLDVVVRTDEL 658
V++ AVAELAF+CLQ +DLRP M + + L I+++ E V+DV
Sbjct: 533 TRVRQTVIAVAELAFQCLQSDKDLRPCMSHVQDTLTRIQNN-GFGSEMDVVDV------- 584
Query: 659 VLLKKGP-YPTSPDSVAEKWVS 679
K GP SPDSV KW S
Sbjct: 585 --NKSGPLVAQSPDSVIVKWDS 604
>At1g25390 putative protein
Length = 629
Score = 443 bits (1140), Expect = e-124
Identities = 278/694 (40%), Positives = 398/694 (57%), Gaps = 89/694 (12%)
Query: 11 ITFIIFFIFYLRHTTSLPSHASLSSCNNTTFNCGHITKLSYPFTGGDRPSYCGPPQFHLN 70
++ ++FF +L + + + C + F CG +PF D PS CG F LN
Sbjct: 4 VSVLLFFFLFLLAAEARSTKRT--GCKD--FTCGE-HDFKFPFFRTDMPSRCG--LFKLN 56
Query: 71 CKNNVPELNISSVSYRVLQVNSVTHSLTLARLD-LWNETCTHHYVNSTFGGTSFSN-GLG 128
C N+PE+ + + V SV+ + T+ +D N++ T T G + S+ L
Sbjct: 57 CSANIPEIQLEKDG-KWYTVKSVSQANTITIIDPRLNQSLT------TGGCSDLSSFSLP 109
Query: 129 NS---KITLFYGCQPTSLFTEKPHNLFYCDSNGYKNNSYTLIGPFPLDPVLKFVQCDYGV 185
+S K+ Y C +S + + Y + G ++ Y +G D+ V
Sbjct: 110 DSPWLKLNTLYKCNNSS----RKNGFSYANCRGEGSSLYYNLGD------------DHDV 153
Query: 186 G----VPILEEQANRFAGNRSLLREVLMEGFNVNYNNPFENDCLECISSGGQQCGFDSDE 241
+ E GN S + F+++ P +C C ++GG+ +
Sbjct: 154 SGCSPIKTPESWVTPKNGNLSDVNAT----FSLHIELP--GNCFRCHNNGGE---CTKVK 204
Query: 242 NEHICICGNGLCPSGNSSTNGGLIGGVVGGVAALCLLGFVACFVVRRRRKNAKKPISNDL 301
N + C+ N + ++ GL G+ G V + +L + + R R+ +S D
Sbjct: 205 NNYRCVGANTEPNNYHAEMRLGL--GIGGSVILIIILVALFAVIHRNYRRKDGSELSRD- 261
Query: 302 YMPPSSTTSGTNTGTLTSTTNSSQSIPSYPSSKTSTMPKSFYFGVQVFTYEELEEATNNF 361
+SK+ +F + +F+Y+EL+ AT+NF
Sbjct: 262 ------------------------------NSKSDVEFSQVFFKIPIFSYKELQAATDNF 291
Query: 362 HTSKELGEGGFGTVYKGDLKDGRVVAVKRHYESNFKRVAQFMNEVEILARLRHKNLVTLY 421
+ LG+GGFGTVY G ++DGR VAVKR YE N++R+ QFMNE+EIL RL HKNLV+LY
Sbjct: 292 SKDRLLGDGGFGTVYYGKVRDGREVAVKRLYEHNYRRLEQFMNEIEILTRLHHKNLVSLY 351
Query: 422 GCTSKHSRELLLVYEYISNGTVADHLHGDRSS-SCLLPWSVRLDIALETAEALAYLHASD 480
GCTS+ SRELLLVYE+I NGTVADHL+G+ + L WS+RL IA+ETA ALAYLHASD
Sbjct: 352 GCTSRRSRELLLVYEFIPNGTVADHLYGENTPHQGFLTWSMRLSIAIETASALAYLHASD 411
Query: 481 VMHRDVKSNNILLDEKFHVKVADFGLSRLFPNDVTHVSTAPQGTPGYVDPEYYQCYQLTD 540
++HRDVK+ NILLD F VKVADFGLSRL P+DVTHVSTAPQGTPGYVDPEY++CY LTD
Sbjct: 412 IIHRDVKTTNILLDRNFGVKVADFGLSRLLPSDVTHVSTAPQGTPGYVDPEYHRCYHLTD 471
Query: 541 KSDVYSFGVVLVELISSLQAVDITRHRNDVNLANMAVNKIQSQELYDLVDPNLGYEKDNS 600
KSDVYSFGVVLVELISS AVDI+R ++++NL+++A+NKIQ+ ++L+D NLGY +
Sbjct: 472 KSDVYSFGVVLVELISSKPAVDISRCKSEINLSSLAINKIQNHATHELIDQNLGYATNEG 531
Query: 601 VKRMTTAVAELAFRCLQQQRDLRPSMDEIVEVLRAIKSDEP-----ETQESKVLDVVVRT 655
V++MTT VAELAF+CLQQ +RP+M+++V L+ I+++E + +E ++
Sbjct: 532 VRKMTTMVAELAFQCLQQDNTMRPTMEQVVHELKGIQNEEQKCPTYDYREETIIPHPSPP 591
Query: 656 D--ELVLLKKGPYPTSPDSVAEKWVSGSSTSTSS 687
D E LLK +P SP SV ++W S S+T +S
Sbjct: 592 DWGEAALLKNMKFPRSPVSVTDQWTSKSTTPNTS 625
>At2g23450 protein kinase like protein
Length = 708
Score = 288 bits (738), Expect = 5e-78
Identities = 175/453 (38%), Positives = 255/453 (55%), Gaps = 67/453 (14%)
Query: 264 LIGGVVGGVAALCLLGFVACFVVRRRRKNAKKPISNDLYMPPSSTTSGTNTGTLTSTTNS 323
++GG VGG L L F F +RRR
Sbjct: 286 IVGGTVGGAFLLAALAFF--FFCKRRR--------------------------------- 310
Query: 324 SQSIPSYPSSKTSTMPKSFYFGVQVFTYEELEEATNNFHTSKELGEGGFGTVYKGDLKDG 383
S + S+ S+K + V F Y+E+E+AT+ F ++LG G +GTVY+G L++
Sbjct: 311 STPLRSHLSAKRLLSEAAGNSSVAFFPYKEIEKATDGFSEKQKLGIGAYGTVYRGKLQND 370
Query: 384 RVVAVKRHYESNFKRVAQFMNEVEILARLRHKNLVTLYGCTSKHSRELLLVYEYISNGTV 443
VA+KR + + + Q MNE+++L+ + H NLV L GC + + +LVYEY+ NGT+
Sbjct: 371 EWVAIKRLRHRDSESLDQVMNEIKLLSSVSHPNLVRLLGCCIEQG-DPVLVYEYMPNGTL 429
Query: 444 ADHLHGDRSSSCLLPWSVRLDIALETAEALAYLHAS---DVMHRDVKSNNILLDEKFHVK 500
++HL DR S LPW++RL +A +TA+A+AYLH+S + HRD+KS NILLD F+ K
Sbjct: 430 SEHLQRDRGSG--LPWTLRLTVATQTAKAIAYLHSSMNPPIYHRDIKSTNILLDYDFNSK 487
Query: 501 VADFGLSRLFPNDVTHVSTAPQGTPGYVDPEYYQCYQLTDKSDVYSFGVVLVELISSLQA 560
VADFGLSRL + +H+STAPQGTPGY+DP+Y+QC+ L+DKSDVYSFGVVL E+I+ L+
Sbjct: 488 VADFGLSRLGMTESSHISTAPQGTPGYLDPQYHQCFHLSDKSDVYSFGVVLAEIITGLKV 547
Query: 561 VDITRHRNDVNLANMAVNKIQSQELYDLVDPNLGYEKDNSVKRMTTAVAELAFRCLQQQR 620
VD TR ++NLA +AV+KI S + +++DP L + D VAELAFRCL
Sbjct: 548 VDFTRPHTEINLAALAVDKIGSGCIDEIIDPILDLDLDAWTLSSIHTVAELAFRCLAFHS 607
Query: 621 DLRPSMDEIVEVLRAIK---------SDEP--------ETQESKVLDVVVRTDELVLLKK 663
D+RP+M E+ + L I+ D P E V + + +V+ +K
Sbjct: 608 DMRPTMTEVADELEQIRLSGWIPSMSLDSPAGSLRSSDRGSERSVKQSSIGSRRVVIPQK 667
Query: 664 GP---------YPTSPDSVAEKWVSGSSTSTSS 687
P +SP SV + W+S S+ +++
Sbjct: 668 QPDCLASVEEISDSSPISVQDPWLSAQSSPSTN 700
>At1g69910 protein kinase like protein
Length = 636
Score = 278 bits (711), Expect = 7e-75
Identities = 224/682 (32%), Positives = 333/682 (47%), Gaps = 107/682 (15%)
Query: 6 LSSPIITFIIFFIFYLRHTTSLPSHASLSSCNNTTFNCGHITKLS-YPFTGGDRPSYCGP 64
+S P IF L ++ PS AS +SC+++ F+C YPF+ CG
Sbjct: 1 MSQPPWRCFSLLIFVLTIFSTKPSSAS-TSCSSS-FHCPPFNSSPPYPFSASPG---CGH 55
Query: 65 PQFHLNCKNNVPELNISSVSYRVLQVNSVTHSLTLARLDLWNET---CTHHYVNST---- 117
P F + C ++ + I ++++ +L +S++ SLTL+ + N C+ +S+
Sbjct: 56 PNFQIQCSSSRATITIKNLTFSILHYSSISSSLTLSPITNTNRNTNNCSSLRFSSSPNRF 115
Query: 118 --FGGTSFS-NGLGNSKITLFYGCQPTSLFTEKPHNLFYC--DSNGYKNNSYTLIGPFPL 172
G+ F + S+++L C P +L N C D KN L G
Sbjct: 116 IDLTGSPFRVSDSSCSRLSLLRPCSPFTL-----PNCSRCPWDCKLLKNPGRILHGCEST 170
Query: 173 DPVLKFVQCDYGVGVPILEEQANRFAGNRSLLREVLMEGFNVNYN---NPFENDCLECIS 229
L C G + L++ RF GF V ++ +P+ C +C
Sbjct: 171 HGSLSEQGCQ-GDLLGFLQDFFTRF-------------GFEVEWDESQDPYFIKCRDCQI 216
Query: 230 SGGQQCGFDSDENEHICICGNGLCPS------GNSSTNGGLIGGVVGGVAALCLLGFVAC 283
G CGF+S IC + N N + ++ + L L+ VA
Sbjct: 217 KNGV-CGFNSTHPNQDFICFHKSRSELVTQRDNNKRVNHIAVLSLIFALTCLLLVFSVAV 275
Query: 284 FVVRRRRKNAKKPISNDLYMPPSSTTSGTNTGTLTSTTNSSQSIPSYPSSKTSTMPKSFY 343
+ R RR + I+ + P++ +S
Sbjct: 276 AIFRSRRASFLSSINEE-----------------------------DPAALFLRHHRSAA 306
Query: 344 FGVQVFTYEELEEATNNFHTSKELGEGGFGTVYKGDLKDGRVVAVK-RHYESNFKRVA-- 400
VFT+EELE ATN F +++G+GGFG+VY G L DG+++AVK H+ A
Sbjct: 307 LLPPVFTFEELESATNKFDPKRKIGDGGFGSVYLGQLSDGQLLAVKFLHHHHGATAAATE 366
Query: 401 --------QFMNEVEILARLRHKNLVTLYGCTSKHSRELLLVYEYISNGTVADHLHGDRS 452
F NE+ IL+ + H NLV L+G S R LLLV++Y++NGT+ADHLHG
Sbjct: 367 HCKAFSMKSFCNEILILSSINHPNLVKLHGYCSD-PRGLLLVHDYVTNGTLADHLHGRGP 425
Query: 453 SSCLLPWSVRLDIALETAEALAYLH---ASDVMHRDVKSNNILLDEKFHVKVADFGLSRL 509
+ W VRLDIAL+TA A+ YLH V+HRD+ S+NI +++ +KV DFGLSRL
Sbjct: 426 K---MTWRVRLDIALQTALAMEYLHFDIVPPVVHRDITSSNIFVEKDMKIKVGDFGLSRL 482
Query: 510 FPNDVT---------HVSTAPQGTPGYVDPEYYQCYQLTDKSDVYSFGVVLVELISSLQA 560
T +V T PQGTPGY+DP+Y++ ++LT+KSDVYS+GVVL+ELI+ ++A
Sbjct: 483 LVFSETTVNSATSSDYVCTGPQGTPGYLDPDYHRSFRLTEKSDVYSYGVVLMELITGMKA 542
Query: 561 VDITRHRNDVNLANMAVNKIQSQELYDLVDPNLGYEKDN----SVKRMTTAVAELAFRCL 616
VD R + D+ LA++ V+KIQ L ++DP L + D+ S AVAELAFRC+
Sbjct: 543 VDQRREKRDMALADLVVSKIQMGLLDQVIDPLLALDGDDVAAVSDGFGVAAVAELAFRCV 602
Query: 617 QQQRDLRPSMDEIVEVLRAIKS 638
+D RP EIV+ LR I+S
Sbjct: 603 ATDKDDRPDAKEIVQELRRIRS 624
>At5g66790 unknown protein
Length = 622
Score = 265 bits (677), Expect = 6e-71
Identities = 142/308 (46%), Positives = 204/308 (66%), Gaps = 8/308 (2%)
Query: 346 VQVFTYEELEEATNNFHTSKELGEGGFGTVYKGDLKDGRVVAVKRHYESNFKRVAQFMNE 405
V +TY+E+E+AT++F LG G +GTVY G+ + VA+KR + + Q +NE
Sbjct: 299 VPFYTYKEIEKATDSFSDKNMLGTGAYGTVYAGEFPNSSCVAIKRLKHKDTTSIDQVVNE 358
Query: 406 VEILARLRHKNLVTLYGCTSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDI 465
+++L+ + H NLV L GC E LVYE++ NGT+ HL +R L W +RL I
Sbjct: 359 IKLLSSVSHPNLVRLLGCCFADG-EPFLVYEFMPNGTLYQHLQHERGQPPL-SWQLRLAI 416
Query: 466 ALETAEALAYLHAS---DVMHRDVKSNNILLDEKFHVKVADFGLSRLFPN---DVTHVST 519
A +TA A+A+LH+S + HRD+KS+NILLD +F+ K++DFGLSRL + + +H+ST
Sbjct: 417 ACQTANAIAHLHSSVNPPIYHRDIKSSNILLDHEFNSKISDFGLSRLGMSTDFEASHIST 476
Query: 520 APQGTPGYVDPEYYQCYQLTDKSDVYSFGVVLVELISSLQAVDITRHRNDVNLANMAVNK 579
APQGTPGY+DP+Y+Q +QL+DKSDVYSFGVVLVE+IS + +D TR ++VNLA++AV++
Sbjct: 477 APQGTPGYLDPQYHQDFQLSDKSDVYSFGVVLVEIISGFKVIDFTRPYSEVNLASLAVDR 536
Query: 580 IQSQELYDLVDPNLGYEKDNSVKRMTTAVAELAFRCLQQQRDLRPSMDEIVEVLRAIKSD 639
I + D++DP L E + + +AELAFRCL R++RP+M EI E L IK
Sbjct: 537 IGRGRVVDIIDPCLNKEINPKMFASIHNLAELAFRCLSFHRNMRPTMVEITEDLHRIKLM 596
Query: 640 EPETQESK 647
T+ K
Sbjct: 597 HYGTESGK 604
>At3g53840 protein kinase-like protein
Length = 640
Score = 254 bits (650), Expect = 9e-68
Identities = 168/443 (37%), Positives = 236/443 (52%), Gaps = 57/443 (12%)
Query: 221 ENDCLECISS---------GGQQC----GFDSDENEHICICGNGLCPSGNSS---TNGGL 264
+ DC + ++S G ++C GF D +C G C S +N L
Sbjct: 228 QGDCRDLLNSVCSNDSTNLGQKRCFCKKGFQWDSVNAVCE-GPVRCSKRKSCKRWSNLPL 286
Query: 265 IGGVVGGVAALCLLGFVACFVVRRRRKNAKKPISNDLYMPPSSTTSGTNTGTLTSTTNSS 324
+GG+ GGV A+ + GF+ +V ++ + + S
Sbjct: 287 LGGLAGGVGAILIAGFITKTIVSKQNRRI------------------AGNQSWASVRKLH 328
Query: 325 QSIPSYPSSKTSTMPKSFYFGVQVFTYEELEEATNNFHTSKELGEGGFGTVYKGDLKDGR 384
+++ S S+ ++FT +E+ +AT+NF S LG GGFG V+KG+L DG
Sbjct: 329 RNLLSINSTGLD----------RIFTGKEIVKATDNFAKSNLLGFGGFGEVFKGNLDDGT 378
Query: 385 VVAVKRHYESNFKRVAQFMNEVEILARLRHKNLVTLYGCTSKHSRELLLVYEYISNGTVA 444
VAVKR N K + Q +NEV+IL ++ HKNLV L GC + +L VYE++ NGT+
Sbjct: 379 TVAVKRAKLGNEKSIYQIVNEVQILCQVSHKNLVKLLGCCIELEMPVL-VYEFVPNGTLF 437
Query: 445 DHLHGDRSSSC----LLPWSVRLDIALETAEALAYLHASD---VMHRDVKSNNILLDEKF 497
+H++G LP RL IA +TA+ L YLH+S + HRDVKS+NILLDE
Sbjct: 438 EHIYGGGGGGGGLYDHLPLRRRLMIAHQTAQGLDYLHSSSSPPIYHRDVKSSNILLDENL 497
Query: 498 HVKVADFGLSRLFPNDVTHVSTAPQGTPGYVDPEYYQCYQLTDKSDVYSFGVVLVELISS 557
VKVADFGLSRL +DV+HV+T QGT GY+DPEYY +QLTDKSDVYSFGVVL EL++
Sbjct: 498 DVKVADFGLSRLGVSDVSHVTTCAQGTLGYLDPEYYLNFQLTDKSDVYSFGVVLFELLTC 557
Query: 558 LQAVDITRHRNDVNLANMAVNKIQSQELYDLVDPNLGYEKDNSVKRMTTAVAELAFRCLQ 617
+A+D R DVNL ++ L D++DP +G A+ LA C++
Sbjct: 558 KKAIDFNREEEDVNLVVFVRKALKEGRLMDVIDPVIGIGATEKEIESMKALGVLAELCVK 617
Query: 618 QQRDLRPSMD----EIVEVLRAI 636
+ R RP+M EI +L I
Sbjct: 618 ETRQCRPTMQVAAKEIENILHGI 640
>At5g02070 putative protein kinase
Length = 657
Score = 254 bits (648), Expect = 1e-67
Identities = 143/301 (47%), Positives = 194/301 (63%), Gaps = 14/301 (4%)
Query: 347 QVFTYEELEEATNNFHTSKELGEGGFGTVYKGDLKDGRVVAVKRHYESNFKRVAQFMNEV 406
++FT E+ +ATNNF +G GGFG V+K L+DG + A+KR +N K Q +NEV
Sbjct: 349 RIFTGREITKATNNFSKDNLIGTGGFGEVFKAVLEDGTITAIKRAKLNNTKGTDQILNEV 408
Query: 407 EILARLRHKNLVTLYGCTSKHSREL-LLVYEYISNGTVADHLHGDRSSSCL-LPWSVRLD 464
IL ++ H++LV L GC EL LL+YE+I NGT+ +HLHG + L W RL
Sbjct: 409 RILCQVNHRSLVRLLGCCV--DLELPLLIYEFIPNGTLFEHLHGSSDRTWKPLTWRRRLQ 466
Query: 465 IALETAEALAYLHAS---DVMHRDVKSNNILLDEKFHVKVADFGLSRLFP-----NDVTH 516
IA +TAE LAYLH++ + HRDVKS+NILLDEK + KV+DFGLSRL N+ +H
Sbjct: 467 IAYQTAEGLAYLHSAAQPPIYHRDVKSSNILLDEKLNAKVSDFGLSRLVDLTETANNESH 526
Query: 517 VSTAPQGTPGYVDPEYYQCYQLTDKSDVYSFGVVLVELISSLQAVDITRHRNDVNLANMA 576
+ T QGT GY+DPEYY+ +QLTDKSDVYSFGVVL+E+++S +A+D TR DVNL M
Sbjct: 527 IFTGAQGTLGYLDPEYYRNFQLTDKSDVYSFGVVLLEMVTSKKAIDFTREEEDVNLV-MY 585
Query: 577 VNKIQSQE-LYDLVDPNLGYEKDNSVKRMTTAVAELAFRCLQQQRDLRPSMDEIVEVLRA 635
+NK+ QE L + +DP L + + + LA CL ++R RPSM E+ + +
Sbjct: 586 INKMMDQERLTECIDPLLKKTANKIDMQTIQQLGNLASACLNERRQNRPSMKEVADEIEY 645
Query: 636 I 636
I
Sbjct: 646 I 646
>At3g46290 receptor protein kinase -like
Length = 830
Score = 253 bits (647), Expect = 2e-67
Identities = 168/414 (40%), Positives = 243/414 (58%), Gaps = 51/414 (12%)
Query: 255 SGNSSTNGGLI-GGVVGGVAALCLLGFVACFVVRRRRKNAKKPISNDLYMPPS--STTSG 311
S +S +N GLI G +G + A+ LG +CFV+ ++RK + S +MP S T+ G
Sbjct: 397 SSSSKSNLGLIVGSAIGSLLAVVFLG--SCFVLYKKRKRGQDGHSKT-WMPFSINGTSMG 453
Query: 312 T---NTGTLTS-TTNSSQSIPSYPSSKTSTMPKSFYFGVQVFTYEELEEATNNFHTSKEL 367
+ N TLTS TTN++ IP + +++ATNNF S+ +
Sbjct: 454 SKYSNGTTLTSITTNANYRIP----------------------FAAVKDATNNFDESRNI 491
Query: 368 GEGGFGTVYKGDLKDGRVVAVKRHYESNFKRVAQFMNEVEILARLRHKNLVTLYGCTSKH 427
G GGFG VYKG+L DG VAVKR + + +A+F E+E+L++ RH++LV+L G ++
Sbjct: 492 GVGGFGKVYKGELNDGTKVAVKRGNPKSQQGLAEFRTEIEMLSQFRHRHLVSLIGYCDEN 551
Query: 428 SRELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDIALETAEALAYLHASD---VMHR 484
+ E++L+YEY+ NGTV HL+G S L W RL+I + A L YLH D V+HR
Sbjct: 552 N-EMILIYEYMENGTVKSHLYGSGLPS--LTWKQRLEICIGAARGLHYLHTGDSKPVIHR 608
Query: 485 DVKSNNILLDEKFHVKVADFGLSRLFPN-DVTHVSTAPQGTPGYVDPEYYQCYQLTDKSD 543
DVKS NILLDE F KVADFGLS+ P D THVSTA +G+ GY+DPEY++ QLTDKSD
Sbjct: 609 DVKSANILLDENFMAKVADFGLSKTGPELDQTHVSTAVKGSFGYLDPEYFRRQQLTDKSD 668
Query: 544 VYSFGVVLVELISSLQAVDITRHRNDVNLANMAVNKIQSQELYDLVDPNL-GYEKDNSVK 602
VYSFGVVL E++ + +D T R VNLA A+ + +L ++D +L G + +S++
Sbjct: 669 VYSFGVVLFEVLCARPVIDPTLPREMVNLAEWAMKWQKKGQLDQIIDQSLRGNIRPDSLR 728
Query: 603 RMTTAVAELAFRCLQQQRDLRPSMDEI-------VEVLRAIKSDEPETQESKVL 649
+ AE +CL RPSM ++ +++ A+ EPE + ++
Sbjct: 729 KF----AETGEKCLADYGVDRPSMGDVLWNLEYALQLQEAVIDGEPEDNSTNMI 778
>At1g69730 putative protein kinase
Length = 792
Score = 253 bits (647), Expect = 2e-67
Identities = 142/356 (39%), Positives = 220/356 (60%), Gaps = 25/356 (7%)
Query: 348 VFTYEELEEATNNFHTSKELGEGGFGTVYKGDLKDGRVVAVKRHYESNFKRVAQFMNEVE 407
VF+ ELE+AT NF +++ LG+GG GTVYKG L DGR+VAVK+ + ++ +F+NEV
Sbjct: 434 VFSSRELEKATENFSSNRILGQGGQGTVYKGMLVDGRIVAVKKSKVVDEDKLEEFINEVV 493
Query: 408 ILARLRHKNLVTLYGCTSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDIAL 467
IL+++ H+N+V L GC + ++ +LVYE+I NG + +HLH + + + W++RL IA+
Sbjct: 494 ILSQINHRNIVKLLGCCLE-TKVPVLVYEFIPNGNLFEHLHDEFDENIMATWNIRLRIAI 552
Query: 468 ETAEALAYLHAS---DVMHRDVKSNNILLDEKFHVKVADFGLSRLFPNDVTHVSTAPQGT 524
+ A AL+YLH+S + HRDVKS NI+LDEK+ KV+DFG SR D TH++T GT
Sbjct: 553 DIAGALSYLHSSASSPIYHRDVKSTNIMLDEKYRAKVSDFGTSRTVTVDHTHLTTVVSGT 612
Query: 525 PGYVDPEYYQCYQLTDKSDVYSFGVVLVELISSLQAVDITRHRNDVNLANMAVNKIQSQE 584
GY+DPEY+Q Q TDKSDVYSFGVVLVELI+ +++ R + + LA + ++ +
Sbjct: 613 VGYMDPEYFQSSQFTDKSDVYSFGVVLVELITGEKSISFLRSQENRTLATYFILAMKENK 672
Query: 585 LYDLVDPNLGYEKDNSVKRMTTAVAELAFRCLQQQRDLRPSMDEIVEVLRAIK------- 637
L+D++D + +D + TA A++A +CL + RPSM E+ L +I+
Sbjct: 673 LFDIIDARI---RDGCMLSQVTATAKVARKCLNLKGRKRPSMREVSMELDSIRMPCGDMQ 729
Query: 638 -------SDEPETQESKVLDVVVRTDELVLLKKGPYPTSPDSVAEKWVSGSSTSTS 686
++E E Q +++ ++R + + + T+P S + SS+S S
Sbjct: 730 LQECVSENEEGEEQNKGLVEDIMRAES----RSKEFVTAPASHYNNIAAASSSSWS 781
>At5g59700 receptor-like protein kinase precursor - like
Length = 829
Score = 246 bits (629), Expect = 2e-65
Identities = 163/423 (38%), Positives = 246/423 (57%), Gaps = 51/423 (12%)
Query: 254 PSGNSSTN----GGLIGGVVGGVAALCLLGFVACFVVRRRRKNAKKPISNDLYMPPSS-- 307
PSG+SST G +IG +G + AL +LG F V +++ + ++ ++P SS
Sbjct: 393 PSGSSSTTKKNVGMIIGLTIGSLLALVVLGG---FFVLYKKRGRDQDGNSKTWIPLSSNG 449
Query: 308 TTSGTNTGTLTS-TTNSSQSIPSYPSSKTSTMPKSFYFGVQVFTYEELEEATNNFHTSKE 366
TTS +N TL S +NSS IP ++EATN+F ++
Sbjct: 450 TTSSSNGTTLASIASNSSYRIPLVA----------------------VKEATNSFDENRA 487
Query: 367 LGEGGFGTVYKGDLKDGRVVAVKRHYESNFKRVAQFMNEVEILARLRHKNLVTLYGCTSK 426
+G GGFG VYKG+L DG VAVKR + + +A+F E+E+L++ RH++LV+L G +
Sbjct: 488 IGVGGFGKVYKGELHDGTKVAVKRANPKSQQGLAEFRTEIEMLSQFRHRHLVSLIGYCDE 547
Query: 427 HSRELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDIALETAEALAYLHASD---VMH 483
++ E++LVYEY+ NGT+ HL+G S L W RL+I + +A L YLH D V+H
Sbjct: 548 NN-EMILVYEYMENGTLKSHLYG--SGLLSLSWKQRLEICIGSARGLHYLHTGDAKPVIH 604
Query: 484 RDVKSNNILLDEKFHVKVADFGLSRLFPN-DVTHVSTAPQGTPGYVDPEYYQCYQLTDKS 542
RDVKS NILLDE KVADFGLS+ P D THVSTA +G+ GY+DPEY++ QLT+KS
Sbjct: 605 RDVKSANILLDENLMAKVADFGLSKTGPEIDQTHVSTAVKGSFGYLDPEYFRRQQLTEKS 664
Query: 543 DVYSFGVVLVELISSLQAVDITRHRNDVNLANMAVNKIQSQELYDLVDPNL-GYEKDNSV 601
DVYSFGVV+ E++ + +D T R VNLA A+ + +L ++DP+L G + +S+
Sbjct: 665 DVYSFGVVMFEVLCARPVIDPTLTREMVNLAEWAMKWQKKGQLEHIIDPSLRGKIRPDSL 724
Query: 602 KRMTTAVAELAFRCLQQQRDLRPSMDEI-------VEVLRAIKSDEPETQESKVLDVVVR 654
++ E +CL RPSM ++ +++ A+ +PE + + ++ +R
Sbjct: 725 RKF----GETGEKCLADYGVDRPSMGDVLWNLEYALQLQEAVVDGDPEDSTNMIGELPLR 780
Query: 655 TDE 657
++
Sbjct: 781 FND 783
>At1g06840 receptor protein kinase, putative
Length = 939
Score = 244 bits (624), Expect = 9e-65
Identities = 156/407 (38%), Positives = 222/407 (54%), Gaps = 56/407 (13%)
Query: 254 PSGNSSTNGGLIGGVVGGVAALCLLGFVACFVVRRRRKNAKKPISNDLYMPPSSTTSGTN 313
PSG S NG + G V+G VAA L + ++ R+R ++
Sbjct: 539 PSGLS--NGAVAGIVLGSVAAAVTLTAIIALIIMRKRMRGYSAVAR-------------- 582
Query: 314 TGTLTSTTNSSQSIPSYPSSKTSTMPKSFYFGVQVFTYEELEEATNNFHTSKELGEGGFG 373
K S+ GV+ FTY EL AT+NF++S ++G+GG+G
Sbjct: 583 -------------------RKRSSKASLKIEGVKSFTYAELALATDNFNSSTQIGQGGYG 623
Query: 374 TVYKGDLKDGRVVAVKRHYESNFKRVAQFMNEVEILARLRHKNLVTLYGCTSKHSRELLL 433
VYKG L G VVA+KR E + + +F+ E+E+L+RL H+NLV+L G + E +L
Sbjct: 624 KVYKGTLGSGTVVAIKRAQEGSLQGEKEFLTEIELLSRLHHRNLVSLLGFCDEEG-EQML 682
Query: 434 VYEYISNGTVADHLHGDRSSSCLLPWSVRLDIALETAEALAYLHAS---DVMHRDVKSNN 490
VYEY+ NGT+ D++ L +++RL IAL +A+ + YLH + HRD+K++N
Sbjct: 683 VYEYMENGTLRDNISVKLKEP--LDFAMRLRIALGSAKGILYLHTEANPPIFHRDIKASN 740
Query: 491 ILLDEKFHVKVADFGLSRLFP------NDVTHVSTAPQGTPGYVDPEYYQCYQLTDKSDV 544
ILLD +F KVADFGLSRL P HVST +GTPGY+DPEY+ +QLTDKSDV
Sbjct: 741 ILLDSRFTAKVADFGLSRLAPVPDMEGISPQHVSTVVKGTPGYLDPEYFLTHQLTDKSDV 800
Query: 545 YSFGVVLVELISSLQAVDITRHRNDVNLANMAVNKIQSQELYDLVDPNLGYEKDNSVKRM 604
YS GVVL+EL + +Q IT +N V N+A +S + VD + D +++
Sbjct: 801 YSLGVVLLELFTGMQ--PITHGKNIVREINIA---YESGSILSTVDKRMSSVPDECLEKF 855
Query: 605 TTAVAELAFRCLQQQRDLRPSMDEIVEVLRAIKSDEPETQESKVLDV 651
T LA RC +++ D RPSM E+V L I PE+ +K D+
Sbjct: 856 AT----LALRCCREETDARPSMAEVVRELEIIWELMPESHVAKTADL 898
>At1g79680 unknown protein
Length = 769
Score = 244 bits (622), Expect = 2e-64
Identities = 134/301 (44%), Positives = 186/301 (61%), Gaps = 7/301 (2%)
Query: 347 QVFTYEELEEATNNFHTSKELGEGGFGTVYKGDLKDGRVVAVKRHYESNFKRVAQFMNEV 406
+VF ELE+AT NF ++ LGEGG GTVYKG L DGR+VAVK+ + ++ +F+NEV
Sbjct: 419 RVFNSRELEKATENFSLTRILGEGGQGTVYKGMLVDGRIVAVKKSKVVDEDKLEEFINEV 478
Query: 407 EILARLRHKNLVTLYGCTSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDIA 466
IL+++ H+N+V L GC + +L VYE+I NG + +HLH D + W VRL IA
Sbjct: 479 VILSQINHRNIVKLLGCCLETDVPIL-VYEFIPNGNLFEHLHDDSDDYTMTTWEVRLRIA 537
Query: 467 LETAEALAYLHA---SDVMHRDVKSNNILLDEKFHVKVADFGLSRLFPNDVTHVSTAPQG 523
++ A AL+YLH+ S + HRD+KS NI+LDEK KV+DFG SR D TH++T G
Sbjct: 538 VDIAGALSYLHSAASSPIYHRDIKSTNIMLDEKHRAKVSDFGTSRTVTVDHTHLTTVVSG 597
Query: 524 TPGYVDPEYYQCYQLTDKSDVYSFGVVLVELISSLQAVDITRHRNDVNLANMAVNKIQSQ 583
T GY+DPEY+Q Q TDKSDVYSFGVVL ELI+ ++V R + LA ++
Sbjct: 598 TVGYMDPEYFQSSQFTDKSDVYSFGVVLAELITGEKSVSFLRSQEYRTLATYFTLAMKEN 657
Query: 584 ELYDLVDPNLGYEKDNSVKRMTTAVAELAFRCLQQQRDLRPSMDEIVEVLRAIKSDEPET 643
L D++D + +D TA A++A +CL + RPSM ++ L I+S +
Sbjct: 658 RLSDIIDARI---RDGCKLNQVTAAAKIARKCLNMKGRKRPSMRQVSMELEKIRSYSEDM 714
Query: 644 Q 644
Q
Sbjct: 715 Q 715
>At4g00330 unknown protein
Length = 411
Score = 241 bits (614), Expect = 1e-63
Identities = 143/351 (40%), Positives = 209/351 (58%), Gaps = 23/351 (6%)
Query: 302 YMPPSSTTSGTNTGTLTSTTNSSQSIPSYPSSKTSTMPKSFYFGVQVFTYEELEEATNNF 361
Y+ S ++S N + S+T SS SY ++ + + FT++E+ +AT NF
Sbjct: 68 YIDNSKSSSDNNIRSRRSSTGSSSVQRSYGNANETEHTR--------FTFDEIYDATKNF 119
Query: 362 HTSKELGEGGFGTVYKGDLKDGRVVAVKRHYESNFK----RVAQFMNEVEILARLRHKNL 417
S +G+GGFGTVYK L+DG+ AVKR +S A+FM+E++ LA++ H +L
Sbjct: 120 SPSFRIGQGGFGTVYKVKLRDGKTFAVKRAKKSMHDDRQGADAEFMSEIQTLAQVTHLSL 179
Query: 418 VTLYGCTSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDIALETAEALAYLH 477
V YG H+ E +LV EY++NGT+ DHL D L + RLDIA + A A+ YLH
Sbjct: 180 VKYYGFVV-HNDEKILVVEYVANGTLRDHL--DCKEGKTLDMATRLDIATDVAHAITYLH 236
Query: 478 A---SDVMHRDVKSNNILLDEKFHVKVADFGLSRLFPND---VTHVSTAPQGTPGYVDPE 531
++HRD+KS+NILL E + KVADFG +RL P+ THVST +GT GY+DPE
Sbjct: 237 MYTQPPIIHRDIKSSNILLTENYRAKVADFGFARLAPDTDSGATHVSTQVKGTAGYLDPE 296
Query: 532 YYQCYQLTDKSDVYSFGVVLVELISSLQAVDITRHRNDVNLANMAVNKIQSQELYDLVDP 591
Y YQLT+KSDVYSFGV+LVEL++ + ++++R + + A+ K S + ++DP
Sbjct: 297 YLTTYQLTEKSDVYSFGVLLVELLTGRRPIELSRGQKERITIRWAIKKFTSGDTISVLDP 356
Query: 592 NLGYEKDNSVKRMTTAVAELAFRCLQQQRDLRPSMDEIVEVLRAIKSDEPE 642
L E++++ V E+AF+CL R RPSM + E+L I+ D E
Sbjct: 357 KL--EQNSANNLALEKVLEMAFQCLAPHRRSRPSMKKCSEILWGIRKDYRE 405
>At1g16150
Length = 779
Score = 240 bits (613), Expect = 2e-63
Identities = 134/301 (44%), Positives = 190/301 (62%), Gaps = 8/301 (2%)
Query: 347 QVFTYEELEEATNNFHTSKELGEGGFGTVYKGDLKDGRVVAVKRHYESNFKRVAQFMNEV 406
++F+ ELE+AT+NF+T++ LG+GG GTVYKG L DGR+VAVKR + +V +F+NEV
Sbjct: 428 KIFSSNELEKATDNFNTNRVLGQGGQGTVYKGMLVDGRIVAVKRSKAMDEDKVEEFINEV 487
Query: 407 EILARLRHKNLVTLYGCTSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDIA 466
+LA++ H+N+V L GC + + +LVYE++ NG + L D ++ W VRL IA
Sbjct: 488 VVLAQINHRNIVKLLGCCLE-TEVPVLVYEFVPNGDLCKRLR-DECDDYIMTWEVRLHIA 545
Query: 467 LETAEALAYLHAS---DVMHRDVKSNNILLDEKFHVKVADFGLSRLFPNDVTHVSTAPQG 523
+E A AL+YLH++ + HRD+K+ NILLDEK+ VKV+DFG SR D TH++T G
Sbjct: 546 IEIAGALSYLHSAASFPIYHRDIKTTNILLDEKYQVKVSDFGTSRSVTIDQTHLTTQVAG 605
Query: 524 TPGYVDPEYYQCYQLTDKSDVYSFGVVLVELISSLQAVDITRHRNDVNLANMAVNKIQSQ 583
T GYVDPEY+Q + TDKSDVYSFGVVLVELI+ + + A V ++
Sbjct: 606 TFGYVDPEYFQSSKFTDKSDVYSFGVVLVELITGKNPSSRVQSEENRGFAAHFVAAVKEN 665
Query: 584 ELYDLVDPNLGYEKDNSVKRMTTAVAELAFRCLQQQRDLRPSMDEIVEVLRAIKSDEPET 643
D+VD + KD AVA+LA RCL ++ RP+M E+ L I+S ++
Sbjct: 666 RFLDIVDERI---KDECNLDQVMAVAKLAKRCLNRKGKKRPNMREVSVELERIRSSSYKS 722
Query: 644 Q 644
+
Sbjct: 723 E 723
>At1g79670 putative protein
Length = 751
Score = 240 bits (612), Expect = 2e-63
Identities = 138/302 (45%), Positives = 192/302 (62%), Gaps = 9/302 (2%)
Query: 347 QVFTYEELEEATNNFHTSKELGEGGFGTVYKGDLKDGRVVAVKRHYESNFKRVAQFMNEV 406
++F+ +ELE+AT+NF+ ++ LG+GG GTVYKG L DGR+VAVKR + +V +F+NEV
Sbjct: 407 KIFSSKELEKATDNFNMNRVLGQGGQGTVYKGMLVDGRIVAVKRSKVLDEDKVEEFINEV 466
Query: 407 EILARLRHKNLVTLYGCTSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDIA 466
+L+++ H+N+V L GC + + +LVYE+I NG + LH D S + W VRL I+
Sbjct: 467 GVLSQINHRNIVKLMGCCLE-TEVPILVYEHIPNGDLFKRLHHD-SDDYTMTWDVRLRIS 524
Query: 467 LETAEALAYLHAS---DVMHRDVKSNNILLDEKFHVKVADFGLSRLFPNDVTHVSTAPQG 523
+E A ALAYLH++ V HRDVK+ NILLDEK+ KV+DFG SR D TH++T G
Sbjct: 525 VEIAGALAYLHSAASTPVYHRDVKTTNILLDEKYRAKVSDFGTSRSINVDQTHLTTLVAG 584
Query: 524 TPGYVDPEYYQCYQLTDKSDVYSFGVVLVELISSLQAVDITRHRNDVNLANMAVNKIQSQ 583
T GY+DPEY+Q Q TDKSDVYSFGVVLVELI+ + + R + L + ++
Sbjct: 585 TFGYLDPEYFQTSQFTDKSDVYSFGVVLVELITGEKPFSVMRPEENRGLVSHFNEAMKQN 644
Query: 584 ELYDLVDPNLGYEKDNSVKRMTTAVAELAFRCLQQQRDLRPSMDEIVEVLRAIKSDEPET 643
+ D+VD + K+ AVA+LA RCL + RP+M E+ L I+S PE
Sbjct: 645 RVLDIVDSRI---KEGCTLEQVLAVAKLARRCLSLKGKKRPNMREVSVELERIRS-SPED 700
Query: 644 QE 645
E
Sbjct: 701 LE 702
>At1g16130 unknown protein
Length = 748
Score = 238 bits (606), Expect = 1e-62
Identities = 133/295 (45%), Positives = 187/295 (63%), Gaps = 8/295 (2%)
Query: 347 QVFTYEELEEATNNFHTSKELGEGGFGTVYKGDLKDGRVVAVKRHYESNFKRVAQFMNEV 406
++F+ ELE+AT+NF+ ++ LG+GG GTVYKG L DGR+VAVKR + RV +F+NEV
Sbjct: 402 RIFSSHELEKATDNFNKNRVLGQGGQGTVYKGMLVDGRIVAVKRSKAVDEDRVEEFINEV 461
Query: 407 EILARLRHKNLVTLYGCTSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDIA 466
+LA++ H+N+V L GC + + +LVYE++ NG + LH D S + W VRL IA
Sbjct: 462 VVLAQINHRNIVKLLGCCLE-TEVPVLVYEFVPNGDLCKRLH-DESDDYTMTWEVRLHIA 519
Query: 467 LETAEALAYLHAS---DVMHRDVKSNNILLDEKFHVKVADFGLSRLFPNDVTHVSTAPQG 523
+E A AL+YLH++ + HRD+K+ NILLDE+ KV+DFG SR D TH++T G
Sbjct: 520 IEIAGALSYLHSAASFPIYHRDIKTTNILLDERNRAKVSDFGTSRSVTIDQTHLTTQVAG 579
Query: 524 TPGYVDPEYYQCYQLTDKSDVYSFGVVLVELISSLQAVDITRHRNDVNLANMAVNKIQSQ 583
T GYVDPEY+Q + T+KSDVYSFGVVLVEL++ + R + LA V ++
Sbjct: 580 TFGYVDPEYFQSSKFTEKSDVYSFGVVLVELLTGEKPSSRVRSEENRGLAAHFVEAVKEN 639
Query: 584 ELYDLVDPNLGYEKDNSVKRMTTAVAELAFRCLQQQRDLRPSMDEIVEVLRAIKS 638
+ D+VD + KD +VA LA RCL ++ RP+M E+ L I+S
Sbjct: 640 RVLDIVDDRI---KDECNMDQVMSVANLARRCLNRKGKKRPNMREVSIELEMIRS 691
>At4g31100 serine/threonine-specific protein kinase - like
Length = 656
Score = 234 bits (597), Expect = 1e-61
Identities = 131/308 (42%), Positives = 193/308 (62%), Gaps = 8/308 (2%)
Query: 347 QVFTYEELEEATNNFHTSKELGEGGFGTVYKGDLKDGRVVAVKRHYESNFKRVAQFMNEV 406
++FT +ELE+AT NF ++ LG GG GTVYKG L DGR VAVK+ + ++ +F+NEV
Sbjct: 300 RIFTSKELEKATENFSENRVLGHGGQGTVYKGMLVDGRTVAVKKSKVIDEDKLQEFINEV 359
Query: 407 EILARLRHKNLVTLYGCTSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDIA 466
IL+++ H+++V L GC + + +LVYE+I NG + H+H + + + W +RL IA
Sbjct: 360 VILSQINHRHVVKLLGCCLE-TEVPILVYEFIINGNLFKHIHEEEADDYTMIWGMRLRIA 418
Query: 467 LETAEALAYLHA---SDVMHRDVKSNNILLDEKFHVKVADFGLSRLFPNDVTHVSTAPQG 523
++ A AL+YLH+ S + HRD+KS NILLDEK+ KVADFG SR D TH +T G
Sbjct: 419 VDIAGALSYLHSAASSPIYHRDIKSTNILLDEKYRAKVADFGTSRSVTIDQTHWTTVISG 478
Query: 524 TPGYVDPEYYQCYQLTDKSDVYSFGVVLVELISSLQAVDITRHRND-VNLANMAVNKIQS 582
T GYVDPEYY+ Q T+KSDVYSFGV+L ELI+ + V + ++ + + LA ++
Sbjct: 479 TVGYVDPEYYRSSQYTEKSDVYSFGVILAELITGDKPVIMVQNTQEIIALAEHFRVAMKE 538
Query: 583 QELYDLVDPNLGYEKDNSVKRMTTAVAELAFRCLQQQRDLRPSMDEIVEVLRAIKSDEPE 642
+ L D++D + +D+S AVA LA +CL + RP+M E+ L I + +
Sbjct: 539 RRLSDIMDARI---RDDSKPEQVMAVANLAMKCLSSRGRNRPNMREVFTELERICTSPED 595
Query: 643 TQESKVLD 650
+Q +D
Sbjct: 596 SQVQNRID 603
>At5g49760 receptor protein kinase-like
Length = 953
Score = 234 bits (596), Expect = 2e-61
Identities = 149/388 (38%), Positives = 221/388 (56%), Gaps = 40/388 (10%)
Query: 255 SGNSSTNGGLIGGVVGGVAALCLLGFVACFVVRRRRKNAKKPISNDLYMPPSSTTSGTNT 314
S SS LIG VVG V L LL + +R++++ + T N
Sbjct: 551 SSKSSNKSILIGAVVGVVVLLLLLTIAGIYALRQKKRAER-------------ATGQNNP 597
Query: 315 GTLTSTTNSSQSIPSYPSSKTSTMPKSFYFGVQVFTYEELEEATNNFHTSKELGEGGFGT 374
T+ SS P G + FT+EEL++ T+NF + ++G GG+G
Sbjct: 598 FAKWDTSKSSIDAPQL-------------MGAKAFTFEELKKCTDNFSEANDVGGGGYGK 644
Query: 375 VYKGDLKDGRVVAVKRHYESNFKRVAQFMNEVEILARLRHKNLVTLYG-CTSKHSRELLL 433
VY+G L +G+++A+KR + + + +F E+E+L+R+ HKN+V L G C ++ E +L
Sbjct: 645 VYRGILPNGQLIAIKRAQQGSLQGGLEFKTEIELLSRVHHKNVVRLLGFCFDRN--EQML 702
Query: 434 VYEYISNGTVADHLHGDRSSSCLLPWSVRLDIALETAEALAYLHA---SDVMHRDVKSNN 490
VYEYISNG++ D L G S L W+ RL IAL + + LAYLH ++HRD+KSNN
Sbjct: 703 VYEYISNGSLKDSLSG--KSGIRLDWTRRLKIALGSGKGLAYLHELADPPIIHRDIKSNN 760
Query: 491 ILLDEKFHVKVADFGLSRLFPN-DVTHVSTAPQGTPGYVDPEYYQCYQLTDKSDVYSFGV 549
ILLDE KVADFGLS+L + + THV+T +GT GY+DPEYY QLT+KSDVY FGV
Sbjct: 761 ILLDENLTAKVADFGLSKLVGDPEKTHVTTQVKGTMGYLDPEYYMTNQLTEKSDVYGFGV 820
Query: 550 VLVELISSLQAVDITRHRNDVNLANMAVNKIQS-QELYDLVDPNLGYEKDNSVKRMTTAV 608
VL+EL++ ++ R + V +NK +S +L +L+D + ++K V
Sbjct: 821 VLLELLTGRSPIE--RGKYVVREVKTKMNKSRSLYDLQELLDTTI-IASSGNLKGFEKYV 877
Query: 609 AELAFRCLQQQRDLRPSMDEIVEVLRAI 636
+LA RC++++ RPSM E+V+ + I
Sbjct: 878 -DLALRCVEEEGVNRPSMGEVVKEIENI 904
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.134 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,451,187
Number of Sequences: 26719
Number of extensions: 761073
Number of successful extensions: 6584
Number of sequences better than 10.0: 1025
Number of HSP's better than 10.0 without gapping: 878
Number of HSP's successfully gapped in prelim test: 147
Number of HSP's that attempted gapping in prelim test: 2350
Number of HSP's gapped (non-prelim): 1285
length of query: 687
length of database: 11,318,596
effective HSP length: 106
effective length of query: 581
effective length of database: 8,486,382
effective search space: 4930587942
effective search space used: 4930587942
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 64 (29.3 bits)
Medicago: description of AC147875.4


