

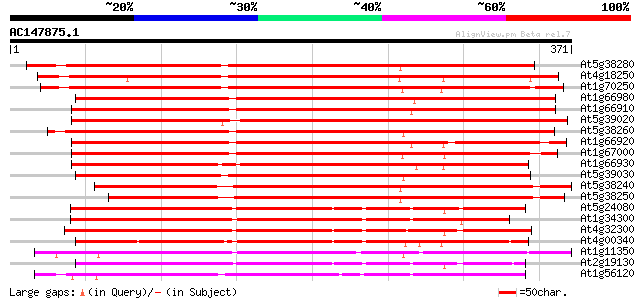
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147875.1 + phase: 0
(371 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g38280 receptor serine/threonine kinase PR5K 385 e-107
At4g18250 receptor serine/threonine kinase-like protein 384 e-107
At1g70250 hypothetical protein 378 e-105
At1g66980 putative kinase 376 e-105
At1g66910 receptor serine/threonine kinase PR5K, putative 375 e-104
At5g39020 receptor protein kinase - like protein 374 e-104
At5g38260 receptor serine/threonine protein kinase - like 374 e-104
At1g66920 receptor serine/threonine kinase PR5K, putative 368 e-102
At1g67000 hypothetical protein 366 e-101
At1g66930 receptor serine/threonine kinase PR5K, putative 358 3e-99
At5g39030 receptor protein kinase - like protein 355 3e-98
At5g38240 receptor serine/threonine protein kinase -like 340 6e-94
At5g38250 receptor serine/threonine protein kinase - like 327 8e-90
At5g24080 receptor-like protein kinase 255 2e-68
At1g34300 putative protein 244 4e-65
At4g32300 S-receptor kinase -like protein 240 1e-63
At4g00340 receptor-like protein kinase 225 3e-59
At1g11350 putative receptor-like protein kinase gi|4008010; simi... 218 4e-57
At2g19130 putative receptor-like protein kinase 217 9e-57
At1g56120 receptor-like protein kinase, putative 210 9e-55
>At5g38280 receptor serine/threonine kinase PR5K
Length = 665
Score = 385 bits (988), Expect = e-107
Identities = 191/340 (56%), Positives = 252/340 (73%), Gaps = 14/340 (4%)
Query: 12 LIPQTILRERTELVDNNVEVFMRSHNLSMPRRYSYAEVKMITNYFREKLGQGGYGVVYKA 71
+I +T +E D NVE ++M +RYSY VK +TN F LG+GG+G VYK
Sbjct: 295 IIVRTKNMRNSEWNDQNVEA------VAMLKRYSYTRVKKMTNSFAHVLGKGGFGTVYKG 348
Query: 72 SLYNS-RQVAVKVISETKGNGEEFINEVASISRTSHMNIVSLLGYCYEENKRALIYEFMP 130
L +S R VAVK++ ++GNGEEFINEVAS+SRTSH+NIVSLLG+CYE+NKRA+IYEFMP
Sbjct: 349 KLADSGRDVAVKILKVSEGNGEEFINEVASMSRTSHVNIVSLLGFCYEKNKRAIIYEFMP 408
Query: 131 KGSLDKFIYKSEFPNAICDFDWNTLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLD 190
GSLDK+I N +W L+ +A+GI++GLEYLH C +RI+H DIKPQNIL+D
Sbjct: 409 NGSLDKYISA----NMSTKMEWERLYDVAVGISRGLEYLHNRCVTRIVHFDIKPQNILMD 464
Query: 191 EDFCPKISDFGLAKICQRKDSIVSILGARGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLI 250
E+ CPKISDFGLAK+C+ K+SI+S+L RGT GY+APE+FS+ FG VS++SDVYSYGM++
Sbjct: 465 ENLCPKISDFGLAKLCKNKESIISMLHMRGTFGYIAPEMFSKNFGAVSHKSDVYSYGMVV 524
Query: 251 LEMIGGR--KNYDTGGSCTSEMYFPDWIYKDLEQGN-TLLNCSTISEEENDMIRKITLVS 307
LEMIG + + + GS MYFP+W+YKD E+G T + +I++EE + +K+ LV+
Sbjct: 525 LEMIGAKNIEKVEYSGSNNGSMYFPEWVYKDFEKGEITRIFGDSITDEEEKIAKKLVLVA 584
Query: 308 LWCIQTKPSDRPPMNKVIEMLQGPLSSVSYPPKPVLYSPE 347
LWCIQ PSDRPPM KVIEML+G L ++ PP P+L+SPE
Sbjct: 585 LWCIQMNPSDRPPMIKVIEMLEGNLEALQVPPNPLLFSPE 624
>At4g18250 receptor serine/threonine kinase-like protein
Length = 687
Score = 384 bits (985), Expect = e-107
Identities = 192/360 (53%), Positives = 262/360 (72%), Gaps = 25/360 (6%)
Query: 19 RERTELVDNNVEVFMRSHNLSMPRRYSYAEVKMITNYFREKLGQGGYGVVYKASLYNS-- 76
+ ++EL D N+E + M +RYS+ +VK +TN F +G+GG+G VYK L ++
Sbjct: 324 KRKSELNDENIEAVV------MLKRYSFEKVKKMTNSFDHVIGKGGFGTVYKGKLPDASG 377
Query: 77 RQVAVKVISETKGNGEEFINEVASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDK 136
R +A+K++ E+KGNGEEFINE+ S+SR SH+NIVSL G+CYE ++RA+IYEFMP GSLDK
Sbjct: 378 RDIALKILKESKGNGEEFINELVSMSRASHVNIVSLFGFCYEGSQRAIIYEFMPNGSLDK 437
Query: 137 FIYKSEFPNAICDFDWNTLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPK 196
FI + N +W TL+ IA+G+A+GLEYLH C S+I+H DIKPQNIL+DED CPK
Sbjct: 438 FISE----NMSTKIEWKTLYNIAVGVARGLEYLHNSCVSKIVHFDIKPQNILIDEDLCPK 493
Query: 197 ISDFGLAKICQRKDSIVSILGARGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGG 256
ISDFGLAK+C++K+SI+S+L ARGT+GY+APE+FS+ +GGVS++SDVYSYGM++LEMIG
Sbjct: 494 ISDFGLAKLCKKKESIISMLDARGTVGYIAPEMFSKNYGGVSHKSDVYSYGMVVLEMIGA 553
Query: 257 --RKNYDTGGSCTSEMYFPDWIYKDLEQGNT--LLNCSTISEEEND-MIRKITLVSLWCI 311
R+ +T + S MYFPDW+Y+DLE+ T LL I EEE + +++++TLV LWCI
Sbjct: 554 TKREEVETSATDKSSMYFPDWVYEDLERKETMRLLEDHIIEEEEEEKIVKRMTLVGLWCI 613
Query: 312 QTKPSDRPPMNKVIEMLQGP-LSSVSYPPKPVL-------YSPERPELQVSDMSSSDLYE 363
QT PSDRPPM KV+EML+G L ++ PPKP+L + Q S +S+ L E
Sbjct: 614 QTNPSDRPPMRKVVEMLEGSRLEALQVPPKPLLNLHVVTDWETSEDSQQTSRLSTQSLLE 673
>At1g70250 hypothetical protein
Length = 676
Score = 378 bits (970), Expect = e-105
Identities = 190/351 (54%), Positives = 249/351 (70%), Gaps = 18/351 (5%)
Query: 21 RTELVDNNVEVFMRSHNLSMPRRYSYAEVKMITNYFREKLGQGGYGVVYKASLYN-SRQV 79
+++L + N+E + M +R+SY +VK +T F LG+GG+G VYK L + SR V
Sbjct: 309 KSDLNEKNMEAVV------MLKRFSYVQVKKMTKSFENVLGKGGFGTVYKGKLPDGSRDV 362
Query: 80 AVKVISETKGNGEEFINEVASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIY 139
AVK++ E+ +GE+FINE+AS+SRTSH NIVSLLG+CYE K+A+IYE MP GSLDKFI
Sbjct: 363 AVKILKESNEDGEDFINEIASMSRTSHANIVSLLGFCYEGRKKAIIYELMPNGSLDKFIS 422
Query: 140 KSEFPNAICDFDWNTLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISD 199
K N +W TL+ IA+G++ GLEYLH C SRI+H DIKPQNIL+D D CPKISD
Sbjct: 423 K----NMSAKMEWKTLYNIAVGVSHGLEYLHSHCVSRIVHFDIKPQNILIDGDLCPKISD 478
Query: 200 FGLAKICQRKDSIVSILGARGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRK- 258
FGLAK+C+ +SI+S+L ARGTIGY+APE+FS+ FGGVS++SDVYSYGM++LEMIG R
Sbjct: 479 FGLAKLCKNNESIISMLHARGTIGYIAPEVFSQNFGGVSHKSDVYSYGMVVLEMIGARNI 538
Query: 259 -NYDTGGSCTSEMYFPDWIYKDLEQGN--TLLNCSTISEEENDMIRKITLVSLWCIQTKP 315
GS + MYFPDWIYKDLE+G + L EE+ +++K+ LV LWCIQT P
Sbjct: 539 GRAQNAGSSNTSMYFPDWIYKDLEKGEIMSFLADQITEEEDEKIVKKMVLVGLWCIQTNP 598
Query: 316 SDRPPMNKVIEMLQGPLSSVSYPPKPVLYSPERPELQVSDMSSSDLYETNS 366
DRPPM+KV+EML+G L ++ PPKP+L P + D+ ET+S
Sbjct: 599 YDRPPMSKVVEMLEGSLEALQIPPKPLLC---LPAITAPITVDEDIQETSS 646
>At1g66980 putative kinase
Length = 1109
Score = 376 bits (966), Expect = e-105
Identities = 180/322 (55%), Positives = 240/322 (73%), Gaps = 8/322 (2%)
Query: 44 YSYAEVKMITNYFREKLGQGGYGVVYKASLYNSRQVAVKVISETKGNGEEFINEVASISR 103
Y+YA+VK IT F E +G+GG+G+VYK +L + R VAVKV+ +TKGNGE+FINEVA++SR
Sbjct: 786 YTYAQVKRITKSFAEVVGRGGFGIVYKGTLSDGRVVAVKVLKDTKGNGEDFINEVATMSR 845
Query: 104 TSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIAIGIA 163
TSH+NIVSLLG+C E +KRA+IYEF+ GSLDKFI N DW L+RIA+G+A
Sbjct: 846 TSHLNIVSLLGFCSEGSKRAIIYEFLENGSLDKFILGKTSVN----MDWTALYRIALGVA 901
Query: 164 KGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGARGTIG 223
GLEYLH C +RI+H DIKPQN+LLD+ FCPK+SDFGLAK+C++K+SI+S+L RGTIG
Sbjct: 902 HGLEYLHHSCKTRIVHFDIKPQNVLLDDSFCPKVSDFGLAKLCEKKESILSMLDTRGTIG 961
Query: 224 YMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSC---TSEMYFPDWIYKDL 280
Y+APE+ SR +G VS++SDVYSYGML+LE+IG R +C TS MYFP+W+Y+DL
Sbjct: 962 YIAPEMISRVYGNVSHKSDVYSYGMLVLEIIGARNKEKANQACASNTSSMYFPEWVYRDL 1021
Query: 281 EQGNTLLNCST-ISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQGPLSSVSYPP 339
E + + I+ EE+++ +K+TLV LWCIQ P DRP MN+V+EM++G L ++ PP
Sbjct: 1022 ESCKSGRHIEDGINSEEDELAKKMTLVGLWCIQPSPVDRPAMNRVVEMMEGSLEALEVPP 1081
Query: 340 KPVLYSPERPELQVSDMSSSDL 361
+PVL L S + S D+
Sbjct: 1082 RPVLQQIPISNLHESSILSEDV 1103
>At1g66910 receptor serine/threonine kinase PR5K, putative
Length = 655
Score = 375 bits (962), Expect = e-104
Identities = 181/323 (56%), Positives = 242/323 (74%), Gaps = 7/323 (2%)
Query: 42 RRYSYAEVKMITNYFREKLGQGGYGVVYKASLYNSRQVAVKVISETKGNGEEFINEVASI 101
+ YSYA+V IT F E +G+GG+G VY+ +LY+ R VAVKV+ E++GNGE+FINEVAS+
Sbjct: 325 KHYSYAQVTSITKSFAEVIGKGGFGTVYRGTLYDGRSVAVKVLKESQGNGEDFINEVASM 384
Query: 102 SRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIAIG 161
S+TSH+NIV+LLG+C E KRA+IYEFM GSLDKFI + DW L+ IA+G
Sbjct: 385 SQTSHVNIVTLLGFCSEGYKRAIIYEFMENGSLDKFISSKKSST----MDWRELYGIALG 440
Query: 162 IAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGARGT 221
+A+GLEYLH GC +RI+H DIKPQN+LLD++ PK+SDFGLAK+C+RK+SI+S++ RGT
Sbjct: 441 VARGLEYLHHGCRTRIVHFDIKPQNVLLDDNLSPKVSDFGLAKLCERKESILSLMDTRGT 500
Query: 222 IGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGG--SCTSEMYFPDWIYKD 279
IGY+APE+FSR +G VS++SDVYSYGML+L++IG R T S TS MYFP+WIY+D
Sbjct: 501 IGYIAPEVFSRVYGRVSHKSDVYSYGMLVLDIIGARNKTSTEDTTSSTSSMYFPEWIYRD 560
Query: 280 LEQGNTLLNCST-ISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQGPLSSVSYP 338
LE+ + + T IS EE+++ +K+TLV LWCIQ P DRP MN+V+EM++G L ++ P
Sbjct: 561 LEKAHNGKSIETAISNEEDEIAKKMTLVGLWCIQPWPLDRPAMNRVVEMMEGNLDALEVP 620
Query: 339 PKPVLYSPERPELQVSDMSSSDL 361
P+PVL LQ S S D+
Sbjct: 621 PRPVLQQIPTATLQESSTFSEDI 643
>At5g39020 receptor protein kinase - like protein
Length = 813
Score = 374 bits (960), Expect = e-104
Identities = 182/331 (54%), Positives = 247/331 (73%), Gaps = 9/331 (2%)
Query: 42 RRYSYAEVKMITNYFREKLGQGGYGVVYKASLYNSRQVAVKVISETKGNGEEFINEVASI 101
++Y YAE+K IT F +G+GG+G VY+ +L N R VAVKV+ + KGNG++FINEV S+
Sbjct: 484 KQYIYAELKKITKSFSHTVGKGGFGTVYRGNLSNGRTVAVKVLKDLKGNGDDFINEVTSM 543
Query: 102 SRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIY--KSEFPNAICDFDWNTLFRIA 159
S+TSH+NIVSLLG+CYE +KRA+I EF+ GSLD+FI KS PN TL+ IA
Sbjct: 544 SQTSHVNIVSLLGFCYEGSKRAIISEFLEHGSLDQFISRNKSLTPNV------TTLYGIA 597
Query: 160 IGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGAR 219
+GIA+GLEYLH GC +RI+H DIKPQNILLD++FCPK++DFGLAK+C++++SI+S++ R
Sbjct: 598 LGIARGLEYLHYGCKTRIVHFDIKPQNILLDDNFCPKVADFGLAKLCEKRESILSLIDTR 657
Query: 220 GTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSCTSEMYFPDWIYKD 279
GTIGY+APE+ SR +GG+S++SDVYSYGML+L+MIG R +T S YFPDWIYKD
Sbjct: 658 GTIGYIAPEVVSRMYGGISHKSDVYSYGMLVLDMIGARNKVETTTCNGSTAYFPDWIYKD 717
Query: 280 LEQGN-TLLNCSTISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQGPLSSVSYP 338
LE G+ T + I+EE+N +++K+ LVSLWCI+ PSDRPPMNKV+EM++G L ++ P
Sbjct: 718 LENGDQTWIIGDEINEEDNKIVKKMILVSLWCIRPCPSDRPPMNKVVEMIEGSLDALELP 777
Query: 339 PKPVLYSPERPELQVSDMSSSDLYETNSVTV 369
PKP + L+ S +S E + T+
Sbjct: 778 PKPSRHISTELVLESSSLSDGQEAEKQTQTL 808
>At5g38260 receptor serine/threonine protein kinase - like
Length = 638
Score = 374 bits (959), Expect = e-104
Identities = 181/338 (53%), Positives = 251/338 (73%), Gaps = 13/338 (3%)
Query: 26 DNNVEVFMRSHNLSMPRRYSYAEVKMITNYFREKLGQGGYGVVYKASLYNSRQVAVKVIS 85
DNN++ L ++YSYAEV+ IT F LG+GG+G VY +L + R+VAVK++
Sbjct: 299 DNNLK------GLVQLKQYSYAEVRKITKLFSHTLGKGGFGTVYGGNLCDGRKVAVKILK 352
Query: 86 ETKGNGEEFINEVASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPN 145
+ K NGE+FINEVAS+S+TSH+NIVSLLG+CYE +KRA++YEF+ GSLD+F+ + + N
Sbjct: 353 DFKSNGEDFINEVASMSQTSHVNIVSLLGFCYEGSKRAIVYEFLENGSLDQFLSEKKSLN 412
Query: 146 AICDFDWNTLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKI 205
D +TL+RIA+G+A+GL+YLH GC +RI+H DIKPQNILLD+ FCPK+SDFGLAK+
Sbjct: 413 ----LDVSTLYRIALGVARGLDYLHHGCKTRIVHFDIKPQNILLDDTFCPKVSDFGLAKL 468
Query: 206 CQRKDSIVSILGARGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKN--YDTG 263
C++++SI+S+L ARGTIGY+APE+FS +G VS++SDVYSYGML+LEMIG + +T
Sbjct: 469 CEKRESILSLLDARGTIGYIAPEVFSGMYGRVSHKSDVYSYGMLVLEMIGAKNKEIEETA 528
Query: 264 GSCTSEMYFPDWIYKDLEQG-NTLLNCSTISEEENDMIRKITLVSLWCIQTKPSDRPPMN 322
S +S YFPDWIYK+LE G +T IS E+ ++ +K+TLV LWCIQ P +RPPMN
Sbjct: 529 ASNSSSAYFPDWIYKNLENGEDTWKFGDEISREDKEVAKKMTLVGLWCIQPSPLNRPPMN 588
Query: 323 KVIEMLQGPLSSVSYPPKPVLYSPERPELQVSDMSSSD 360
+++EM++G L + PPKP ++ P Q+S S +
Sbjct: 589 RIVEMMEGSLDVLEVPPKPSIHYSAEPLPQLSSFSEEN 626
>At1g66920 receptor serine/threonine kinase PR5K, putative
Length = 609
Score = 368 bits (944), Expect = e-102
Identities = 184/333 (55%), Positives = 248/333 (74%), Gaps = 19/333 (5%)
Query: 42 RRYSYAEVKMITNYFREKLGQGGYGVVYKASLYNSRQVAVKVISETKGN-GEEFINEVAS 100
++YSY +VK ITN F E +G+GG+G+VY+ +L + R VAVKV+ + KGN GE+FINEVAS
Sbjct: 287 KQYSYEQVKRITNSFAEVVGRGGFGIVYRGTLSDGRMVAVKVLKDLKGNNGEDFINEVAS 346
Query: 101 ISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIAI 160
+S+TSH+NIV+LLG+C E KRA+IYEFM GSLDKFI + DW L+ IA+
Sbjct: 347 MSQTSHVNIVTLLGFCSEGYKRAIIYEFMENGSLDKFISSKKSST----MDWRELYGIAL 402
Query: 161 GIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGARG 220
G+A+GLEYLH GC +RI+H DIKPQN+LLD++ PK+SDFGLAK+C+RK+SI+S++ RG
Sbjct: 403 GVARGLEYLHHGCRTRIVHFDIKPQNVLLDDNLSPKVSDFGLAKLCERKESILSLMDTRG 462
Query: 221 TIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGG--SCTSEMYFPDWIYK 278
TIGY+APE+FSR +G VS++SDVYSYGML+L++IG R T S TS MYFP+WIYK
Sbjct: 463 TIGYIAPEVFSRVYGSVSHKSDVYSYGMLVLDIIGARNKTSTEDTTSSTSSMYFPEWIYK 522
Query: 279 DLEQGNT---LLNCSTISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQGPLSSV 335
DLE+G+ ++N S EE+++ +K+TLV LWCIQ P DRP MN+V+EM++G L ++
Sbjct: 523 DLEKGDNGRLIVNRS----EEDEIAKKMTLVGLWCIQPWPLDRPAMNRVVEMMEGNLDAL 578
Query: 336 SYPPKPVLYSPERPELQVSDMSSSDLYETNSVT 368
PP+PVL P L SS + E NS++
Sbjct: 579 EVPPRPVLQCSVVPHL-----DSSWISEENSIS 606
>At1g67000 hypothetical protein
Length = 717
Score = 366 bits (940), Expect = e-101
Identities = 186/334 (55%), Positives = 247/334 (73%), Gaps = 24/334 (7%)
Query: 42 RRYSYAEVKMITNYFREKLGQGGYGVVYKASLYNSRQVAVKVISETKG-NGEEFINEVAS 100
+ Y+YAEVK +T F E +G+GG+G+VY +L +S VAVKV+ ++KG +GE+FINEVAS
Sbjct: 369 KHYTYAEVKKMTKSFTEVVGRGGFGIVYSGTLSDSSMVAVKVLKDSKGTDGEDFINEVAS 428
Query: 101 ISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIAI 160
+S+TSH+NIVSLLG+C E ++RA+IYEF+ GSLDKFI N D TL+ IA+
Sbjct: 429 MSQTSHVNIVSLLGFCCEGSRRAIIYEFLGNGSLDKFISDKSSVN----LDLKTLYGIAL 484
Query: 161 GIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGARG 220
G+A+GLEYLH GC +RI+H DIKPQN+LLD++ CPK+SDFGLAK+C++K+SI+S+L RG
Sbjct: 485 GVARGLEYLHYGCKTRIVHFDIKPQNVLLDDNLCPKVSDFGLAKLCEKKESILSLLDTRG 544
Query: 221 TIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRK--NYDTGG-SCTSEMYFPDWIY 277
TIGY+APE+ SR +G VS++SDVYSYGML+LEMIG RK +D S S +YFP+WIY
Sbjct: 545 TIGYIAPEMISRLYGSVSHKSDVYSYGMLVLEMIGARKKERFDQNSRSDGSSIYFPEWIY 604
Query: 278 KDLEQGNTL---------LNCSTISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEML 328
KDLE+ N L + IS EE ++ RK+TLV LWCIQ+ PSDRPPMNKV+EM+
Sbjct: 605 KDLEKANIKDIEKTENGGLIENGISSEEEEIARKMTLVGLWCIQSSPSDRPPMNKVVEMM 664
Query: 329 QGPLSSVSYPPKPVLYSPERPELQVSDMSSSDLY 362
+G L ++ PP+PVL Q+S S SD +
Sbjct: 665 EGSLDALEVPPRPVLQ-------QISASSVSDSF 691
>At1g66930 receptor serine/threonine kinase PR5K, putative
Length = 876
Score = 358 bits (919), Expect = 3e-99
Identities = 175/308 (56%), Positives = 239/308 (76%), Gaps = 10/308 (3%)
Query: 42 RRYSYAEVKMITNYFREKLGQGGYGVVYKASLYNSRQVAVKVISETKGNG-EEFINEVAS 100
+ Y+YA+VK +T F E +G+GG+G+VY+ +L + R VAVKV+ E+KGN E+FINEV+S
Sbjct: 536 KHYTYAQVKRMTKSFAEVVGRGGFGIVYRGTLCDGRMVAVKVLKESKGNNSEDFINEVSS 595
Query: 101 ISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIAI 160
+S+TSH+NIVSLLG+C E ++RA+IYEF+ GSLDKFI SE + I D L+ IA+
Sbjct: 596 MSQTSHVNIVSLLGFCSEGSRRAIIYEFLENGSLDKFI--SEKTSVILDL--TALYGIAL 651
Query: 161 GIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGARG 220
G+A+GLEYLH GC +RI+H DIKPQN+LLD++ PK+SDFGLAK+C++K+S++S++ RG
Sbjct: 652 GVARGLEYLHYGCKTRIVHFDIKPQNVLLDDNLSPKVSDFGLAKLCEKKESVMSLMDTRG 711
Query: 221 TIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSCT--SEMYFPDWIYK 278
TIGY+APE+ SR +G VS++SDVYSYGML+ EMIG RK G + S MYFP+WIYK
Sbjct: 712 TIGYIAPEMISRVYGSVSHKSDVYSYGMLVFEMIGARKKERFGQNSANGSSMYFPEWIYK 771
Query: 279 DLEQGNT--LLNCST-ISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQGPLSSV 335
DLE+ + L + IS EE ++ +K+TLV LWCIQ+ PSDRPPMNKV+EM++G L ++
Sbjct: 772 DLEKADNGDLEHIEIGISSEEEEIAKKMTLVGLWCIQSSPSDRPPMNKVVEMMEGSLDAL 831
Query: 336 SYPPKPVL 343
PP+PVL
Sbjct: 832 EVPPRPVL 839
>At5g39030 receptor protein kinase - like protein
Length = 806
Score = 355 bits (910), Expect = 3e-98
Identities = 168/304 (55%), Positives = 233/304 (76%), Gaps = 7/304 (2%)
Query: 44 YSYAEVKMITNYFREKLGQGGYGVVYKASLYNSRQVAVKVISETKGNGEEFINEVASISR 103
Y+YAE+K IT F +G+GG+G VY +L N R+VAVKV+ + KG+ E+FINEVAS+S+
Sbjct: 488 YTYAELKKITKSFSYIIGKGGFGTVYGGNLSNGRKVAVKVLKDLKGSAEDFINEVASMSQ 547
Query: 104 TSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIAIGIA 163
TSH+NIVSLLG+C+E +KRA++YEF+ GSLD+F+ + N D TL+ IA+GIA
Sbjct: 548 TSHVNIVSLLGFCFEGSKRAIVYEFLENGSLDQFMSR----NKSLTQDVTTLYGIALGIA 603
Query: 164 KGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGARGTIG 223
+GLEYLH GC +RI+H DIKPQNILLD + CPK+SDFGLAK+C++++S++S++ RGTIG
Sbjct: 604 RGLEYLHYGCKTRIVHFDIKPQNILLDGNLCPKVSDFGLAKLCEKRESVLSLMDTRGTIG 663
Query: 224 YMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKN--YDTGGSCTSEMYFPDWIYKDLE 281
Y+APE+FSR +G VS++SDVYS+GML+++MIG R +T S S YFPDWIYKDLE
Sbjct: 664 YIAPEVFSRMYGRVSHKSDVYSFGMLVIDMIGARSKEIVETVDSAASSTYFPDWIYKDLE 723
Query: 282 QG-NTLLNCSTISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQGPLSSVSYPPK 340
G T + I++EE ++ +K+ +V LWCIQ PSDRP MN+V+EM++G L ++ PPK
Sbjct: 724 DGEQTWIFGDEITKEEKEIAKKMIVVGLWCIQPCPSDRPSMNRVVEMMEGSLDALEIPPK 783
Query: 341 PVLY 344
P ++
Sbjct: 784 PSMH 787
>At5g38240 receptor serine/threonine protein kinase -like
Length = 588
Score = 340 bits (873), Expect = 6e-94
Identities = 167/318 (52%), Positives = 235/318 (73%), Gaps = 17/318 (5%)
Query: 57 REKLGQGGYGVVYKASLYNSRQVAVKVISETKGNGEEFINEVASISRTSHMNIVSLLGYC 116
+E +G+GG+G VYK +L + R+VAVK++ ++ GN E+FINEVASIS+TSH+NIVSLLG+C
Sbjct: 284 QEVVGRGGFGTVYKGNLRDGRKVAVKILKDSNGNCEDFINEVASISQTSHVNIVSLLGFC 343
Query: 117 YEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIAIGIAKGLEYLHQGCSSR 176
+E++KRA++YEF+ GSLD+ + D +TL+ IA+G+A+G+EYLH GC R
Sbjct: 344 FEKSKRAIVYEFLENGSLDQS----------SNLDVSTLYGIALGVARGIEYLHFGCKKR 393
Query: 177 ILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGARGTIGYMAPEIFSRAFGG 236
I+H DIKPQN+LLDE+ PK++DFGLAK+C++++SI+S+L RGTIGY+APE+FSR +G
Sbjct: 394 IVHFDIKPQNVLLDENLKPKVADFGLAKLCEKQESILSLLDTRGTIGYIAPELFSRVYGN 453
Query: 237 VSYRSDVYSYGMLILEMIGGR--KNYDTGGSCTSEMYFPDWIYKDLEQGNTL-LNCSTIS 293
VS++SDVYSYGML+LEM G R + S S YFPDWI+KDLE G+ + L ++
Sbjct: 454 VSHKSDVYSYGMLVLEMTGARNKERVQNADSNNSSAYFPDWIFKDLENGDYVKLLADGLT 513
Query: 294 EEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQGPLSSVSYPPKPVLYSPERPELQV 353
EE D+ +K+ LV LWCIQ +PSDRP MNKV+ M++G L S+ PPKP+L+ P +Q
Sbjct: 514 REEEDIAKKMILVGLWCIQFRPSDRPSMNKVVGMMEGNLDSLDPPPKPLLHMP----MQN 569
Query: 354 SDMSSSDLYETNSVTVSK 371
++ SS E +S S+
Sbjct: 570 NNAESSQPSEEDSSIYSE 587
>At5g38250 receptor serine/threonine protein kinase - like
Length = 579
Score = 327 bits (837), Expect = 8e-90
Identities = 164/305 (53%), Positives = 223/305 (72%), Gaps = 17/305 (5%)
Query: 66 GVVYKASLYNSRQVAVKVISETKGNGEEFINEVASISRTSHMNIVSLLGYCYEENKRALI 125
G + L + R+VAVKV+ ++KGN E+FINEVAS+S+TSH+NIV+LLG+CYE +KRA+I
Sbjct: 285 GTLRGGRLRDGRKVAVKVLKDSKGNCEDFINEVASMSQTSHVNIVTLLGFCYEGSKRAII 344
Query: 126 YEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQ 185
YEF+ GSLD+ + + D +TL+ IA+G+A+GLEYLH GC +RI+H DIKPQ
Sbjct: 345 YEFLENGSLDQSL----------NLDVSTLYGIALGVARGLEYLHYGCKTRIVHFDIKPQ 394
Query: 186 NILLDEDFCPKISDFGLAKICQRKDSIVSILGARGTIGYMAPEIFSRAFGGVSYRSDVYS 245
N+LLDE+ PK++DFGLAK+C++++SI+S+L RGTIGY+APE+FSR +G VS++SDVYS
Sbjct: 395 NVLLDENLRPKVADFGLAKLCEKQESILSLLDTRGTIGYIAPELFSRMYGSVSHKSDVYS 454
Query: 246 YGMLILEMIGGR--KNYDTGGSCTSEMYFPDWIYKDLEQ-GNTLLNCSTISEEENDMIRK 302
YGML+LEMIG R + S YFPDWIYKDLE NT L ++ EE +K
Sbjct: 455 YGMLVLEMIGARNKERVQNADPNNSSAYFPDWIYKDLENFDNTRLLGDGLTREEEKNAKK 514
Query: 303 ITLVSLWCIQTKPSDRPPMNKVIEMLQGPLSSVSYPPKPVLYSPERPELQVSDMSSSDLY 362
+ LV LWCIQ +PSDRP MNKV+EM++G L S+ PPKP+L+ P +Q ++ SS L
Sbjct: 515 MILVGLWCIQFRPSDRPSMNKVVEMMEGSLDSLDPPPKPLLHMP----MQNNNAESSQLS 570
Query: 363 ETNSV 367
+S+
Sbjct: 571 VESSI 575
>At5g24080 receptor-like protein kinase
Length = 872
Score = 255 bits (652), Expect = 2e-68
Identities = 134/306 (43%), Positives = 193/306 (62%), Gaps = 13/306 (4%)
Query: 41 PRRYSYAEVKMITNYFREKLGQGGYGVVYKASLYNSRQVAVKVISETKGNGE-EFINEVA 99
P ++Y +++ TN F + LG GG+G VYK ++ VAVK + +GE EFI EV
Sbjct: 517 PVSFTYRDLQNCTNNFSQLLGSGGFGTVYKGTVAGETLVAVKRLDRALSHGEREFITEVN 576
Query: 100 SISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIA 159
+I HMN+V L GYC E++ R L+YE+M GSLDK+I+ SE + DW T F IA
Sbjct: 577 TIGSMHHMNLVRLCGYCSEDSHRLLVYEYMINGSLDKWIFSSEQTANL--LDWRTRFEIA 634
Query: 160 IGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGAR 219
+ A+G+ Y H+ C +RI+H DIKP+NILLD++FCPK+SDFGLAK+ R+ S V + R
Sbjct: 635 VATAQGIAYFHEQCRNRIIHCDIKPENILLDDNFCPKVSDFGLAKMMGREHSHV-VTMIR 693
Query: 220 GTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSCTSEMYFPDWIYKD 279
GT GY+APE S ++ ++DVYSYGML+LE++GGR+N D + ++P W YK+
Sbjct: 694 GTRGYLAPEWVSNR--PITVKADVYSYGMLLLEIVGGRRNLDMSYD-AEDFFYPGWAYKE 750
Query: 280 LEQGNTL----LNCSTISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQGPLSSV 335
L G +L ++EEE + K V+ WCIQ + S RP M +V+++L+G +
Sbjct: 751 LTNGTSLKAVDKRLQGVAEEEE--VVKALKVAFWCIQDEVSMRPSMGEVVKLLEGTSDEI 808
Query: 336 SYPPKP 341
+ PP P
Sbjct: 809 NLPPMP 814
>At1g34300 putative protein
Length = 829
Score = 244 bits (624), Expect = 4e-65
Identities = 123/294 (41%), Positives = 187/294 (62%), Gaps = 10/294 (3%)
Query: 41 PRRYSYAEVKMITNYFREKLGQGGYGVVYKASLYNSRQVAVKVISETKGNGEEFINEVAS 100
P +++Y E++ T F+EKLG GG+G VY+ L N VAVK + + ++F EVA+
Sbjct: 471 PVQFTYKELQRCTKSFKEKLGAGGFGTVYRGVLTNRTVVAVKQLEGIEQGEKQFRMEVAT 530
Query: 101 ISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIAI 160
IS T H+N+V L+G+C + R L+YEFM GSLD F++ ++ + W F IA+
Sbjct: 531 ISSTHHLNLVRLIGFCSQGRHRLLVYEFMRNGSLDNFLFTTDSAKFL---TWEYRFNIAL 587
Query: 161 GIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGARG 220
G AKG+ YLH+ C I+H DIKP+NIL+D++F K+SDFGLAK+ KD+ ++ RG
Sbjct: 588 GTAKGITYLHEECRDCIVHCDIKPENILVDDNFAAKVSDFGLAKLLNPKDNRYNMSSVRG 647
Query: 221 TIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSCTSEMYFPDWIYKDL 280
T GY+APE + ++ +SDVYSYGM++LE++ G++N+D T+ F W Y++
Sbjct: 648 TRGYLAPEWLANL--PITSKSDVYSYGMVLLELVSGKRNFDVSEK-TNHKKFSIWAYEEF 704
Query: 281 EQGNTLLNCST-ISEEEN---DMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQG 330
E+GNT T +SE++ + + ++ S WCIQ +P RP M KV++ML+G
Sbjct: 705 EKGNTKAILDTRLSEDQTVDMEQVMRMVKTSFWCIQEQPLQRPTMGKVVQMLEG 758
>At4g32300 S-receptor kinase -like protein
Length = 821
Score = 240 bits (612), Expect = 1e-63
Identities = 132/316 (41%), Positives = 198/316 (61%), Gaps = 16/316 (5%)
Query: 37 NLS-MPRRYSYAEVKMITNYFREKLGQGGYGVVYKASLYNSRQVAVKVISETKGNGEEFI 95
NLS MP R++Y +++ TN F KLGQGG+G VY+ +L + ++AVK + +EF
Sbjct: 475 NLSGMPIRFAYKDLQSATNNFSVKLGQGGFGSVYEGTLPDGSRLAVKKLEGIGQGKKEFR 534
Query: 96 NEVASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTL 155
EV+ I H+++V L G+C E R L YEF+ KGSL+++I++ + + + DW+T
Sbjct: 535 AEVSIIGSIHHLHLVRLRGFCAEGAHRLLAYEFLSKGSLERWIFRKKDGDVL--LDWDTR 592
Query: 156 FRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSI 215
F IA+G AKGL YLH+ C +RI+H DIKP+NILLD++F K+SDFGLAK+ R+ S V
Sbjct: 593 FNIALGTAKGLAYLHEDCDARIVHCDIKPENILLDDNFNAKVSDFGLAKLMTREQSHV-F 651
Query: 216 LGARGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSCTSEMYFPDW 275
RGT GY+APE + +S +SDVYSYGM++LE+IGGRKNYD + + +FP +
Sbjct: 652 TTMRGTRGYLAPEWITNY--AISEKSDVYSYGMVLLELIGGRKNYDP-SETSEKCHFPSF 708
Query: 276 IYKDLEQGNTL------LNCSTISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQ 329
+K +E+G + + +++E +++ +LWCIQ RP M+KV++ML+
Sbjct: 709 AFKKMEEGKLMDIVDGKMKNVDVTDER---VQRAMKTALWCIQEDMQTRPSMSKVVQMLE 765
Query: 330 GPLSSVSYPPKPVLYS 345
G V P + S
Sbjct: 766 GVFPVVQPPSSSTMGS 781
>At4g00340 receptor-like protein kinase
Length = 797
Score = 225 bits (573), Expect = 3e-59
Identities = 131/311 (42%), Positives = 193/311 (61%), Gaps = 20/311 (6%)
Query: 44 YSYAEVKMITNYFREKLGQGGYGVVYKASLYNSRQ-VAVKVISETKGNGE-EFINEVASI 101
+S+ E++ TN F +K+G GG+G V+K +L S VAVK + E G+GE EF EV +I
Sbjct: 451 FSFKELQSATNGFSDKVGHGGFGAVFKGTLPGSSTFVAVKRL-ERPGSGESEFRAEVCTI 509
Query: 102 SRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIAIG 161
H+N+V L G+C E R L+Y++MP+GSL ++ ++ P + W T FRIA+G
Sbjct: 510 GNIQHVNLVRLRGFCSENLHRLLVYDYMPQGSLSSYLSRTS-PKLL---SWETRFRIALG 565
Query: 162 IAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGARGT 221
AKG+ YLH+GC I+H DIKP+NILLD D+ K+SDFGLAK+ R S V + RGT
Sbjct: 566 TAKGIAYLHEGCRDCIIHCDIKPENILLDSDYNAKVSDFGLAKLLGRDFSRV-LATMRGT 624
Query: 222 IGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNY----DTGGSCTSE---MYFPD 274
GY+APE S ++ ++DVYS+GM +LE+IGGR+N DT G +E +FP
Sbjct: 625 WGYVAPEWISGL--PITTKADVYSFGMTLLELIGGRRNVIVNSDTLGEKETEPEKWFFPP 682
Query: 275 WIYKDLEQGN--TLLNCSTISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQGPL 332
W +++ QGN ++++ E + + ++ V++WCIQ RP M V++ML+G +
Sbjct: 683 WAAREIIQGNVDSVVDSRLNGEYNTEEVTRMATVAIWCIQDNEEIRPAMGTVVKMLEG-V 741
Query: 333 SSVSYPPKPVL 343
V+ PP P L
Sbjct: 742 VEVTVPPPPKL 752
>At1g11350 putative receptor-like protein kinase gi|4008010; similar
to EST gb|AI999419.1
Length = 830
Score = 218 bits (555), Expect = 4e-57
Identities = 139/365 (38%), Positives = 201/365 (54%), Gaps = 19/365 (5%)
Query: 17 ILRERTELVDNNV--EVFMRSHNLSMPRRYSYAEVKMITNYFR--EKLGQGGYGVVYKAS 72
+L ER E + +N + + + L + + + + TN F KLGQGG+G VYK
Sbjct: 471 LLNERMEALSSNDVGAILVNQYKLKELPLFEFQVLAVATNNFSITNKLGQGGFGAVYKGR 530
Query: 73 LYNSRQVAVKVISETKGNG-EEFINEVASISRTSHMNIVSLLGYCYEENKRALIYEFMPK 131
L +AVK +S T G G EEF+NEV IS+ H N+V LLG+C E +R L+YEFMP+
Sbjct: 531 LQEGLDIAVKRLSRTSGQGVEEFVNEVVVISKLQHRNLVRLLGFCIEGEERMLVYEFMPE 590
Query: 132 GSLDKFIYKSEFPNAICDFDWNTLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLDE 191
LD +++ + DW T F I GI +GL YLH+ +I+H D+K NILLDE
Sbjct: 591 NCLDAYLFDPVKQRLL---DWKTRFNIIDGICRGLMYLHRDSRLKIIHRDLKASNILLDE 647
Query: 192 DFCPKISDFGLAKICQRKDSIVSILGARGTIGYMAPEIFSRAFGGV-SYRSDVYSYGMLI 250
+ PKISDFGLA+I Q + VS + GT GYMAPE A GG+ S +SDV+S G+++
Sbjct: 648 NLNPKISDFGLARIFQGNEDEVSTVRVVGTYGYMAPEY---AMGGLFSEKSDVFSLGVIL 704
Query: 251 LEMIGGRKN---YDTGGSCTSEMYFPDWIYKDLEQGNTLLNCSTISEEENDMIRKITLVS 307
LE++ GR+N Y+ G + Y W + + L++ E + IR+ V
Sbjct: 705 LEIVSGRRNSSFYNDGQNPNLSAY--AWKLWNTGEDIALVDPVIFEECFENEIRRCVHVG 762
Query: 308 LWCIQTKPSDRPPMNKVIEMLQGPLSSVSYPPKPVLYSPERPELQVSDMSSSD-LYETNS 366
L C+Q +DRP + VI ML S++ P +P + P R +V SD N+
Sbjct: 763 LLCVQDHANDRPSVATVIWMLSSENSNLPEPKQPA-FIPRRGTSEVESSGQSDPRASINN 821
Query: 367 VTVSK 371
V+++K
Sbjct: 822 VSLTK 826
>At2g19130 putative receptor-like protein kinase
Length = 828
Score = 217 bits (552), Expect = 9e-57
Identities = 119/305 (39%), Positives = 180/305 (59%), Gaps = 16/305 (5%)
Query: 44 YSYAEVKMITNYFREKLGQGGYGVVYKASLYNSRQVAVKVISETKGNGEEFINEVASISR 103
+SY E++ T F +KLG GG+G V+K +L +S +AVK + ++F EV +I
Sbjct: 483 FSYRELQNATKNFSDKLGGGGFGSVFKGALPDSSDIAVKRLEGISQGEKQFRTEVVTIGT 542
Query: 104 TSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIAIGIA 163
H+N+V L G+C E +K+ L+Y++MP GSLD ++ ++ I W F+IA+G A
Sbjct: 543 IQHVNLVRLRGFCSEGSKKLLVYDYMPNGSLDSHLFLNQVEEKIV-LGWKLRFQIALGTA 601
Query: 164 KGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGARGTIG 223
+GL YLH C I+H DIKP+NILLD FCPK++DFGLAK+ R S V + RGT G
Sbjct: 602 RGLAYLHDECRDCIIHCDIKPENILLDSQFCPKVADFGLAKLVGRDFSRV-LTTMRGTRG 660
Query: 224 YMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSCTSEMYFPDWIYKDLEQG 283
Y+APE S ++ ++DVYSYGM++ E++ GR+N + + +FP W L +
Sbjct: 661 YLAPEWISGV--AITAKADVYSYGMMLFELVSGRRNTEQSEN-EKVRFFPSWAATILTKD 717
Query: 284 NTL-------LNCSTISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQGPLSSVS 336
+ L + EE + + V+ WCIQ + S RP M++V+++L+G L V+
Sbjct: 718 GDIRSLVDPRLEGDAVDIEE---VTRACKVACWCIQDEESHRPAMSQVVQILEGVL-EVN 773
Query: 337 YPPKP 341
PP P
Sbjct: 774 PPPFP 778
>At1g56120 receptor-like protein kinase, putative
Length = 1045
Score = 210 bits (535), Expect = 9e-55
Identities = 124/332 (37%), Positives = 197/332 (58%), Gaps = 21/332 (6%)
Query: 17 ILRERTELVDNNVEVFMRSHNLSM---PRRYSYAEVKMITNYF--REKLGQGGYGVVYKA 71
++R+R + ++ E+ LSM P ++Y+E+K T F KLG+GG+G VYK
Sbjct: 672 VIRKRRKPYTDDEEI------LSMDVKPYTFTYSELKNATQDFDLSNKLGEGGFGAVYKG 725
Query: 72 SLYNSRQVAVKVISETKGNGE-EFINEVASISRTSHMNIVSLLGYCYEENKRALIYEFMP 130
+L + R+VAVK +S G+ +F+ E+ +IS H N+V L G C+E + R L+YE++P
Sbjct: 726 NLNDGREVAVKQLSIGSRQGKGQFVAEIIAISSVLHRNLVKLYGCCFEGDHRLLVYEYLP 785
Query: 131 KGSLDKFIYKSEFPNAICDFDWNTLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLD 190
GSLD+ + F + DW+T + I +G+A+GL YLH+ S RI+H D+K NILLD
Sbjct: 786 NGSLDQAL----FGDKSLHLDWSTRYEICLGVARGLVYLHEEASVRIIHRDVKASNILLD 841
Query: 191 EDFCPKISDFGLAKICQRKDSIVSILGARGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLI 250
+ PK+SDFGLAK+ K + +S A GTIGY+APE R G ++ ++DVY++G++
Sbjct: 842 SELVPKVSDFGLAKLYDDKKTHISTRVA-GTIGYLAPEYAMR--GHLTEKTDVYAFGVVA 898
Query: 251 LEMIGGRKNYDTGGSCTSEMYFPDWIYKDLEQGNTL-LNCSTISEEENDMIRKITLVSLW 309
LE++ GRKN D + Y +W + E+ + L +SE + ++++ ++L
Sbjct: 899 LELVSGRKNSDENLE-EGKKYLLEWAWNLHEKNRDVELIDDELSEYNMEEVKRMIGIALL 957
Query: 310 CIQTKPSDRPPMNKVIEMLQGPLSSVSYPPKP 341
C Q+ + RPPM++V+ ML G KP
Sbjct: 958 CTQSSYALRPPMSRVVAMLSGDAEVNDATSKP 989
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.136 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,521,459
Number of Sequences: 26719
Number of extensions: 383230
Number of successful extensions: 4730
Number of sequences better than 10.0: 1013
Number of HSP's better than 10.0 without gapping: 842
Number of HSP's successfully gapped in prelim test: 171
Number of HSP's that attempted gapping in prelim test: 1048
Number of HSP's gapped (non-prelim): 1106
length of query: 371
length of database: 11,318,596
effective HSP length: 101
effective length of query: 270
effective length of database: 8,619,977
effective search space: 2327393790
effective search space used: 2327393790
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC147875.1


