

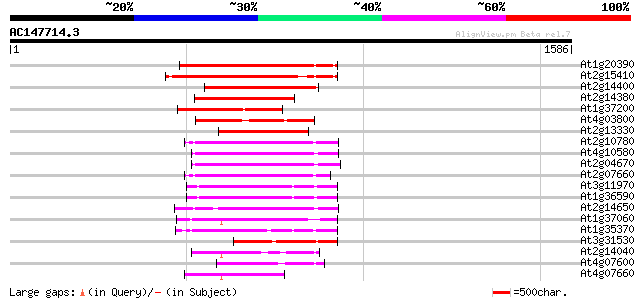
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147714.3 - phase: 0 /pseudo
(1586 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g20390 hypothetical protein 405 e-113
At2g15410 putative retroelement pol polyprotein 367 e-101
At2g14400 putative retroelement pol polyprotein 317 3e-86
At2g14380 putative retroelement pol polyprotein 288 1e-77
At1g37200 hypothetical protein 279 1e-74
At4g03800 268 1e-71
At2g13330 F14O4.9 266 7e-71
At2g10780 pseudogene 250 4e-66
At4g10580 putative reverse-transcriptase -like protein 245 2e-64
At2g04670 putative retroelement pol polyprotein 244 2e-64
At2g07660 putative retroelement pol polyprotein 242 1e-63
At3g11970 hypothetical protein 233 7e-61
At1g36590 hypothetical protein 233 7e-61
At2g14650 putative retroelement pol polyprotein 233 9e-61
At1g37060 Athila retroelment ORF 1, putative 229 1e-59
At1g35370 hypothetical protein 224 3e-58
At3g31530 hypothetical protein 223 7e-58
At2g14040 putative retroelement pol polyprotein 216 8e-56
At4g07600 194 3e-49
At4g07660 putative athila transposon protein 186 9e-47
>At1g20390 hypothetical protein
Length = 1791
Score = 405 bits (1042), Expect = e-113
Identities = 199/448 (44%), Positives = 294/448 (65%), Gaps = 3/448 (0%)
Query: 480 ELINLGTEENKREIKVGAALEEGVKRKIFQLL*EYPDIFAWSYEDMPGLDPKIVEHRIPT 539
E++N+ + R + VGA + ++ ++ LL FAWS EDM G+DP I H +
Sbjct: 712 EMVNIDESDPTRCVGVGAEISPSIRLELIALLKRNSKTFAWSIEDMKGIDPAITAHELNV 771
Query: 540 KPECPPVRQKLRRTHPDMALKIKSEV*KQIDAGFLMTIEYPEWVANIVPVPKKDGKVRMC 599
P PV+QK R+ P+ A + EV K + AG ++ ++YPEW+AN V V KK+GK R+C
Sbjct: 772 DPTFKPVKQKRRKLGPERARAVNEEVEKLLKAGQIIEVKYPEWLANPVVVKKKNGKWRVC 831
Query: 600 VDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSFITP 659
VD+ DLNKA PKD++PLPHID LV+ T+ + + SFMD FSGYNQI M +D+EKTSF+T
Sbjct: 832 VDYTDLNKACPKDSYPLPHIDRLVEATSGNGLLSFMDAFSGYNQILMHKDDQEKTSFVTD 891
Query: 660 WGTFCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHVEYLTKM 719
GT+CYKVM FGL NAGATYQR + + D I + VEVY+DDM+VKS E HVE+L+K
Sbjct: 892 RGTYCYKVMSFGLKNAGATYQRFVNKMLADQIGRTVEVYIDDMLVKSLKPEDHVEHLSKC 951
Query: 720 FERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREMPAPQTEKQVRGF 779
F+ L Y ++LNP KCTFGV SG+ LG++V+++GIE +P ++RAI E+P+P+ ++V+
Sbjct: 952 FDVLNTYGMKLNPTKCTFGVTSGEFLGYVVTKRGIEANPKQIRAILELPSPRNAREVQRL 1011
Query: 780 LRRLNYISRFISHMTATCGPIFKLLRKNQPIVWNDECQGAFDSIKNYLLEPPILVPPMEG 839
R+ ++RFIS T C P + LL++ W+ + + AF+ +K+YL PPILV P G
Sbjct: 1012 TGRIAALNRFISRSTDKCLPFYNLLKRRAQFDWDKDSEEAFEKLKDYLSTPPILVKPEVG 1071
Query: 840 KPLIMYLSVFDESVGCVLGQQDETGKKEHAIYYLSKKFTDCETRYMMLEKTCCAPAWAAK 899
+ L +Y++V D +V VL ++D ++ I+Y SK + ETRY ++EK A +A+
Sbjct: 1072 ETLYLYIAVSDHAVSSVLVREDR--GEQRPIFYTSKSLVEAETRYPVIEKAALAVVTSAR 1129
Query: 900 RLRHYLVNHTTWLISRMDPIKYIFEKPA 927
+LR Y +HT +++ P++ P+
Sbjct: 1130 KLRPYFQSHTIAVLTD-QPLRVALHSPS 1156
>At2g15410 putative retroelement pol polyprotein
Length = 1787
Score = 367 bits (941), Expect = e-101
Identities = 190/486 (39%), Positives = 293/486 (60%), Gaps = 30/486 (6%)
Query: 442 FEFPVYEAEDEEGDDIPYEITRLLEQERKAIQPHQEEIELINLGTEENKREIKVGAALEE 501
FEF V + I Y + + Q+ + P + + +N+ + R + + L
Sbjct: 686 FEFAVVDKP------IIYNVILVPTQDERP-NPQKGTVVQVNIDESDPSRCVGIRIDLPS 738
Query: 502 GVKRKIFQLL*EYPDIFAWSYEDMPGLDPKIVEHRIPTKPECPPVRQKLRRTHPDMALKI 561
++ ++ L + FAWS EDMPG+D + H + P P++QK R+ PD +
Sbjct: 739 ELQNELVNFLRQNAATFAWSVEDMPGIDSAVTCHELNVDPTYKPLKQKRRKLGPDRTKDV 798
Query: 562 KSEV*KQIDAGFLMTIEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLPHIDV 621
EV K +DAG ++ + YP+W+ N V V KK+GK R+C+DF DLNKA PKD+FPLPHID
Sbjct: 799 NEEVKKLLDAGSIVEVRYPDWLRNPVVVKKKNGKWRVCIDFTDLNKACPKDSFPLPHIDR 858
Query: 622 LVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPFGLINAGATYQR 681
LV+ TA +++ SFMD FSGYNQI M DREKT FIT GT+CYKVMPFGL NAGATY R
Sbjct: 859 LVEATAGNELLSFMDAFSGYNQILMHQNDREKTVFITDQGTYCYKVMPFGLKNAGATYPR 918
Query: 682 GMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRS 741
+ +F D + +EVY+DDM+VKS E+H+ +L + F+ L +Y ++LNP+KCTFGV S
Sbjct: 919 LVNQMFTDQLDHSMEVYIDDMLVKSLRAEEHITHLRQCFQVLNRYNMKLNPSKCTFGVTS 978
Query: 742 GKLLGFIVSQKGIEVDPDKVRAIREMPAPQTEKQVRGFLRRLNYISRFISHMTATCGPIF 801
G+ LG++V+++GIE +P ++ AI ++P+P+ ++V+ + R+ ++RFIS T C P +
Sbjct: 979 GEFLGYLVTRRGIEANPKQISAIIDLPSPRNTREVQRLIGRIAALNRFISRSTDKCLPFY 1038
Query: 802 KLLRKNQPIVWNDECQGAFDSIKNYLLEPPILVPPMEGKPLIMYLSVFDESVGCVLGQQD 861
+LLR N+ W+++C+ EG+ L +Y++V +V VL ++D
Sbjct: 1039 QLLRANKRFEWDEKCE--------------------EGETLYLYIAVSTSAVSGVLVRED 1078
Query: 862 ETGKKEHAIYYLSKKFTDCETRYMMLEKTCCAPAWAAKRLRHYLVNHTTWLISRMDPIKY 921
++H I+Y+SK E RY LEK A +A++LR Y ++T +++ P++
Sbjct: 1079 R--GEQHPIFYVSKTLDGAELRYPTLEKLAFAVVISARKLRPYFKSYTVEVLTN-QPLRT 1135
Query: 922 IFEKPA 927
I P+
Sbjct: 1136 ILHSPS 1141
>At2g14400 putative retroelement pol polyprotein
Length = 1466
Score = 317 bits (812), Expect = 3e-86
Identities = 152/323 (47%), Positives = 226/323 (69%), Gaps = 2/323 (0%)
Query: 551 RRTHPDMALKIKSEV*KQIDAGFLMTIEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASP 610
R+ + A + EV K + G + ++YP+W+AN V V KK+GK R+C+DF DLNKA P
Sbjct: 470 RKLGVERAKAVNDEVDKLLKIGSIREVQYPDWLANTVVVKKKNGKDRVCIDFTDLNKACP 529
Query: 611 KDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPF 670
KD+FPLPHID LV++TA +++ SFMD FSGYNQI M+PED+EKT FIT G +CYKVMPF
Sbjct: 530 KDSFPLPHIDRLVESTAGNELLSFMDAFSGYNQIMMNPEDQEKTLFITDRGIYCYKVMPF 589
Query: 671 GLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHVEYLTKMFERLRKYKLRL 730
GL NAGATY R + +F + + K +EVY+DDM++KS +E HV++L + F L +Y+++L
Sbjct: 590 GLRNAGATYPRLVNKMFSEHVGKTMEVYIDDMLIKSLKKEDHVKHLEECFAILNQYQMKL 649
Query: 731 NPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREMPAPQTEKQVRGFLRRLNYISRFI 790
NP KCTFGV SG+ LG+IV+++GIE +P+++ A MP+P+ K+V+ R+ ++RFI
Sbjct: 650 NPAKCTFGVPSGEFLGYIVTKRGIEANPNQINAFLNMPSPKNFKEVQRLTGRIAALNRFI 709
Query: 791 SHMTATCGPIFKLLRKNQPIVWNDECQGAFDSIKNYLLEPPILVPPMEGKPLIMYLSVFD 850
S T P +++L+ N+ +W+++C+ AF +K YL PP+L P + L +Y+SV +
Sbjct: 710 SRSTDKSLPFYQILKGNKEFLWDEKCEEAFGQLKAYLTSPPVLFKPELDEKLYLYVSVSN 769
Query: 851 ESVGCVLGQQDETGKKEHAIYYL 873
+V VL ++D +K IYY+
Sbjct: 770 HAVSGVLIREDRGEQK--PIYYI 790
>At2g14380 putative retroelement pol polyprotein
Length = 764
Score = 288 bits (738), Expect = 1e-77
Identities = 137/282 (48%), Positives = 196/282 (68%)
Query: 524 DMPGLDPKIVEHRIPTKPECPPVRQKLRRTHPDMALKIKSEV*KQIDAGFLMTIEYPEWV 583
DM G+DP++ H + P V+QK R+ P+ + + EV K +DAG ++ ++YPEW+
Sbjct: 476 DMVGIDPEVACHELNVDPTFKLVKQKRRKLGPERSKAVNDEVDKLLDAGSIVEVKYPEWL 535
Query: 584 ANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQ 643
AN V V KK+ K R+C+DF DLNKA PKD+FPLPHID +V+ T +++ SFMD FSGYNQ
Sbjct: 536 ANPVVVKKKNDKWRVCIDFTDLNKACPKDSFPLPHIDRMVEATTGNELLSFMDAFSGYNQ 595
Query: 644 IKMSPEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMI 703
I M +D+EKTSFI GT+CYKVMPFGL N GA YQR + +F + K +EVY+DDM+
Sbjct: 596 IPMHKDDQEKTSFIIDRGTYCYKVMPFGLKNVGARYQRLVNQMFAPQLGKTMEVYIDDML 655
Query: 704 VKSKDEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRA 763
VKS H+++L FE L KY ++LNP KC FGV SG+ LG+IV+++GIE +P ++RA
Sbjct: 656 VKSTRSADHIDHLKACFETLNKYNMKLNPAKCLFGVTSGEFLGYIVTKRGIEANPKQIRA 715
Query: 764 IREMPAPQTEKQVRGFLRRLNYISRFISHMTATCGPIFKLLR 805
I ++ +P+ +K+V+ R+ ++RFI+ T P ++LLR
Sbjct: 716 ILDLQSPRNKKEVQRLTGRIAGLNRFIARSTDKSLPFYQLLR 757
>At1g37200 hypothetical protein
Length = 1564
Score = 279 bits (713), Expect = 1e-74
Identities = 133/298 (44%), Positives = 197/298 (65%), Gaps = 4/298 (1%)
Query: 474 PHQEEIELINLGTEENKREIKVGAALEEGVKRKIFQLL*EYPDIFAWSYEDMPGLDPKIV 533
P + I + + + K+ + VG L+ ++ L E D FAWS ++ G+ ++
Sbjct: 614 PQKSSITQVYIDESDPKQCVGVGQDLDPAIREDFITFLKENKDSFAWSSANLQGISLEVT 673
Query: 534 EHRIPTKPECPPVRQKLRRTHPDMALKIKSEV*KQIDAGFLMTIEYPEWVANIVPVPKKD 593
H + P P++QK R+ P+ A ++ EV + + G + ++YP+W+AN V V K+
Sbjct: 674 SHELNVDPTYRPIKQKRRKLGPERAKAVQDEVDRLLKIGSIREVKYPDWLANPVVVKNKN 733
Query: 594 GKVRMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREK 653
GK R+C+DF DLNKA PKD+FPLPHID LV TA ++ SFMD FSGYNQI M P+D+EK
Sbjct: 734 GKWRVCIDFMDLNKACPKDSFPLPHIDRLVKATAGHELLSFMDAFSGYNQILMRPDDQEK 793
Query: 654 TSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHV 713
T+FIT CYKVMPFGL N GATYQR + +F D + K +E+Y+DDM+VKS E+ H+
Sbjct: 794 TAFITD----CYKVMPFGLKNTGATYQRLVNRMFADQLGKTMELYIDDMLVKSAHEKDHL 849
Query: 714 EYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREMPAPQ 771
L + F+ L K++++LNP KC+FGV SG+ LG++V+Q+GIE +P ++ A +MP+P+
Sbjct: 850 PQLRECFKILNKFEMKLNPEKCSFGVPSGEFLGYLVTQRGIEANPKQIAAFIDMPSPK 907
Score = 42.7 bits (99), Expect = 0.002
Identities = 24/62 (38%), Positives = 34/62 (54%), Gaps = 3/62 (4%)
Query: 861 DETGKKEHAIYYLSKKFTDCETRYMMLEKTCCAPAWAAKRLRHYLVNHTTWLISRMDPIK 920
D G+K IYY+SK D ET Y +EK A +A++LR YL +H ++ PI+
Sbjct: 927 DHEGQKP--IYYVSKTLIDAETHYRAMEKLALAVVMSARKLRQYLQSHPI-VVMTSQPIR 983
Query: 921 YI 922
I
Sbjct: 984 TI 985
>At4g03800
Length = 637
Score = 268 bits (686), Expect = 1e-71
Identities = 136/337 (40%), Positives = 212/337 (62%), Gaps = 21/337 (6%)
Query: 525 MPGLDPKIVEHRIPTKPECPPVRQKLRRTHPDMALKIKSEV*KQIDAGFLMTIEYPEWVA 584
MP +DP I+ H + P P++QK R+ + A + ++ K + G + ++YP+WVA
Sbjct: 321 MPDIDPSIICHELNVDPRFKPLKQKRRKLGVERAKAVNGKIDKLLKIGSIREVQYPDWVA 380
Query: 585 NIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQI 644
V V KK+GK R+C+DF DLNKA PKD+FPLPHID LV++TA +++ +FMD F GYNQI
Sbjct: 381 ITVVVKKKNGKDRVCIDFTDLNKACPKDSFPLPHIDRLVESTAGNELLTFMDAFLGYNQI 440
Query: 645 KMSPEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIV 704
M+PED+EKTSFIT + GATYQ + +F++ + K +EV +DD +V
Sbjct: 441 MMNPEDQEKTSFIT---------------DRGATYQWLVNKMFNEHLRKTMEVSIDDTLV 485
Query: 705 KSKDEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAI 764
KS +E HV++L + FE L +Y+++LN KCTFGV SG+ LG+IV+++GIE +P+++ A
Sbjct: 486 KSLKKEDHVKHLGECFEILNQYQMKLNLAKCTFGVPSGEFLGYIVTKRGIEANPNQINAF 545
Query: 765 REMPAPQTEKQVRGFLRRLNYISRFISHMTATCGPIFKLLRKNQPIVWNDECQGAFDSIK 824
+ P+ + K+V+ R S T P +++L+ N +W+++C+ AF +K
Sbjct: 546 LKTPSLRNFKEVQ------RLTGRIASRSTDKSLPFYQILKGNNGFLWDEKCEEAFRQLK 599
Query: 825 NYLLEPPILVPPMEGKPLIMYLSVFDESVGCVLGQQD 861
YL PP+L P + L +Y+ V + +V VL ++D
Sbjct: 600 AYLTTPPVLSKPEADEKLYLYVFVSNHAVSGVLVRED 636
>At2g13330 F14O4.9
Length = 889
Score = 266 bits (680), Expect = 7e-71
Identities = 126/255 (49%), Positives = 179/255 (69%)
Query: 591 KKDGKVRMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPED 650
+K+GK R+C+DFRDLNKA PKD+FPLPHID LV+ T + + SFMD F GYNQI M +
Sbjct: 289 EKNGKWRVCIDFRDLNKACPKDSFPLPHIDRLVEATVEHEKLSFMDAFYGYNQILMRRDG 348
Query: 651 REKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEE 710
+EKT+FIT GT+ YKVMPFGL NAG TYQR + +F D + K +EVY+DDM+VKS E+
Sbjct: 349 QEKTAFITDRGTYYYKVMPFGLKNAGTTYQRLVNRMFVDQLGKTIEVYIDDMLVKSAHEK 408
Query: 711 QHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREMPAP 770
HV L + F+ L K++++LNP KC+F V+S + L ++V+++GIE +P ++ A EMP+P
Sbjct: 409 DHVPQLRECFKILIKFEMKLNPEKCSFEVQSREFLEYLVTERGIEANPKQIAAFIEMPSP 468
Query: 771 QTEKQVRGFLRRLNYISRFISHMTATCGPIFKLLRKNQPIVWNDECQGAFDSIKNYLLEP 830
+ ++V+ R+ ++ FIS C P ++ LRK + WN +C+ AF +K YL EP
Sbjct: 469 KMAREVQRLTGRIPALNGFISRSANKCVPFYQPLRKGKEFDWNKDCEQAFKQLKAYLTEP 528
Query: 831 PILVPPMEGKPLIMY 845
PIL P +G+PL +Y
Sbjct: 529 PILAKPEKGEPLYLY 543
>At2g10780 pseudogene
Length = 1611
Score = 250 bits (639), Expect = 4e-66
Identities = 149/438 (34%), Positives = 235/438 (53%), Gaps = 16/438 (3%)
Query: 494 KVGAALEEGVKRKIFQLL*EYPDIFAWSYEDMPGLDPKIVE-HRIPTKPECPPVRQKLRR 552
+VGA+ E K ++ E+ D+FA + G+ P + I +P P+ + R
Sbjct: 615 EVGASAE----LKDIPIVNEFSDVFA----AVSGVPPDRSDPFTIELEPGTTPISKAPYR 666
Query: 553 THPDMALKIKSEV*KQIDAGFLMTIEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASPKD 612
P K+K ++ + +D GF+ P W A ++ V KKDG R+C+D+R LNK + K+
Sbjct: 667 MAPAEMAKLKKQLEELLDKGFIRPSSSP-WGAPVLFVKKKDGSFRLCIDYRGLNKVTVKN 725
Query: 613 NFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPFGL 672
+PLP ID L+D ++ FS +D SGY+QI + P D KT+F T + F + VMPFGL
Sbjct: 726 KYPLPRIDELMDQLGGAQWFSKIDLASGYHQIPIEPTDVRKTAFRTRYDHFEFVVMPFGL 785
Query: 673 INAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHVEYLTKMFERLRKYKLRLNP 732
NA A + + M +F D + + V ++++D++V SK E H E+L + ERLR+++L
Sbjct: 786 TNAPAAFMKMMNGVFRDFLDEFVIIFINDILVYSKSWEAHQEHLRAVLERLREHELFAKL 845
Query: 733 NKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREMPAPQTEKQVRGFLRRLNYISRFISH 792
+KC+F RS LG ++S +G+ VDP+K+R+I+E P P+ ++R FL Y RF+
Sbjct: 846 SKCSFWQRSVGFLGHVISDQGVSVDPEKIRSIKEWPRPRNATEIRSFLGLAGYYRRFVMS 905
Query: 793 MTATCGPIFKLLRKNQPIVWNDECQGAFDSIKNYLLEPPILVPPMEGKPLIMYLSVFDES 852
+ P+ +L K+ W+DEC+ +F +K L P+LV P EG+P +Y
Sbjct: 906 FASMAQPLTRLTGKDTAFNWSDECEKSFLELKAMLTNAPVLVLPEEGEPYTVYTDASIVG 965
Query: 853 VGCVLGQQDETGKKEHAIYYLSKKFTDCETRYMMLEKTCCAPAWAAKRLRHYLVNHTTWL 912
+GCVL Q K I Y S++ E Y + A + K R YL +
Sbjct: 966 LGCVLMQ------KGSVIAYASRQLRKHEKNYPTHDLEMAAVVFFLKIWRSYLYGAKVQI 1019
Query: 913 ISRMDPIKYIFEKPAVLL 930
+ +KYIF +P + L
Sbjct: 1020 YTDHKSLKYIFTQPELNL 1037
>At4g10580 putative reverse-transcriptase -like protein
Length = 1240
Score = 245 bits (625), Expect = 2e-64
Identities = 142/419 (33%), Positives = 223/419 (52%), Gaps = 12/419 (2%)
Query: 513 EYPDIFAWSYEDMPGLDPKIVE-HRIPTKPECPPVRQKLRRTHPDMALKIKSEV*KQIDA 571
E+ D+F + + GL P + I +P P+ + R P ++K ++ +
Sbjct: 462 EFQDVF----QSLQGLPPSQSDPFTIELEPGTAPLSKAPYRMAPAEMAELKKQLKDLLGK 517
Query: 572 GFLMTIEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQSKV 631
GF+ P W A ++ V KKDG R+C+D+R+LN+ + K+ +PLP ID L+D +
Sbjct: 518 GFIRPSTSP-WGAPVLFVKKKDGSFRLCIDYRELNRVTVKNRYPLPRIDELLDQLRGATC 576
Query: 632 FSFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMI 691
FS +D SGY+QI ++ D KT+F T +G F + VMPFGL NA A + R M ++F + +
Sbjct: 577 FSKIDLTSGYHQIPIAEADVRKTAFRTRYGHFEFVVMPFGLTNAPAVFMRLMNSVFQEFL 636
Query: 692 HKEVEVYVDDMIVKSKDEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQ 751
+ V +++DD++V SK E+ +L ++ E+LR+ KL +KC+F R LG IVS
Sbjct: 637 DEFVIIFIDDILVYSKSPEEQEVHLRRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSA 696
Query: 752 KGIEVDPDKVRAIREMPAPQTEKQVRGFLRRLNYISRFISHMTATCGPIFKLLRKNQPIV 811
+G+ VDP+K+ AIR+ P P ++R FL Y RF+ + P+ KL K+ P V
Sbjct: 697 EGVSVDPEKIEAIRDWPRPTNATEIRSFLGWAGYYRRFVKGFASMAQPMTKLTGKDVPFV 756
Query: 812 WNDECQGAFDSIKNYLLEPPILVPPMEGKPLIMYLSVFDESVGCVLGQQDETGKKEHAIY 871
W+ EC+ F S+K L P+L P G+P ++Y +GCVL Q + I
Sbjct: 757 WSQECEEGFVSLKEMLTSTPVLALPEHGQPYMVYTDASRVGLGCVLMQHGK------VIA 810
Query: 872 YLSKKFTDCETRYMMLEKTCCAPAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKPAVLL 930
Y S++ E Y + A +A K R YL + + +KYIF +P + L
Sbjct: 811 YASRQLMKHEGNYPTHDLEMAAVIFALKIWRSYLYGGKVQVFTDHKSLKYIFTQPELNL 869
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 244 bits (624), Expect = 2e-64
Identities = 142/422 (33%), Positives = 224/422 (52%), Gaps = 12/422 (2%)
Query: 513 EYPDIFAWSYEDMPGLDPKIVE-HRIPTKPECPPVRQKLRRTHPDMALKIKSEV*KQIDA 571
E+ D+F + + GL P + I +P P+ + R P ++K ++ +
Sbjct: 488 EFEDVF----QSLQGLPPSRSDPFTIELEPGTAPLSKAPYRMAPAEMTELKKQLEDLLGK 543
Query: 572 GFLMTIEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQSKV 631
GF+ P W A ++ V KKDG R+C+D+R LN + K+ +PLP ID L+D +
Sbjct: 544 GFIRPSTSP-WGAPVLFVKKKDGSFRLCIDYRGLNWVTVKNKYPLPRIDELLDQLRGATC 602
Query: 632 FSFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMI 691
FS +D SGY+QI ++ D KT+F T +G F + VMPF L NA A + R M ++F + +
Sbjct: 603 FSKIDLTSGYHQIPIAEADVRKTAFRTRYGHFEFVVMPFALTNAPAAFMRLMNSVFQEFL 662
Query: 692 HKEVEVYVDDMIVKSKDEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQ 751
+ V +++DD++V SK E+H +L ++ E+LR+ KL +KC+F R LG IVS
Sbjct: 663 DEFVIIFIDDILVYSKSPEEHEVHLRRVMEKLREQKLFAKLSKCSFWQREIGFLGHIVSA 722
Query: 752 KGIEVDPDKVRAIREMPAPQTEKQVRGFLRRLNYISRFISHMTATCGPIFKLLRKNQPIV 811
+G+ VDP+K+ AIR+ P P ++R FLR Y RF+ + P+ KL K+ P V
Sbjct: 723 EGVSVDPEKIEAIRDWPRPTNATEIRSFLRLTGYYRRFVKGFASMAQPMTKLTGKDVPFV 782
Query: 812 WNDECQGAFDSIKNYLLEPPILVPPMEGKPLIMYLSVFDESVGCVLGQQDETGKKEHAIY 871
W+ EC+ F S+K L P+L P G+P ++Y +GCVL Q+ + I
Sbjct: 783 WSPECEEGFVSLKEMLTSTPVLALPEHGQPYMVYTDASRVGLGCVLMQRGK------VIA 836
Query: 872 YLSKKFTDCETRYMMLEKTCCAPAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKPAVLLG 931
Y S++ E Y + +A K R YL + + +KYIF +P + L
Sbjct: 837 YASRQLRKHEGNYPTHDLEMAVVIFALKIWRSYLYGGKVQVFTDHKSLKYIFNQPELNLR 896
Query: 932 RL 933
++
Sbjct: 897 QM 898
>At2g07660 putative retroelement pol polyprotein
Length = 949
Score = 242 bits (618), Expect = 1e-63
Identities = 143/414 (34%), Positives = 226/414 (54%), Gaps = 16/414 (3%)
Query: 494 KVGAALEEGVKRKIFQLL*EYPDIFAWSYEDMPGLDPKIVE-HRIPTKPECPPVRQKLRR 552
+VGA+ E K ++ E+ D+FA + G+ P + I +P P+ + R
Sbjct: 110 EVGASAE----LKDILIVNEFSDVFA----AVSGVPPDRSDPFTIELEPGTTPISKAPYR 161
Query: 553 THPDMALKIKSEV*KQIDAGFLMTIEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASPKD 612
P ++K ++ + + GF+ P W A ++ V KKDG R+C+D+R LNK + K+
Sbjct: 162 MAPAEMAELKKQLEELLAKGFIRPSSSP-WGAPVLFVKKKDGSFRLCIDYRGLNKVTVKN 220
Query: 613 NFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPFGL 672
+PLP ID L+D ++ FS +D SGY+QI + P D KT+F T +G F + VMPFGL
Sbjct: 221 KYPLPRIDELMDQLGGAQWFSKIDLASGYHQIPIEPTDVRKTAFRTRYGHFEFVVMPFGL 280
Query: 673 INAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHVEYLTKMFERLRKYKLRLNP 732
NA A + + M +F D + + V +++DD++V SK E H E+L + ERLR+++L
Sbjct: 281 TNAPAAFMKMMNGVFRDFLDEFVIIFIDDILVHSKSWEAHQEHLRAVLERLREHELFAKL 340
Query: 733 NKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREMPAPQTEKQVRGFLRRLNYISRFISH 792
+K +F RS LG ++S +G+ VDP+K+R+I+E P P+ ++R FL Y RF+
Sbjct: 341 SKFSFWQRSVGFLGHVISDQGVSVDPEKIRSIKEWPRPRNATEIRSFLGLAGYYRRFVMS 400
Query: 793 MTATCGPIFKLLRKNQPIVWNDECQGAFDSIKNYLLEPPILVPPMEGKPLIMYLSVFDES 852
+ P+ +L K+ W+DEC+ +F +K L+ P+LV P EG+P +Y
Sbjct: 401 FASMAQPLTRLTGKDTAFNWSDECEKSFLELKAMLINAPVLVLPEEGEPYTVYTDASIVG 460
Query: 853 VGCVLGQQDETGKKEHAIYYLSKKFTDCETRYMMLEKTCCAPAWAAKRLRHYLV 906
+GCVL Q K I Y S++ E Y + A +A K R YL+
Sbjct: 461 LGCVLMQ------KGSVIAYASRQLRKHEKNYPTHDLEMAAVVFALKIWRSYLI 508
>At3g11970 hypothetical protein
Length = 1499
Score = 233 bits (594), Expect = 7e-61
Identities = 144/426 (33%), Positives = 223/426 (51%), Gaps = 10/426 (2%)
Query: 500 EEGVKRKIFQLL*EYPDIFAWSYEDMPGLDPKIVEHRIPTKPECPPVRQKLRRTHPDMAL 559
E G + + ++L EYPDIF P + H+I PV Q+ R
Sbjct: 558 ELGEESVVEEVLNEYPDIFIEPTALPPFREKH--NHKIKLLEGSNPVNQRPYRYSIHQKN 615
Query: 560 KIKSEV*KQIDAGFLMTIEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLPHI 619
+I V + G + P + + +V V KKDG R+CVD+R+LN + KD+FP+P I
Sbjct: 616 EIDKLVEDLLTNGTVQASSSP-YASPVVLVKKKDGTWRLCVDYRELNGMTVKDSFPIPLI 674
Query: 620 DVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPFGLINAGATY 679
+ L+D + +FS +D +GY+Q++M P+D +KT+F T G F Y VMPFGL NA AT+
Sbjct: 675 EDLMDELGGAVIFSKIDLRAGYHQVRMDPDDIQKTAFKTHSGHFEYLVMPFGLTNAPATF 734
Query: 680 QRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHVEYLTKMFERLRKYKLRLNPNKCTFGV 739
Q M +F + K V V+ DD++V S E+H ++L ++FE +R KL +KC F V
Sbjct: 735 QGLMNFIFKPFLRKFVLVFFDDILVYSSSLEEHRQHLKQVFEVMRANKLFAKLSKCAFAV 794
Query: 740 RSGKLLGFIVSQKGIEVDPDKVRAIREMPAPQTEKQVRGFLRRLNYISRFISHMTATCGP 799
+ LG +S +GIE DP K++A++E P P T KQ+RGFL Y RF+ GP
Sbjct: 795 PKVEYLGHFISAQGIETDPAKIKAVKEWPQPTTLKQLRGFLGLAGYYRRFVRSFGVIAGP 854
Query: 800 IFKLLRKNQPIVWNDECQGAFDSIKNYLLEPPILVPPMEGKPLIMYLSVFDESVGCVLGQ 859
+ L K W Q AF+ +K L + P+L P+ K ++ + +G VL Q
Sbjct: 855 L-HALTKTDAFEWTAVAQQAFEDLKAALCQAPVLSLPLFDKQFVVETDACGQGIGAVLMQ 913
Query: 860 QDETGKKEHAIYYLSKKFTDCETRYMMLEKTCCAPAWAAKRLRHYLVNHTTWLISRMDPI 919
+ H + Y+S++ + + EK A +A ++ RHYL+ + + +
Sbjct: 914 EG------HPLAYISRQLKGKQLHLSIYEKELLAVIFAVRKWRHYLLQSHFIIKTDQRSL 967
Query: 920 KYIFEK 925
KY+ E+
Sbjct: 968 KYLLEQ 973
>At1g36590 hypothetical protein
Length = 1499
Score = 233 bits (594), Expect = 7e-61
Identities = 144/426 (33%), Positives = 223/426 (51%), Gaps = 10/426 (2%)
Query: 500 EEGVKRKIFQLL*EYPDIFAWSYEDMPGLDPKIVEHRIPTKPECPPVRQKLRRTHPDMAL 559
E G + + ++L EYPDIF P + H+I PV Q+ R
Sbjct: 558 ELGEESVVEEVLNEYPDIFIEPTALPPFREKH--NHKIKLLEGSNPVNQRPYRYSIHQKN 615
Query: 560 KIKSEV*KQIDAGFLMTIEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLPHI 619
+I V + G + P + + +V V KKDG R+CVD+R+LN + KD+FP+P I
Sbjct: 616 EIDKLVEDLLTNGTVQASSSP-YASPVVLVKKKDGTWRLCVDYRELNGMTVKDSFPIPLI 674
Query: 620 DVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPFGLINAGATY 679
+ L+D + +FS +D +GY+Q++M P+D +KT+F T G F Y VMPFGL NA AT+
Sbjct: 675 EDLMDELGGAVIFSKIDLRAGYHQVRMDPDDIQKTAFKTHSGHFEYLVMPFGLTNAPATF 734
Query: 680 QRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHVEYLTKMFERLRKYKLRLNPNKCTFGV 739
Q M +F + K V V+ DD++V S E+H ++L ++FE +R KL +KC F V
Sbjct: 735 QGLMNFIFKPFLRKFVLVFFDDILVYSSSLEEHRQHLKQVFEVMRANKLFAKLSKCAFAV 794
Query: 740 RSGKLLGFIVSQKGIEVDPDKVRAIREMPAPQTEKQVRGFLRRLNYISRFISHMTATCGP 799
+ LG +S +GIE DP K++A++E P P T KQ+RGFL Y RF+ GP
Sbjct: 795 PKVEYLGHFISAQGIETDPAKIKAVKEWPQPTTLKQLRGFLGLAGYYRRFVRSFGVIAGP 854
Query: 800 IFKLLRKNQPIVWNDECQGAFDSIKNYLLEPPILVPPMEGKPLIMYLSVFDESVGCVLGQ 859
+ L K W Q AF+ +K L + P+L P+ K ++ + +G VL Q
Sbjct: 855 L-HALTKTDAFEWTAVAQQAFEDLKAALCQAPVLSLPLFDKQFVVETDACGQGIGAVLMQ 913
Query: 860 QDETGKKEHAIYYLSKKFTDCETRYMMLEKTCCAPAWAAKRLRHYLVNHTTWLISRMDPI 919
+ H + Y+S++ + + EK A +A ++ RHYL+ + + +
Sbjct: 914 EG------HPLAYISRQLKGKQLHLSIYEKELLAVIFAVRKWRHYLLQSHFIIKTDQRSL 967
Query: 920 KYIFEK 925
KY+ E+
Sbjct: 968 KYLLEQ 973
>At2g14650 putative retroelement pol polyprotein
Length = 1328
Score = 233 bits (593), Expect = 9e-61
Identities = 146/467 (31%), Positives = 232/467 (49%), Gaps = 24/467 (5%)
Query: 465 LEQERKAIQPHQEEIELINLGTEENKREIKVGAALEEGVKRKIFQLL*EYPDIFAWSYED 524
L+ R + + E L+ G + + A E + K + E+ D+F +
Sbjct: 401 LDCHRGRVSFERPEGSLVYQGVRPTSGSLVISAVQAEKMIEKGCEAYLEFEDVF----QS 456
Query: 525 MPGLDPKIVE-HRIPTKPECPPVRQKLRRTHPDMALKIKSEV*KQIDAGFLMTIEYPEWV 583
+ GL P + I +P P+ + R P ++K ++ + A L
Sbjct: 457 LQGLPPSRSDPFTIELEPGTAPLSKAPYRMAPAEMAELKKQLEDLLGAPVLF-------- 508
Query: 584 ANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQ 643
V KKDG R+C+D+R LN + K+ +PLP ID L+D + FS +D SGY+
Sbjct: 509 -----VKKKDGSFRLCIDYRGLNWVTVKNKYPLPRIDELLDQLRGATCFSKIDLTSGYHL 563
Query: 644 IKMSPEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMI 703
I ++ D KT+F T +G F + VMPFGL NA A + R M ++F +++ + V +++DD++
Sbjct: 564 IPIAEADVRKTAFRTRYGHFEFVVMPFGLTNAPAAFMRLMNSVFQEVLDEFVIIFIDDIL 623
Query: 704 VKSKDEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRA 763
V SK E+H +L ++ E+LR+ KL +KC+F R LG IVS +G+ VDP+K+ A
Sbjct: 624 VYSKSLEEHEVHLRRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEKIEA 683
Query: 764 IREMPAPQTEKQVRGFLRRLNYISRFISHMTATCGPIFKLLRKNQPIVWNDECQGAFDSI 823
IR+ P ++R FL Y RF+ + P+ KL K+ P VW+ EC+ F S+
Sbjct: 684 IRDWHTPTNATEIRSFLGLAGYYRRFVKGFASMAQPMTKLTGKDVPFVWSPECEEGFVSL 743
Query: 824 KNYLLEPPILVPPMEGKPLIMYLSVFDESVGCVLGQQDETGKKEHAIYYLSKKFTDCETR 883
K L P+L P G+P ++Y +GCVL Q+ + I Y S++ E
Sbjct: 744 KEMLTSTPVLALPEHGEPYMVYTDASGVGLGCVLMQRGK------VIAYASRQLRKHEGN 797
Query: 884 YMMLEKTCCAPAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKPAVLL 930
Y + A +A K R YL + + +KYIF +P + L
Sbjct: 798 YPTHDLEMAAVIFALKIWRSYLYGGNVQVFTDHKSLKYIFTQPELNL 844
>At1g37060 Athila retroelment ORF 1, putative
Length = 1734
Score = 229 bits (584), Expect = 1e-59
Identities = 145/472 (30%), Positives = 227/472 (47%), Gaps = 49/472 (10%)
Query: 472 IQPHQEEIELINLGTEENKREIKVGAALEEGVKRKIFQLL*EYPDIFAWSYEDMPGLDPK 531
++P + + + LG I + V I +L+ +Y +S +D+ G+ P
Sbjct: 827 LKPLPKGLRYVFLGLNSTYPVIVNDELTADQVNLLITELM-KYRKAIGYSLDDIKGISPT 885
Query: 532 IVEHRIPTKPECPPVRQKLRRTHPDMALKIKSEV*KQIDAGFLMTIEYPEWVANIVPVPK 591
+ HRI + E + RR +P++ +K E+ K +DAG + I WV+ + VPK
Sbjct: 886 LCTHRIHLENESYSSIEPQRRLNPNLKEVVKKEILKLLDAGVIYPISDSTWVSPVHCVPK 945
Query: 592 KDGKV------------------RMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFS 633
K G RMC+++R LN AS K++FPLP ID +++ A +
Sbjct: 946 KGGMTVVKNSKDELIPTRTITGHRMCIEYRKLNVASRKEHFPLPFIDHMLERLANHPYYC 1005
Query: 634 FMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMIHK 693
F+D +SG+ QI + P D+ KT+F P+GTF YK MPFGL NA AT+QR MT++F D+I +
Sbjct: 1006 FLDSYSGFFQIPIHPNDQGKTTFTCPYGTFAYKRMPFGLCNAPATFQRCMTSIFSDLIEE 1065
Query: 694 EVEVYVDDMIVKSKDEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKG 753
VEV++DD V + L ++ +R + L LN KC F VR G +LG +S++G
Sbjct: 1066 MVEVFMDDFSVYGSSFSSCLLNLCRVLKRCEETNLVLNWEKCHFMVREGIVLGRKISEEG 1125
Query: 754 IEVDPDKVRAIREMPAPQTEKQVRGFLRRLNYISRFISHMTATCGPIFKLLRKNQPIVWN 813
IEVD K+ + ++ P+T K +R FL + FI + P+ +LL K ++
Sbjct: 1126 IEVDKAKIDVMMQLQPPKTVKDIRSFLGHAGFYRIFIKDFSKLARPLTRLLCKETEFAFD 1185
Query: 814 DECQGAFDSIKNYLLEPPILVPPMEGKPLIMYLSVFDESVGCVLGQQDETGKKEHAIYYL 873
DEC AF IK L+ PI+ P P +
Sbjct: 1186 DECLTAFKLIKEALITAPIVQAPNWDFPFEII---------------------------- 1217
Query: 874 SKKFTDCETRYMMLEKTCCAPAWAAKRLRHYLVNHTTWLISRMDPIKYIFEK 925
D + RY EK A +A ++ R YLV + + +++I+ K
Sbjct: 1218 --TMDDAQVRYATTEKELLAVVFAFEKFRSYLVGSKVTIYTDHAALRHIYAK 1267
>At1g35370 hypothetical protein
Length = 1447
Score = 224 bits (571), Expect = 3e-58
Identities = 152/457 (33%), Positives = 235/457 (51%), Gaps = 30/457 (6%)
Query: 469 RKAIQPHQEEIELINLGTEENKREIKVGAALEEGVKRKIFQLL*EYPDIFAWSYEDMPGL 528
R+ + ++EI I+ T + +EE V + I + E+PD+FA D+P
Sbjct: 518 REVVSDEEQEIGSISALTSD---------VVEESVVQNIVE---EFPDVFA-EPTDLPPF 564
Query: 529 DPKIVEHRIPTKPECPPVRQKLRRTHPDMALKIKSEV*KQIDAGFLMTIEYPEWVANIVP 588
K +H+I PV Q+ R +I V I +G + P + + +V
Sbjct: 565 REKH-DHKIKLLEGANPVNQRPYRYVVHQKDEIDKIVQDMIKSGTIQVSSSP-FASPVVL 622
Query: 589 VPKKDGKVRMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSP 648
V KKDG R+CVD+ +LN + KD F +P I+ L+D S VFS +D +GY+Q++M P
Sbjct: 623 VKKKDGTWRLCVDYTELNGMTVKDRFLIPLIEDLMDELGGSVVFSKIDLRAGYHQVRMDP 682
Query: 649 EDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKD 708
+D +KT+F T G F Y VM FGL NA AT+Q M ++F D + K V V+ DD+++ S
Sbjct: 683 DDIQKTAFKTHNGHFEYLVMLFGLTNAPATFQSLMNSVFRDFLRKFVLVFFDDILIYSSS 742
Query: 709 EEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREMP 768
E+H E+L +FE +R +KL F S + LG +S + IE DP K++A++E P
Sbjct: 743 IEEHKEHLRLVFEVMRLHKL--------FAKGSKEHLGHFISAREIETDPAKIQAVKEWP 794
Query: 769 APQTEKQVRGFLRRLNYISRFISHMTATCGPIFKLLRKNQPIVWNDECQGAFDSIKNYLL 828
P T KQVRGFL Y RF+ + GP+ L K W+ E Q AFD++K L
Sbjct: 795 TPTTVKQVRGFLGFAGYYRRFVRNFGVIAGPL-HALTKTDGFCWSLEAQSAFDTLKAVLC 853
Query: 829 EPPILVPPMEGKPLIMYLSVFDESVGCVLGQQDETGKKEHAIYYLSKKFTDCETRYMMLE 888
P+L P+ K ++ + + VL Q K H + Y+S++ + + E
Sbjct: 854 NAPVLALPVFDKQFMVETDACGQGIRAVLMQ------KGHPLAYISRQLKGKQLHLSIYE 907
Query: 889 KTCCAPAWAAKRLRHYLVNHTTWLISRMDPIKYIFEK 925
K A +A ++ RHYL+ + + +KY+ E+
Sbjct: 908 KELLAFIFAVRKWRHYLLPSHFIIKTDQRSLKYLLEQ 944
>At3g31530 hypothetical protein
Length = 831
Score = 223 bits (568), Expect = 7e-58
Identities = 115/295 (38%), Positives = 180/295 (60%), Gaps = 14/295 (4%)
Query: 633 SFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMIH 692
S+ ++ + + M+PED+EKT+F T G FCY+VMPFGL NAGATY+R + +F I
Sbjct: 83 SYCHSWTLFVVMMMNPEDQEKTAFYTEQGIFCYRVMPFGLKNAGATYERFVNKIFALQIG 142
Query: 693 KEVEVYVDDMIVKSKDEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQK 752
K +EVY+DDM+VKS E+ H+ +L + F++L Y ++LNP KC FGVRS
Sbjct: 143 KTMEVYIDDMLVKSMTEKDHISHLRECFKQLNLYNVKLNPAKCRFGVRS----------- 191
Query: 753 GIEVDPDKVRAIREMPAPQTEKQVRGFLRRLNYISRFISHMTATCGPIFKLLRKNQPIVW 812
GIE +P ++ A+ M +PQ +++V+ R+ ++RFIS T C + +LR+N+ W
Sbjct: 192 GIEANPKQIEALLGMASPQNKREVQCLTGRVAALNRFISRSTEKCLAFYDVLRRNKKFEW 251
Query: 813 NDECQGAFDSIKNYLLEPPILVPPMEGKPLIMYLSVFDESVGCVLGQQDETGKKEHAIYY 872
C+ AF +K YL PPIL P+ G+P +Y++V D +V L ++D +K I+Y
Sbjct: 252 TTRCEEAFQELKKYLATPPILAKPVIGEPQYLYVAVSDTAVSGELVREDRGEQK--LIFY 309
Query: 873 LSKKFTDCETRYMMLEKTCCAPAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKPA 927
+S+ FT E+RY +EK A +A++LR Y +H+ ++ M P++ I P+
Sbjct: 310 VSQTFTSAESRYPQMEKLALAVVMSAQKLRPYFQSHSIIVMGSM-PLRVILHSPS 363
>At2g14040 putative retroelement pol polyprotein
Length = 841
Score = 216 bits (550), Expect = 8e-56
Identities = 132/381 (34%), Positives = 195/381 (50%), Gaps = 48/381 (12%)
Query: 513 EYPDIFAWSYEDMPGLDPKIVEHRIPTKPECPPVRQKLRRTHPDMALKIKSEV*KQIDAG 572
+Y +S +D+ G+ P + H I + E + RR + ++ +K E+ K +DAG
Sbjct: 342 KYRKALGYSLDDITGISPTLCMHMIHLEGESITSVEHQRRLNSNLRDVVKKEIMKLLDAG 401
Query: 573 FLMTIEYPEWVANIVPVPKKDGKV------------------RMCVDFRDLNKASPKDNF 614
+ I WV+ + VPKK G + RMC+D+R LN A+ KDNF
Sbjct: 402 IIYPISDSTWVSPVHVVPKKGGVIVIKNEKNELIPTRTVTGHRMCIDYRKLNSATRKDNF 461
Query: 615 PLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPFGLIN 674
PL ID +++ + + F+DG+ G+ QI + P+D+EKT+F P+GTF Y+ MPFGL N
Sbjct: 462 PLSFIDQMLERLSNQPYYCFLDGYLGFFQILIHPDDQEKTTFTCPYGTFAYRRMPFGLCN 521
Query: 675 AGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHVEYLTKMFERLRKYKLRLNPNK 734
A AT+Q M +F DMI +EV++DD +V + E++H L LN K
Sbjct: 522 APATFQHCMKYIFSDMIEDFMEVFIDDFLVLQRCEDKH---------------LVLNWEK 566
Query: 735 CTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREMPAPQTEKQVRGFLRRLNYISRFISHMT 794
F VR G +LG +S+KG+EVD K+ +R FL + RFI +
Sbjct: 567 SHFMVRDGIVLGHKISEKGVEVDRAKIEIMR-------------FLGHAGFYRRFIKDFS 613
Query: 795 ATCGPIFKLLRKNQPIVWNDECQGAFDSIKNYLLEPPILVPPMEGKPLIMYLSVFDESVG 854
P +LL K Q ++D C AF+ +K L+ I+ PP P + D VG
Sbjct: 614 KIARPCTQLLCKEQKSEFDDTCLQAFNIVKESLVSALIVQPPEWELPFEVICDASDYVVG 673
Query: 855 CVLGQQDETGKKEHAIYYLSK 875
VLGQ+ + KK HAIYY S+
Sbjct: 674 AVLGQRKD--KKLHAIYYASR 692
>At4g07600
Length = 630
Score = 194 bits (494), Expect = 3e-49
Identities = 111/304 (36%), Positives = 165/304 (53%), Gaps = 21/304 (6%)
Query: 586 IVPVPKKDGKVRMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIK 645
++P+ G +RMC+D+R+LN AS D+FPLP D +++ A + F+DG+SG+ QI
Sbjct: 346 LIPIRTITG-LRMCIDYRNLNAASRNDHFPLPFTDQMLERLANHPYYCFLDGYSGFFQIP 404
Query: 646 MSPEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVK 705
+ P D EKT+F P+GTF Y+ MPFGL NA AT+QR MT++F D+I + VEV++DD V
Sbjct: 405 IHPNDHEKTTFTCPYGTFAYERMPFGLCNAPATFQRCMTSIFSDIIEEMVEVFMDDFSVY 464
Query: 706 SKDEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIR 765
+ L ++ R + L LN KC F V+ G +LG +S+KGIEVD
Sbjct: 465 GPSFSSCLLNLGRVLTRCEETNLVLNWEKCHFMVKEGIMLGHKISEKGIEVDKG------ 518
Query: 766 EMPAPQTEKQVRGFLRRLNYISRFISHMTATCGPIFKLLRKNQPIVWNDECQGAFDSIKN 825
FL + RFI + P+ +LL K ++++C +F +IK
Sbjct: 519 ------------CFLGHAGFYRRFIKDFSKIARPLTRLLCKETEFEFDEDCPKSFHTIKE 566
Query: 826 YLLEPPILVPPMEGKPLIMYLSVFDESVGCVLGQQDETGKKEHAIYYLSKKFTDCETRYM 885
L+ P++ P + D +VG VLGQ+ + KK H IYY S+ + + RY
Sbjct: 567 ALVSAPVVRATNWDYPFEIMCDALDYAVGAVLGQKID--KKLHVIYYASRTLDEAQGRYA 624
Query: 886 MLEK 889
EK
Sbjct: 625 TTEK 628
>At4g07660 putative athila transposon protein
Length = 724
Score = 186 bits (472), Expect = 9e-47
Identities = 104/300 (34%), Positives = 160/300 (52%), Gaps = 18/300 (6%)
Query: 495 VGAALEEGVKRKIFQLL*EYPDIFAWSYEDMPGLDPKIVEHRIPTKPECPPVRQKLRRTH 554
+ L + + L +Y +S D+ + P + HRI + E + RR +
Sbjct: 299 INVELNDNEVNLLLSELRKYKRAIGYSLSDIKRISPSLCNHRIHLENESYSSIEPQRRLN 358
Query: 555 PDMALKIKSEV*KQIDAGFLMTIEYPEWVANIVPVPKKDGKV------------------ 596
++ +K E+ K +DAG + I WV + VPKK G
Sbjct: 359 LNLKEVVKKEILKLLDAGVIYPISDSTWVFPVHCVPKKGGMTVVKNEKDELIPTRTITGH 418
Query: 597 RMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSF 656
R+C+D+R LN AS KD+FPLP + +++ A F+DG+SG+ QI + P D+EKT+F
Sbjct: 419 RVCIDYRKLNAASRKDHFPLPFTNQMLEGLANHLYNCFLDGYSGFFQIPIHPNDQEKTTF 478
Query: 657 ITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHVEYL 716
P+GTF YK MPFGL NA T+QR MT++F D+I K VEV++DD V + L
Sbjct: 479 TCPYGTFAYKRMPFGLCNAPTTFQRCMTSIFSDLIEKMVEVFMDDFSVYGPSFSSCLLNL 538
Query: 717 TKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREMPAPQTEKQV 776
++ + + L LN KC F V+ G +LG +S+KGIEVD +K++ + ++ P+T K +
Sbjct: 539 GRVLTKWEETNLVLNWEKCYFMVKEGIVLGHKISEKGIEVDKEKIKVMMQLQPPKTVKDI 598
Score = 32.0 bits (71), Expect = 2.9
Identities = 19/68 (27%), Positives = 32/68 (46%), Gaps = 4/68 (5%)
Query: 870 IYYLSKKFTDCETRYMMLEKTCCAPAWAAKRLRHYLVNHTTWLISRMDPIKYIF----EK 925
IYY S+ + + RY EK +A K+ R YLV + + +++++ K
Sbjct: 598 IYYASRTLDEAQGRYATTEKELLVVVFAFKKFRSYLVESKVTVYTDHAALRHMYAKKDTK 657
Query: 926 PAVLLGRL 933
P +L G L
Sbjct: 658 PRLLRGIL 665
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.346 0.153 0.549
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 33,453,276
Number of Sequences: 26719
Number of extensions: 1398051
Number of successful extensions: 5221
Number of sequences better than 10.0: 52
Number of HSP's better than 10.0 without gapping: 49
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 5092
Number of HSP's gapped (non-prelim): 82
length of query: 1586
length of database: 11,318,596
effective HSP length: 113
effective length of query: 1473
effective length of database: 8,299,349
effective search space: 12224941077
effective search space used: 12224941077
T: 11
A: 40
X1: 15 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.7 bits)
S2: 67 (30.4 bits)
Medicago: description of AC147714.3


