

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147714.14 + phase: 0
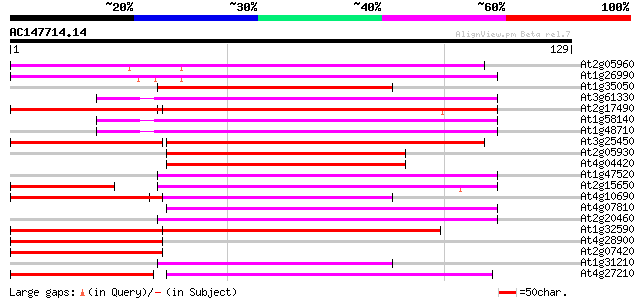
(129 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g05960 putative retroelement pol polyprotein 61 1e-10
At1g26990 polyprotein, putative 60 3e-10
At1g35050 putative protein 58 1e-09
At3g61330 copia-type polyprotein 56 4e-09
At2g17490 putative retroelement pol polyprotein 56 6e-09
At1g58140 hypothetical protein 56 6e-09
At1g48710 hypothetical protein 56 6e-09
At3g25450 hypothetical protein 54 3e-08
At2g05930 copia-like retroelement pol polyprotein 51 1e-07
At4g04420 putative transposon protein 50 4e-07
At1g47520 hypothetical protein 49 5e-07
At2g15650 putative retroelement pol polyprotein 48 1e-06
At4g10690 retrotransposon like protein 48 2e-06
At4g07810 putative polyprotein 47 2e-06
At2g20460 putative retroelement pol polyprotein 46 5e-06
At1g32590 hypothetical protein, 5' partial 46 5e-06
At4g28900 putative protein 46 6e-06
At2g07420 putative retroelement pol polyprotein 45 8e-06
At1g31210 putative reverse transcriptase 45 1e-05
At4g27210 putative protein 45 1e-05
>At2g05960 putative retroelement pol polyprotein
Length = 1200
Score = 61.2 bits (147), Expect = 1e-10
Identities = 39/139 (28%), Positives = 66/139 (47%), Gaps = 30/139 (21%)
Query: 1 MKVKSAFLNGAISEEVYVKQPPGFED---------LKHPDYVYKDEI------------- 38
+ VK+AFL+G + E VYV QP GF L YVY D++
Sbjct: 892 LDVKTAFLHGELKENVYVTQPEGFVTKGSEEKVYKLHKALYVYVDDLLVTGSSPKLIDDF 951
Query: 39 --------EMSMMGELKFFLRIQINQCKDEVYVHQSKYTKDLLMKFKVDDCNEMNTPMHP 90
EMS +G L ++L I++ Q +D + + Q +Y K +L + +++C + PM+
Sbjct: 952 KKGMSKNFEMSDLGRLTYYLGIEVTQEEDGIILKQERYAKKILEEAGINECKSILVPMNS 1011
Query: 91 TCTLSEEDADAKVDQKLFR 109
LS+ + +D + +R
Sbjct: 1012 GLELSKALDEKSIDGQQYR 1030
>At1g26990 polyprotein, putative
Length = 1436
Score = 60.1 bits (144), Expect = 3e-10
Identities = 43/158 (27%), Positives = 67/158 (42%), Gaps = 46/158 (29%)
Query: 1 MKVKSAFLNGAISEEVYVKQPPGFEDLK----------HPDY---------------VYK 35
+ + +A LNG + EE+Y+K PPG+ +++ H D+ VY
Sbjct: 1060 LDISNALLNGDLEEEIYMKLPPGYSEIQGQEVSPNAKCHGDHTLFVKAQDGFFLVVLVYV 1119
Query: 36 DEI---------------------EMSMMGELKFFLRIQINQCKDEVYVHQSKYTKDLLM 74
D+I ++ +GE KFFL I+I + D + + Q KY DLL
Sbjct: 1120 DDILIASTTEAASAELTSQLSSFFQLRDLGEPKFFLGIEIARNADGISLCQRKYVLDLLA 1179
Query: 75 KFKVDDCNEMNTPMHPTCTLSEEDADAKVDQKLFRGII 112
DC + PM P LS++ D K +R I+
Sbjct: 1180 SSDFSDCKPSSIPMEPNQKLSKDTGTLLEDGKQYRRIL 1217
>At1g35050 putative protein
Length = 450
Score = 58.2 bits (139), Expect = 1e-09
Identities = 25/54 (46%), Positives = 37/54 (68%)
Query: 35 KDEIEMSMMGELKFFLRIQINQCKDEVYVHQSKYTKDLLMKFKVDDCNEMNTPM 88
K+E EM +G+ KF L +QI +D ++VHQS YTK +L +F +D N ++TPM
Sbjct: 241 KEEFEMKDLGKTKFCLGLQIEHFQDGIFVHQSNYTKKILKRFNMDKANPLSTPM 294
>At3g61330 copia-type polyprotein
Length = 1352
Score = 56.2 bits (134), Expect = 4e-09
Identities = 29/92 (31%), Positives = 51/92 (54%), Gaps = 3/92 (3%)
Query: 21 PPGFEDLKHPDYVYKDEIEMSMMGELKFFLRIQINQCKDEVYVHQSKYTKDLLMKFKVDD 80
P FE+ K E EM+ +G + ++L I++ Q + +++ Q Y K++L KFK+DD
Sbjct: 1042 PSIFEEFKKE---MTKEFEMTDIGLMSYYLGIEVKQEDNGIFITQEGYAKEVLKKFKIDD 1098
Query: 81 CNEMNTPMHPTCTLSEEDADAKVDQKLFRGII 112
N + TPM LS+++ VD F+ ++
Sbjct: 1099 SNPVCTPMECGIKLSKKEEGEGVDPTTFKSLV 1130
Score = 38.5 bits (88), Expect = 0.001
Identities = 16/24 (66%), Positives = 20/24 (82%)
Query: 1 MKVKSAFLNGAISEEVYVKQPPGF 24
M VKSAFLNG + EEVY++QP G+
Sbjct: 945 MDVKSAFLNGDLEEEVYIEQPQGY 968
>At2g17490 putative retroelement pol polyprotein
Length = 822
Score = 55.8 bits (133), Expect = 6e-09
Identities = 31/81 (38%), Positives = 49/81 (60%), Gaps = 3/81 (3%)
Query: 35 KDEIEMSMMGELKFFLRIQINQCKDEVYVHQSKYTKDLLMKFKVDDCNEMNTPMHPTCTL 94
K+E EMS +G L +FL ++I Q + +++ Q Y + LL KF ++D M+TP+ P
Sbjct: 679 KNEFEMSDLGLLNYFLGMEIVQSLEGIFLSQECYARKLLKKFNMEDSKIMSTPLLPQRKD 738
Query: 95 SEED---ADAKVDQKLFRGII 112
EED AD+KV + L G++
Sbjct: 739 QEEDESLADSKVYRSLVGGLL 759
Score = 42.0 bits (97), Expect = 9e-05
Identities = 18/35 (51%), Positives = 25/35 (71%)
Query: 1 MKVKSAFLNGAISEEVYVKQPPGFEDLKHPDYVYK 35
+ VKSAFLNG + EE+Y +QP GFE+ D+V +
Sbjct: 571 LDVKSAFLNGTLEEEIYAEQPLGFEEEGKEDHVLR 605
>At1g58140 hypothetical protein
Length = 1320
Score = 55.8 bits (133), Expect = 6e-09
Identities = 29/92 (31%), Positives = 51/92 (54%), Gaps = 3/92 (3%)
Query: 21 PPGFEDLKHPDYVYKDEIEMSMMGELKFFLRIQINQCKDEVYVHQSKYTKDLLMKFKVDD 80
P FE+ K E EM+ +G + ++L I++ Q + +++ Q Y K++L KFK+DD
Sbjct: 1010 PSMFEEFKKE---MTKEFEMTDIGLMSYYLGIEVKQEDNGIFITQEGYAKEVLKKFKMDD 1066
Query: 81 CNEMNTPMHPTCTLSEEDADAKVDQKLFRGII 112
N + TPM LS+++ VD F+ ++
Sbjct: 1067 SNPVCTPMECGIKLSKKEEGEGVDPTTFKSLV 1098
Score = 38.5 bits (88), Expect = 0.001
Identities = 16/24 (66%), Positives = 20/24 (82%)
Query: 1 MKVKSAFLNGAISEEVYVKQPPGF 24
M VKSAFLNG + EEVY++QP G+
Sbjct: 913 MDVKSAFLNGDLEEEVYIEQPQGY 936
>At1g48710 hypothetical protein
Length = 1352
Score = 55.8 bits (133), Expect = 6e-09
Identities = 29/92 (31%), Positives = 51/92 (54%), Gaps = 3/92 (3%)
Query: 21 PPGFEDLKHPDYVYKDEIEMSMMGELKFFLRIQINQCKDEVYVHQSKYTKDLLMKFKVDD 80
P FE+ K E EM+ +G + ++L I++ Q + +++ Q Y K++L KFK+DD
Sbjct: 1042 PSMFEEFKKE---MTKEFEMTDIGLMSYYLGIEVKQEDNGIFITQEGYAKEVLKKFKMDD 1098
Query: 81 CNEMNTPMHPTCTLSEEDADAKVDQKLFRGII 112
N + TPM LS+++ VD F+ ++
Sbjct: 1099 SNPVCTPMECGIKLSKKEEGEGVDPTTFKSLV 1130
Score = 38.5 bits (88), Expect = 0.001
Identities = 16/24 (66%), Positives = 20/24 (82%)
Query: 1 MKVKSAFLNGAISEEVYVKQPPGF 24
M VKSAFLNG + EEVY++QP G+
Sbjct: 945 MDVKSAFLNGDLEEEVYIEQPQGY 968
>At3g25450 hypothetical protein
Length = 1343
Score = 53.5 bits (127), Expect = 3e-08
Identities = 24/73 (32%), Positives = 45/73 (60%)
Query: 37 EIEMSMMGELKFFLRIQINQCKDEVYVHQSKYTKDLLMKFKVDDCNEMNTPMHPTCTLSE 96
+ EMS +G+L ++L I++ Q KD + + Q +Y K +L + + CN +NTPM + LS+
Sbjct: 1037 KFEMSDLGKLTYYLGIEVLQSKDGITLKQERYAKKILEEAGMSKCNTVNTPMIASLELSK 1096
Query: 97 EDADAKVDQKLFR 109
+ ++D+ +R
Sbjct: 1097 AQDEKRIDETDYR 1109
Score = 39.3 bits (90), Expect = 6e-04
Identities = 17/35 (48%), Positives = 25/35 (70%)
Query: 1 MKVKSAFLNGAISEEVYVKQPPGFEDLKHPDYVYK 35
+ VK+AFL+G + E+VYV QP GF + + + VYK
Sbjct: 927 LDVKTAFLHGELREDVYVSQPEGFTNKESKEKVYK 961
>At2g05930 copia-like retroelement pol polyprotein
Length = 916
Score = 51.2 bits (121), Expect = 1e-07
Identities = 23/55 (41%), Positives = 36/55 (64%)
Query: 37 EIEMSMMGELKFFLRIQINQCKDEVYVHQSKYTKDLLMKFKVDDCNEMNTPMHPT 91
E +SM+GELK+FL +QINQ + + + QS Y ++L+ +F + N + TPM T
Sbjct: 758 EFRLSMVGELKYFLGLQINQIDEGIAISQSTYDQNLVKRFDMCSSNPVKTPMSTT 812
>At4g04420 putative transposon protein
Length = 1008
Score = 49.7 bits (117), Expect = 4e-07
Identities = 22/55 (40%), Positives = 35/55 (63%)
Query: 37 EIEMSMMGELKFFLRIQINQCKDEVYVHQSKYTKDLLMKFKVDDCNEMNTPMHPT 91
E +SM+GELK+F +QINQ + + + QS Y ++L+ +F + N + TPM T
Sbjct: 850 EFRLSMVGELKYFRGLQINQINEGIAISQSTYAQNLVKRFDMCSSNPVETPMSTT 904
>At1g47520 hypothetical protein
Length = 634
Score = 49.3 bits (116), Expect = 5e-07
Identities = 22/78 (28%), Positives = 43/78 (54%)
Query: 35 KDEIEMSMMGELKFFLRIQINQCKDEVYVHQSKYTKDLLMKFKVDDCNEMNTPMHPTCTL 94
K+ + +G LK+FL +++ + + + + Q KY +LL + DC +TPM P L
Sbjct: 486 KESFRLRELGPLKYFLGLEVARTSEGISLCQRKYALELLTSTGMLDCQHSSTPMIPNLHL 545
Query: 95 SEEDADAKVDQKLFRGII 112
S+ D + D++ +R ++
Sbjct: 546 SKSDGELLEDKEYYRRLV 563
Score = 32.3 bits (72), Expect = 0.068
Identities = 12/28 (42%), Positives = 19/28 (67%)
Query: 1 MKVKSAFLNGAISEEVYVKQPPGFEDLK 28
+ + +AFLNG + E +Y+K P G+ D K
Sbjct: 374 LDISNAFLNGDLEETIYMKLPEGYADSK 401
>At2g15650 putative retroelement pol polyprotein
Length = 1347
Score = 48.1 bits (113), Expect = 1e-06
Identities = 25/80 (31%), Positives = 43/80 (53%), Gaps = 2/80 (2%)
Query: 35 KDEIEMSMMGELKFFLRIQINQCKDEVYVHQSKYTKDLLMKFKVDDCNEMNTPMHPTCTL 94
KDE EM+ +G L +FL +++NQ +++ Q KY L+ KF + + ++TP+ P
Sbjct: 1052 KDEFEMTDLGLLNYFLGMEVNQDDSGIFLSQEKYANKLIDKFGMKESKSVSTPLTPQGKR 1111
Query: 95 SEEDADAK--VDQKLFRGII 112
+ D K D +R I+
Sbjct: 1112 KGVEGDDKEFADPTKYRRIV 1131
Score = 42.7 bits (99), Expect = 5e-05
Identities = 19/24 (79%), Positives = 20/24 (83%)
Query: 1 MKVKSAFLNGAISEEVYVKQPPGF 24
M VKSAFLNG + EEVYV QPPGF
Sbjct: 944 MDVKSAFLNGELEEEVYVTQPPGF 967
>At4g10690 retrotransposon like protein
Length = 1515
Score = 47.8 bits (112), Expect = 2e-06
Identities = 22/56 (39%), Positives = 32/56 (56%)
Query: 33 VYKDEIEMSMMGELKFFLRIQINQCKDEVYVHQSKYTKDLLMKFKVDDCNEMNTPM 88
+ E M MG L +FL IQ + D +++ Q KYT DLL+ + DC+ M TP+
Sbjct: 1133 ILSTEFRMKDMGALHYFLGIQAHYHNDGLFLSQEKYTSDLLVNAGMSDCSSMPTPL 1188
Score = 44.3 bits (103), Expect = 2e-05
Identities = 19/35 (54%), Positives = 25/35 (71%)
Query: 1 MKVKSAFLNGAISEEVYVKQPPGFEDLKHPDYVYK 35
+ VK+AFL+ + E V++ QPPGFED PDYV K
Sbjct: 1027 LDVKNAFLHDELKETVFMTQPPGFEDPSRPDYVCK 1061
>At4g07810 putative polyprotein
Length = 1366
Score = 47.4 bits (111), Expect = 2e-06
Identities = 23/76 (30%), Positives = 42/76 (55%)
Query: 37 EIEMSMMGELKFFLRIQINQCKDEVYVHQSKYTKDLLMKFKVDDCNEMNTPMHPTCTLSE 96
E ++ +G+ ++FL ++I + K+ + + Q KY +LL +F C + TPM LS+
Sbjct: 1110 EFKLKDLGQKRYFLGLEIARSKEGISISQRKYALELLEEFGFLGCKPVPTPMELNLKLSQ 1169
Query: 97 EDADAKVDQKLFRGII 112
ED +D +R +I
Sbjct: 1170 EDGALLLDASHYRKLI 1185
Score = 28.9 bits (63), Expect = 0.75
Identities = 12/28 (42%), Positives = 18/28 (63%)
Query: 1 MKVKSAFLNGAISEEVYVKQPPGFEDLK 28
M V +AFL+G EE+Y++ P G+ K
Sbjct: 996 MDVTNAFLHGDFEEEIYMQLPQGYTPRK 1023
>At2g20460 putative retroelement pol polyprotein
Length = 1461
Score = 46.2 bits (108), Expect = 5e-06
Identities = 21/78 (26%), Positives = 42/78 (52%)
Query: 35 KDEIEMSMMGELKFFLRIQINQCKDEVYVHQSKYTKDLLMKFKVDDCNEMNTPMHPTCTL 94
K ++ +G LK+FL +++ + + + + Q KY +LL + DC + PM P L
Sbjct: 1166 KASFKLRELGPLKYFLGLEVARTSEGISLSQRKYALELLTSADMLDCKPSSIPMTPNIRL 1225
Query: 95 SEEDADAKVDQKLFRGII 112
S+ D D++++R ++
Sbjct: 1226 SKNDGLLLEDKEMYRRLV 1243
Score = 33.9 bits (76), Expect = 0.023
Identities = 12/28 (42%), Positives = 20/28 (70%)
Query: 1 MKVKSAFLNGAISEEVYVKQPPGFEDLK 28
+ + +AFLNG + E +Y+K P G+ D+K
Sbjct: 1054 LDISNAFLNGDLEETIYMKLPDGYADIK 1081
>At1g32590 hypothetical protein, 5' partial
Length = 1263
Score = 46.2 bits (108), Expect = 5e-06
Identities = 17/64 (26%), Positives = 41/64 (63%)
Query: 36 DEIEMSMMGELKFFLRIQINQCKDEVYVHQSKYTKDLLMKFKVDDCNEMNTPMHPTCTLS 95
+E M+ +G++K+FL +++ Q + ++++Q KY +++ K+ ++ CN + P+ P L+
Sbjct: 1003 EEFAMTDLGKMKYFLGVEVIQDERGIFINQRKYAAEIIKKYGMEGCNSVKNPIVPGQKLT 1062
Query: 96 EEDA 99
+ A
Sbjct: 1063 KAGA 1066
Score = 39.3 bits (90), Expect = 6e-04
Identities = 18/35 (51%), Positives = 25/35 (71%)
Query: 1 MKVKSAFLNGAISEEVYVKQPPGFEDLKHPDYVYK 35
+ VKSAFL+G + E+V+V+QP GFE + VYK
Sbjct: 894 LDVKSAFLHGDLKEDVFVEQPKGFEVEEESSKVYK 928
>At4g28900 putative protein
Length = 1415
Score = 45.8 bits (107), Expect = 6e-06
Identities = 20/35 (57%), Positives = 26/35 (74%)
Query: 1 MKVKSAFLNGAISEEVYVKQPPGFEDLKHPDYVYK 35
+ VK+AFL+G + E VY+ QPPGFE+ PDYV K
Sbjct: 985 LDVKNAFLHGDLKETVYMYQPPGFENQDRPDYVCK 1019
>At2g07420 putative retroelement pol polyprotein
Length = 1664
Score = 45.4 bits (106), Expect = 8e-06
Identities = 19/35 (54%), Positives = 26/35 (74%)
Query: 1 MKVKSAFLNGAISEEVYVKQPPGFEDLKHPDYVYK 35
M VK+AFL G + E+VY+K PPG ED+ P+ V+K
Sbjct: 999 MDVKNAFLQGELEEKVYMKPPPGLEDINAPNKVFK 1033
Score = 37.7 bits (86), Expect = 0.002
Identities = 22/68 (32%), Positives = 36/68 (52%), Gaps = 7/68 (10%)
Query: 35 KDEIEMSMMGELKFFLRIQINQCKDEVYVHQSKYTKDLLMKFKVDDCNEMNTPMHPTCTL 94
K ++ +GELK+FL I++ + K+ +++ Q KYT DLL + TP+
Sbjct: 1107 KSVFDIKDLGELKYFLGIEVCRSKEGLFLSQRKYTLDLLAQVGKLGVKPAKTPL------ 1160
Query: 95 SEEDADAK 102
E+D AK
Sbjct: 1161 -EDDYKAK 1167
>At1g31210 putative reverse transcriptase
Length = 1415
Score = 45.1 bits (105), Expect = 1e-05
Identities = 19/54 (35%), Positives = 32/54 (59%)
Query: 35 KDEIEMSMMGELKFFLRIQINQCKDEVYVHQSKYTKDLLMKFKVDDCNEMNTPM 88
K+ M +G ++FL IQI + +++HQ+ Y D+L + + DCN M TP+
Sbjct: 1075 KNRFSMKDLGPPRYFLGIQIEDYANGLFLHQTAYATDILQQAGMSDCNPMPTPL 1128
Score = 32.7 bits (73), Expect = 0.052
Identities = 14/33 (42%), Positives = 22/33 (66%)
Query: 1 MKVKSAFLNGAISEEVYVKQPPGFEDLKHPDYV 33
+ V +AFL+G + E V++ QP GF D + P +V
Sbjct: 967 LDVSNAFLHGELQEPVFMYQPSGFIDPQKPTHV 999
>At4g27210 putative protein
Length = 1318
Score = 44.7 bits (104), Expect = 1e-05
Identities = 22/75 (29%), Positives = 39/75 (51%)
Query: 37 EIEMSMMGELKFFLRIQINQCKDEVYVHQSKYTKDLLMKFKVDDCNEMNTPMHPTCTLSE 96
E M+ MG+L +FL IQ+ + ++ +++ Q KY +DLL+ ++ C + TP+
Sbjct: 931 EFRMTDMGQLHYFLGIQVQRQQNGLFMSQQKYAEDLLIAASMEHCTPLPTPLPVQLDRVP 990
Query: 97 EDADAKVDQKLFRGI 111
+ D FR I
Sbjct: 991 HQEELFSDPTYFRSI 1005
Score = 40.0 bits (92), Expect = 3e-04
Identities = 18/33 (54%), Positives = 23/33 (69%)
Query: 1 MKVKSAFLNGAISEEVYVKQPPGFEDLKHPDYV 33
M VK+AFL+G + E VY+ QP GF D PD+V
Sbjct: 821 MDVKNAFLHGDLKETVYMTQPAGFVDPSKPDHV 853
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.141 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,961,521
Number of Sequences: 26719
Number of extensions: 117890
Number of successful extensions: 372
Number of sequences better than 10.0: 94
Number of HSP's better than 10.0 without gapping: 91
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 223
Number of HSP's gapped (non-prelim): 149
length of query: 129
length of database: 11,318,596
effective HSP length: 88
effective length of query: 41
effective length of database: 8,967,324
effective search space: 367660284
effective search space used: 367660284
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 54 (25.4 bits)
Medicago: description of AC147714.14


